This work analyzes gene regulatory networks involved in GA responses by focusing on a lipase gene (LIP1) expressed in the epidermis during germination. The results support a model in which GA mediates the activation of downstream target genes containing L1 box sequences in their promoters by releasing two HD-ZIP TFs (ATML1 and PDF2) from their inhibitory interaction with DELLA proteins.
Abstract
Gibberellins (GAs) are plant hormones that affect plant growth and regulate gene expression differentially across tissues. To study the molecular mechanisms underlying GA signaling in Arabidopsis thaliana, we focused on a GDSL lipase gene (LIP1) induced by GA and repressed by DELLA proteins. LIP1 contains an L1 box promoter sequence, conserved in the promoters of epidermis-specific genes, that is bound by ATML1, an HD-ZIP transcription factor required for epidermis specification. In this study, we demonstrate that LIP1 is specifically expressed in the epidermis and that its L1 box sequence mediates GA-induced transcription. We show that this sequence is overrepresented in the upstream regulatory regions of GA-induced and DELLA-repressed transcriptomes and that blocking GA signaling in the epidermis represses the expression of L1 box–containing genes and negatively affects seed germination. We show that DELLA proteins interact directly with ATML1 and its paralogue PDF2 and that silencing of both HD-ZIP transcription factors inhibits epidermal gene expression and delays germination. Our results indicate that, upon seed imbibition, increased GA levels reduce DELLA protein abundance and release ATML1/PDF2 to activate L1 box gene expression, thus enhancing germination potential.
INTRODUCTION
Gibberellins (GAs) are key plant hormones important for plant growth and development whose levels are modulated in response to developmental and environmental cues (Davière et al., 2008; Hauvermale et al., 2012; Davière and Achard, 2013). Genetic screenings have identified three main components in GA perception and early GA signaling: the GIBBERELLIN INSENSITIVE DWARF1 (GID1) GA receptors and the SLEEPY1 (SLY1) F-box protein, which are positive regulators of GA signaling, and the DELLA proteins, which play a negative role in GA-regulated expression and are targeted for degradation in the presence of GA. DELLAs are nuclear proteins lacking a DNA binding domain and negatively regulate GA responses through interaction with other transcription factors (TFs). Indeed, DELLAs have been shown to interact with different types of proteins to regulate growth as well as many other physiological processes (Penfield et al., 2005; de Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Hou et al., 2010; Feurtado et al., 2011; Heo et al., 2011; Josse et al., 2011; Zhang et al., 2011; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Hong et al., 2012; Wild et al., 2012; Yu et al., 2012; Locascio et al., 2013a; Park et al., 2013). Binding of GA to the GID1 receptors promotes interaction of the GID1-GA complex with DELLA proteins and recognition of these repressors by the F-box SLY1 protein. Ubiquitination by the SCFSLY E3 ligase complex targets DELLA proteins for degradation and releases the inhibition imposed by these repressors on their interacting TFs, alleviating GA responses. Mutant versions of DELLA proteins lacking the DELLA domain are resistant to degradation and cause a similar phenotype to that in GA-deficient mutants, except that they are unable to respond to GA (Hauvermale et al., 2012; Davière and Achard, 2013; Locascio et al., 2013b).
Tissue-specific GA signaling is important for proper development, since different tissues have different sensitivities and contributions to GA-regulated processes (Penfield et al., 2006; Ubeda-Tomás et al., 2008, 2009; Lee et al., 2010; Heo et al., 2011; Zhang et al., 2011; Duan et al., 2013; Geng et al., 2013; Löfke et al., 2013). Accordingly, recent studies indicate that GAs are not distributed homogenously between tissues and growth regions (Löfke et al., 2013; Shani et al., 2013) and that their distribution can be changed in response to several environmental stimuli (Duan et al., 2013; Geng et al., 2013; Löfke et al., 2013). Several articles have demonstrated the importance of GA signaling in the endodermis in the regulation of root growth and meristem size (Ubeda-Tomás et al., 2008, 2009; Heo et al., 2011) as well as other GA-mediated responses (Zhang et al., 2011). The endodermis of the root elongation zone has also been found to be the primary site for GA-induced DELLA degradation (Ubeda-Tomás et al., 2008; Shani et al., 2013) and for GA accumulation (Shani et al., 2013). Moreover, endodermal abscisic acid signaling promotes lateral root quiescence during salt stress in Arabidopsis thaliana seedlings, a process antagonized by GA (Duan et al., 2013; Geng et al., 2013). However, the tissue-specific molecular mechanisms underlying GA perception and DELLA protein–protein interactions are not well understood.
The plant seed is an excellent system in which to study tissue-specific GA signaling, since GAs are indispensable for seed germination. Mutants unable to synthesize GA never germinate (Ogawa et al., 2003), a phenotype reverted by the loss of function of specific DELLA proteins (Cao et al., 2005). Mechanistically, the ability of the seed to germinate results from a balance between a physical restriction imposed by the embryo-surrounding tissues and the ability of the embryo to grow and protrude (Holdsworth et al., 2008). GAs favor germination by promoting the weakening of seed coat layers and embryo growth (Piskurewicz et al., 2009). Also, the differential contributions of seed tissues to germination may reflect differences in their signaling pathways (Penfield et al., 2004, 2006; Lee et al., 2010). Moreover, genes involved in GA biosynthesis during seed germination have different cell type–specific expression that may change in response to various stimuli, and genes responsive to GA during seed imbibition are not expressed uniformly throughout seed tissues (Yamaguchi et al., 2001; Ogawa et al., 2003; Yamauchi et al., 2004; Iglesias-Fernández et al., 2011, 2013).
Here, we have studied gene regulatory networks involved in GA responses by focusing on LIP1, a gene highly induced by GA (mRNA levels and transcription) and repressed by DELLAs. We previously identified an HD-ZIP TF (ATML1) that binds to an L1 box sequence present in the LIP1 promoter. According to the literature, ATML1 is required for epidermis specification (Abe et al., 2003) and the L1 box is found in the promoters of epidermis-specific genes (Abe et al., 2001). In this study, we demonstrate that the L1 box also mediates GA-induced transcription and that GA signaling in the epidermis is required for epidermal gene expression and seed germination. We show that GAs mediate the activation of downstream L1 box target genes by releasing two HD-ZIP TFs (ATML1 and PDF2) from their inhibitory interaction with DELLA proteins.
RESULTS
An L1 Box Sequence Mediates LIP1 Promoter Induction by GA
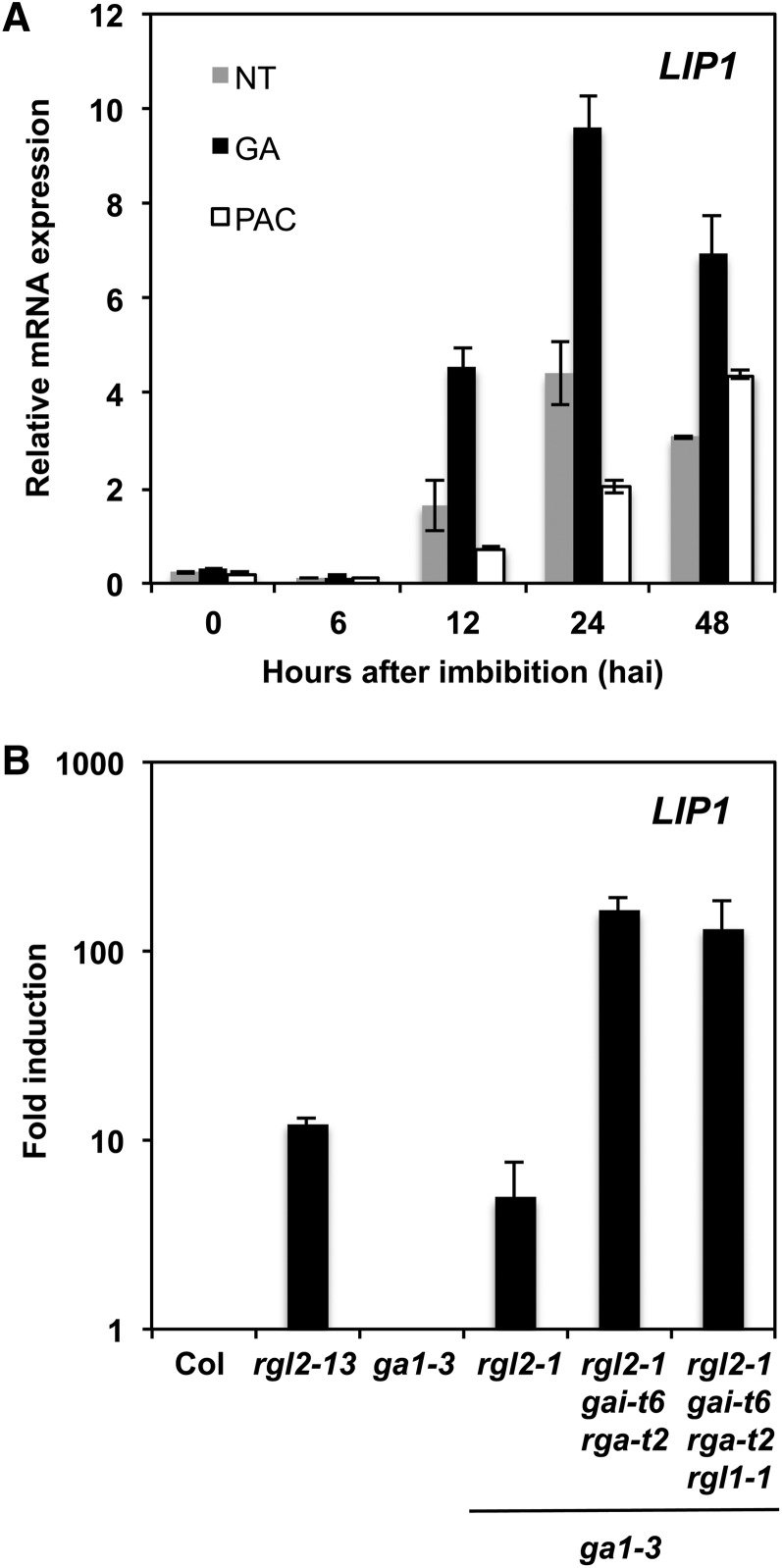
According to microarray data, LIP1 is induced by GA and repressed by DELLAs (Ogawa et al., 2003; Cao et al., 2006). To characterize in more detail LIP1 expression kinetics, we quantified LIP1 mRNAs by quantitative RT-PCR (qRT-PCR) in RNA samples from seeds imbibed in the absence (control) or presence of exogenous GA or paclobutrazol (PAC), an inhibitor of GA biosynthesis. As observed in Figure 1A, LIP1 mRNA abundance increased during seed imbibition, peaking at 24 h after imbibition (hai). In the presence of GA, LIP1 mRNAs increased above the levels detected in control samples. On the contrary, seed imbibition in the presence of PAC delayed LIP1 mRNA accumulation and reduced LIP1 mRNA abundance. Seeds used in these experiments exhibited changes in germination kinetics and gene expression in response to these treatments, as described previously in the literature, indicating that our results are meaningful in the context of seed germination (Supplemental Figures 1A and 1B). These results demonstrate that LIP1 is induced during seed imbibition and that its transcript abundance increases in response to GA. To determine if LIP1 expression was also regulated by DELLAs, we quantified its mRNA levels in several null mutants lacking DELLA genes and/or GA1, which encodes an enzyme that catalyzes the first committed step in the GA biosynthetic pathway (Sun and Kamiya, 1994). As observed in Figure 1B, LIP1 mRNA levels were induced in the rga-like2 (rgl2-13; Tyler et al., 2004) and ga1-3 rgl2-1 (Lee et al., 2002) mutants when compared with their controls (Columbia-0 [Col-0] and ga1-3). The induction levels were further increased when the rgl2-1 allele was combined with the repressor of ga1-3 (rga-t2) and ga-insensitive (gai-t6) null alleles (Yu et al., 2004), indicating that LIP1 expression is under the control of the RGL2 protein, the major DELLA repressor of seed germination (Lee et al., 2002; Tyler et al., 2004), and the RGA and/or GAI DELLA proteins.
Figure 1.
LIP1 Expression in Response to GA and DELLAs.
(A) LIP1 mRNA expression levels during seed imbibition (24 hai) in the absence (nontreated [NT]) or presence of GA (50 μM) or PAC (2 μM) determined by qRT-PCR. Values are relative to ACT8 expression levels (normalized), and averages and se of two replicates are shown.
(B) Fold induction of LIP1 expression. Normalized mRNA values for LIP1 during seed imbibition (24 hai) in various DELLA mutants were divided by normalized values obtained in their corresponding wild-type samples (Col-0 or ga1-3). Averages and se of two replicates are shown.
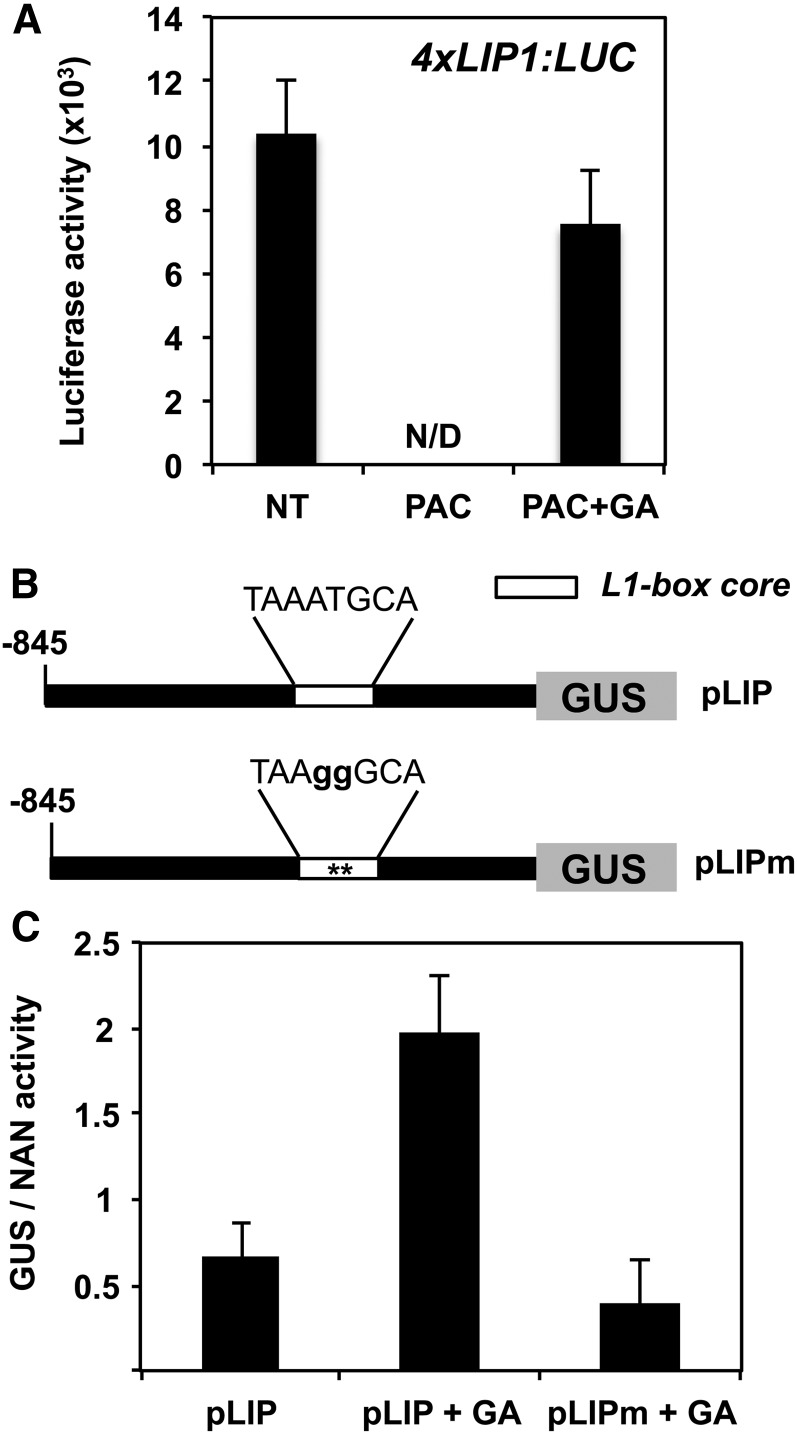
In silico analyses of several Brassicaceae LIP1 promoters identified a highly conserved sequence (LIP1 element) that is sufficient to drive the strong expression of a reporter gene in planta (Castrillo et al., 2011). We used transgenic plants carrying four copies of the LIP1 element fused to the luciferase reporter gene (4xLIP1; Castrillo et al., 2011) to quantify the in vivo response of this element to changes in GA. As observed in Figure 2A, and in agreement with our previous results, high levels of luciferase activity were detected at 24 hai in control conditions but not in the presence of PAC. In addition, luciferase levels in seeds imbibed with PAC but supplemented with GA were almost identical to those in the control (Figure 2A). Luciferase activity from a control construct with a minimal promoter (Chen and Singh, 1999; Castrillo et al., 2011) could be detected only at 48 hai; the activity was 50-fold lower than that of the 4xLIP1 construct and hardly responded to any of the treatments (Supplemental Figures 2A and 2B). These results indicate that transcription driven by the LIP1 element is modulated in response to GA.
Figure 2.
Transcriptional Response of LIP1 Promoter Constructs to GAs.
(A) Seeds from a representative transgenic line (4xLIP:LUC) described by Castrillo et al. (2011) were imbibed in the absence (nontreated [NT]) or presence of PAC (5 μM) or PAC (50 μM) plus GA (5 μM). Averages and se of luciferase activity at 24 hai from at least 20 seeds are shown. N/D, not detected.
(B) Diagram of the LIP1 promoter constructs used for transient expression in Arabidopsis mesophyll protoplasts.
(C) GUS activity in protoplasts transformed with the constructs described in (B) normalized with the NAN activity obtained from a Pro35S:NAN construct. Protoplasts were incubated in a control solution with or without GA (50 µM). Average values and se of four biological replicates are shown.
We previously identified an HD-ZIP TF (ATML1) that binds to the L1 box present in the LIP1 element (Castrillo et al., 2011). A 2-bp mutation within the L1 box sequence abolishes epidermal expression and ATML1 binding in vitro (Abe et al., 2001) and in vivo (Castrillo et al., 2011). To analyze the role of the LIP1 L1 box in GA activation in the context of a full promoter, an ∼1000-bp LIP1 promoter fragment (pLIP) and a mutated version (pLIPm) carrying the above-mentioned 2-bp changes in the L1 box sequence were fused to the β-glucuronidase (GUS) reporter gene (Figure 2B). GUS activity was enhanced by GA in protoplasts transformed with the pLIP construct, while protoplasts transformed with the pLIPm version had similar GUS activity levels to the control (pLIP without treatment; Figure 2C). These results demonstrate that the L1 box sequence mediates LIP1 promoter induction by GA and that the increased levels of LIP1 mRNA observed in response to GA treatment are due to the increased transcriptional activity of the LIP1 promoter.
The L1 Box Sequence Is Overrepresented in the Promoters of Genes Induced by GA and Repressed by DELLAs
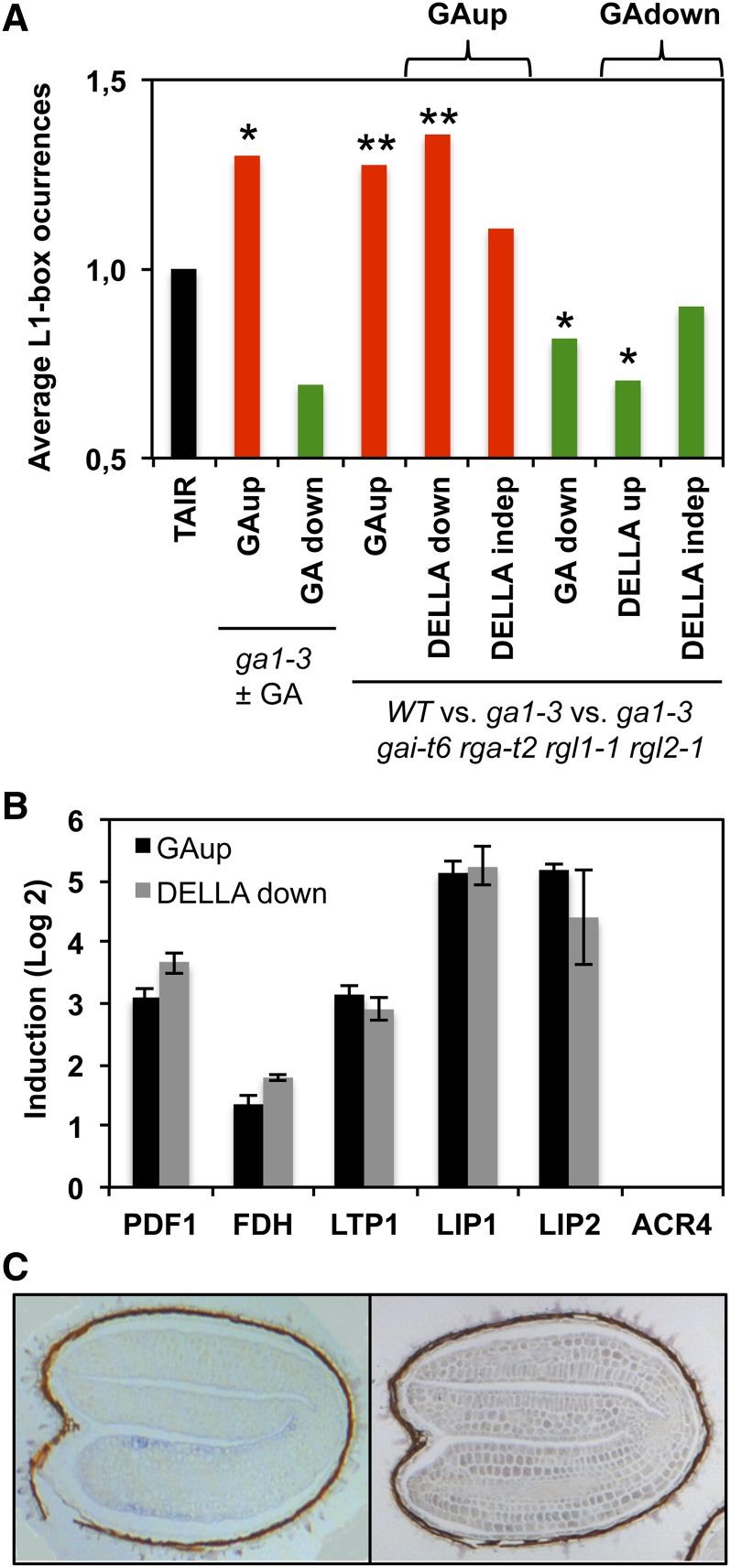
To determine if the L1 box sequence plays a broader role in GA-regulated gene expression, we looked for enrichment of the L1 box core consensus sequence (5′-TAAATGY-3′; Nakamura et al., 2006) in the transcriptomes of seeds of the GA-deficient ga1-3 mutant (unable to produce GA; Koornneef and van der Veen, 1980; Sun and Kamiya, 1994) germinated in the absence or presence of GA (Ogawa et al., 2003). Since DELLA proteins play a central role in GA signaling, we also used whole-genome microarray data comparing the wild type with the ga1-3 mutant and with a ga1-3 mutant lacking four DELLA genes (gai-t6 rga-t2 rgl1-1 rgl2-1; Cao et al., 2006). As observed in Figure 3A, the average occurrence of the L1 box sequence is significantly higher in GA-upregulated (GAup) and DELLA-downregulated (DELLAdown) transcriptomes and lower in GA-downregulated (GAdown) and DELLA-upregulated (DELLAup) transcriptomes when compared with the control (randomly expected occurrence in the Arabidopsis genes represented on the Affymetrix chip used in transcriptomic analyses [TAIR]), while it does not differ significantly from DELLA-independent transcriptomes. When we analyzed a well-known monocot GA-responsive cis-element (GARE) (Skriver et al., 1991; Gubler and Jacobsen, 1992) in these transcriptomes, we did not find significant enrichment for this element (Supplemental Figure 3A) (Ogawa et al., 2003).
Figure 3.
L1 Box Occurrence in Promoters of Genes Differentially Expressed in GA-Related Transcriptomes and Expression of Epidermal Genes.
(A) L1 box sequence content in 500-bp promoters of genes upregulated (GAup) or downregulated (GAdown) in response to GA or in DELLA mutants (DELLAup or DELLAdown) as described by Ogawa et al. (2003) and Cao et al. (2006). DELLAindep, DELLA independent. The average L1 box occurrence in the Arabidopsis genes represented on the Affymetrix chip used in transcriptomic analyses is taken arbitrarily as 1 (TAIR). Asterisks represent significant differences, at P < 0.01 (**) or P < 0.05 (*), from the control (TAIR) using the χ2 test.
(B) Fold induction of epidermal L1 box–containing genes in the GAup and DELLAdown transcriptomes (Cao et al., 2006).
(C) mRNA in situ hybridization of Col-0 seeds at 24 hai. Samples were hybridized with LIP1-derived antisense (left panel) or sense (right panel) probes.
We then queried the microarray data from Cao et al. (2006) for RNA levels of well-known L1 box–containing genes specifically expressed in the epidermis and potential direct targets of ATML1 (hereafter referred to as epidermal genes; Abe et al., 2001, 2003). These included PDF1, encoding a putative extracellular Pro-rich protein (Abe et al., 1999, 2003; Ogawa et al., 2003), FDH/KCS10, encoding a putative 3-ketoacyl-CoA synthase involved in the biosynthesis of long-chain fatty acids (Yephremov et al., 1999; Abe et al., 2001), LTP1, encoding a cell wall–localized lipid transfer protein (Thoma et al., 1994; Abe et al., 2001), and ACR4, encoding a membrane-localized protein with similarity to receptor kinases (Tanaka et al., 2002). We also included LIP1 in these analyses and a second GDSL lipase gene (LIP2) that shares 86% nucleotide identity with LIP1, has an L1 box in its promoter (Supplemental Figure 3B), and was found to be coregulated (r = 0.817) with ATML1. All of these genes, aside from ACR4, were induced and repressed in the GAup and DELLAdown transcriptomes, respectively (Figure 3B). Altogether, these results indicate that the L1 box sequence does serve as a common cis-element for GA responsiveness in a DELLA-dependent pathway, at least in a subset of Arabidopsis epidermal genes.
According to the literature, ATML1 is required for epidermis specification and the L1 box is found in the promoters of epidermis-specific genes (Abe et al., 2001, 2003). To determine if LIP1 was also expressed specifically in the epidermis, we performed mRNA in situ hybridizations with LIP1-specific probes in imbibed seeds. As seen in Figure 3C, the hybridization signal was restricted to the epidermis of the embryonic axis. These results are compatible with LIP1 having an L1 box sequence in its promoter and being under the regulation of ATML1 in the epidermis.
L1 Box Gene Expression Is Reduced by Blocking GA Signaling in the Epidermis
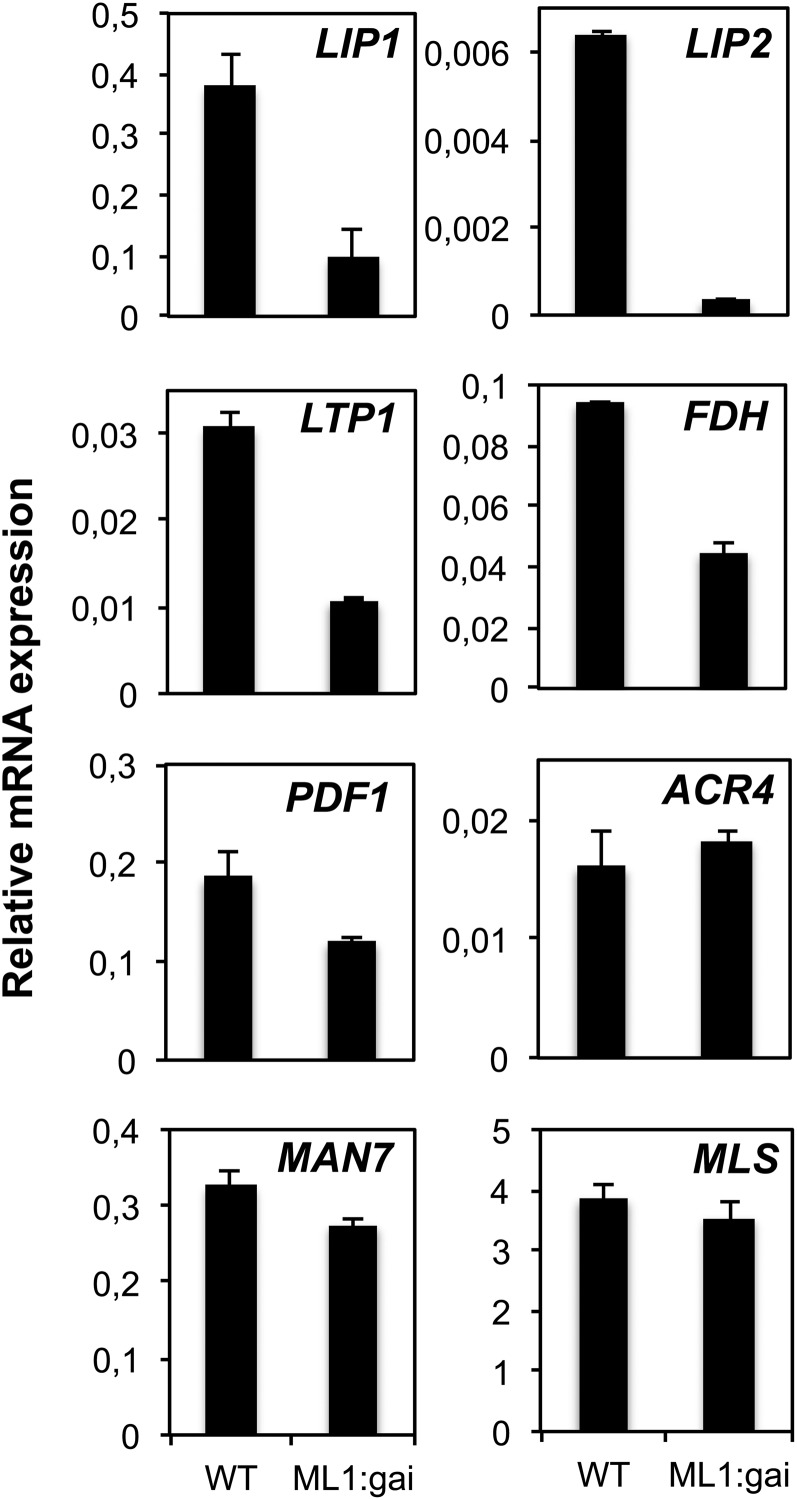
Given that the L1 box sequence confers epidermis-specific expression (Lu et al., 1996; Abe et al., 1999, 2001, 2003; Sessions et al., 1999; Ogawa et al., 2003) and is responsive to GA and DELLAs, we assessed the contribution of DELLA-dependent GA signaling in this tissue to the expression of epidermal genes. To block GA signaling only in the epidermis, we used a transgenic line expressing a GA-resistant version of the GAI protein (gai-1) fused to the green fluorescent protein (GFP) reporter gene under the control of the ATML1 promoter (ProML1:GFP-gai-1; Gallego-Bartolomé et al., 2011b). As observed in Figure 4, the mRNA levels of LIP1 and other epidermal genes during seed imbibition were reduced when GA signaling was blocked in the epidermis (wild type versus ProML1:GFP-gai-1). However, ACR4 mRNA levels, which are not induced by GA or repressed by DELLAs (Figure 3B), were not altered, confirming that this gene is not under the control of DELLAs (Figure 4). Gene expression outside the epidermis did not seem to be affected, since a GA-induced mannanase gene (MAN7) expressed in the micropylar endosperm and the radicle (Iglesias-Fernández et al., 2011, 2013) and reported to be regulated by the DELLAs (Cao et al., 2006) was not affected (Figure 4). Finally, the general metabolic activity of these seeds did not seem to be affected, since the MALATE SYNTHASE (MLS) mRNAs for a glyoxylate cycle gene (Cornah et al., 2004) were not altered in the mutant (Figure 4). These results indicate that GA-regulated L1 box gene expression is also regulated by DELLA proteins in the epidermis and suggest that DELLA repression in this tissue does not affect gene expression related to lipid utilization and gluconeogenesis.
Figure 4.
Quantification of L1 Box Gene Expression after Blocking GA Signaling in the Epidermis.
Quantification by qRT-PCR of mRNA levels of epidermal and control genes (ACR4, MAN7, and MLS) in wild-type Ler and ProML1:GFP-gai-1 (ML1:gai) genotypes. RNAs were isolated from seeds at 24 hai, and their levels are shown relative to those of ACT8. Average values and se for two replicates are shown. Similar results were obtained with an additional biological replicate.
Seed Germination in Arabidopsis Requires GA Signaling in the Epidermis
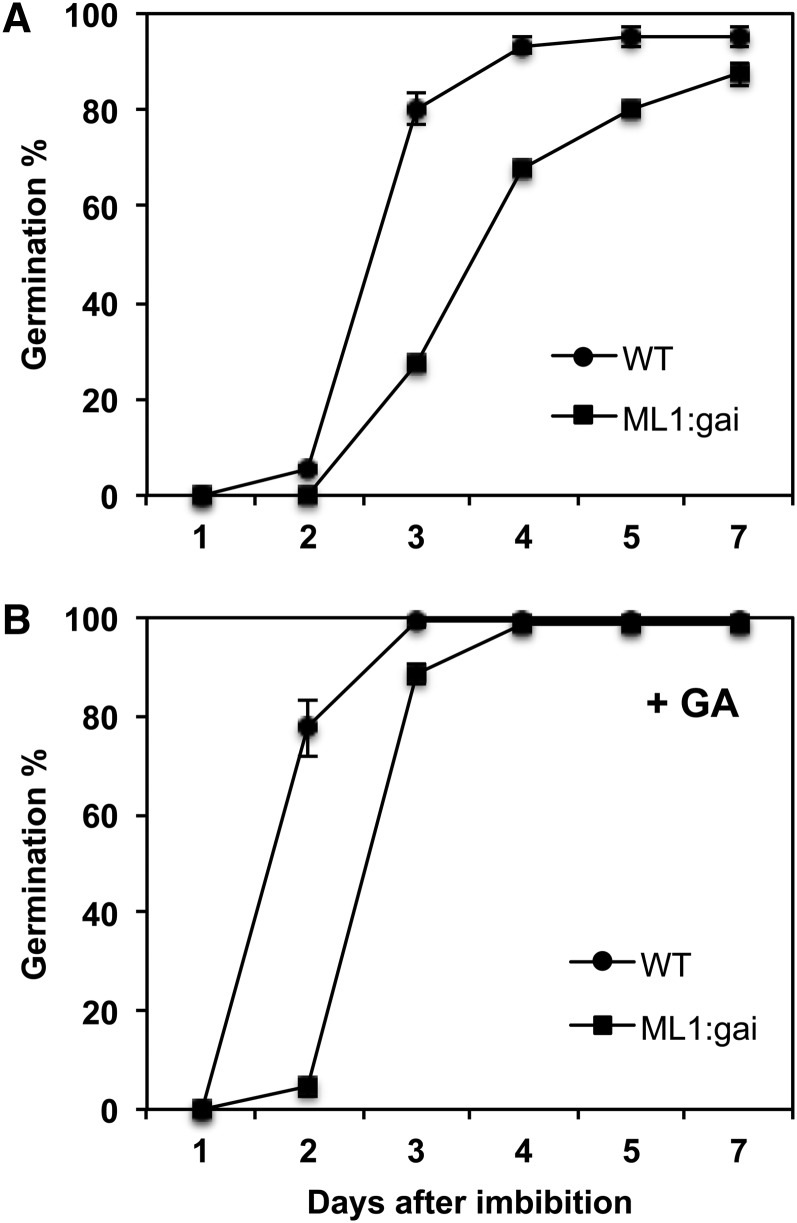
To check if the molecular phenotypes observed had an effect on seed germination, we analyzed the germination kinetics of wild-type versus ProML1:GFP-gai-1 seeds. We observed a delay in ProML1:GFP-gai-1 seed germination (T50 ∼ 84 h) when compared with the wild type (T50 ∼ 60 h; Figure 5A). Although the addition of GA accelerated germination in both genotypes, it did not eliminate the germination delay (T50 ∼ 60 h in ProML1:GFP-gai-1 versus T50 ∼ 40 h in the wild type), indicating that lack of GA production was not the cause of the phenotype (Figure 5B). These results indicate that ProML1:GFP-gai-1 seeds have delayed germination due to blocked GA signaling in the epidermis.
Figure 5.
Blocking GA Signaling in the Epidermis Delays Seed Germination.
(A) Germination of wild-type Ler and ProML1:GFP-gai-1 (ML1:gai) seeds was scored during 7 d after imbibition.
(B) Same as in (A), except that seeds were imbibed in the presence of 5 μM GA.
Each germination percentage shown is an average with se of three replicates.
Physical Interaction between ATML1, PDF2, and DELLA Proteins
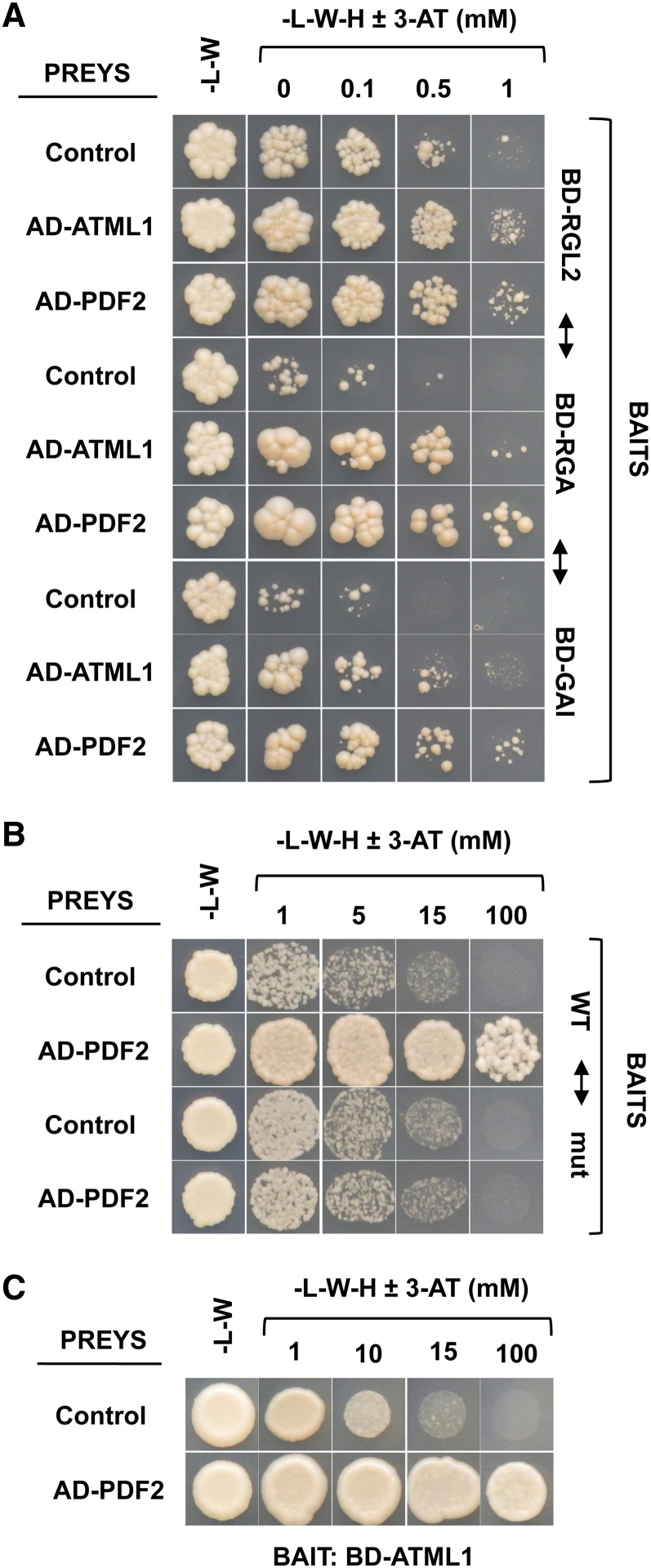
ATML1 is a regulator in the epidermis that binds the L1 box sequence in the promoters of its target genes. Although RGL2 encodes the predominant DELLA repressor of seed germination in Arabidopsis, RGA and GAI also play a role in this repression (Lee et al., 2002; Tyler et al., 2004; Cao et al., 2005). One possibility is that DELLAs block GA signaling and L1 box–mediated gene expression by sequestering ATML1 in a physical interaction. To test this possibility, the coding sequences (CDSs) for these three DELLA proteins were fused to the GAL4 DNA binding domain (BD-DELLAs) and used as bait for yeast two-hybrid experiments. As prey, we fused the ATML1 CDS to the GAL4 activation domain (AD) and used the empty plasmid as a negative control. As shown in Figure 6A, yeast cells carrying BD-RGL2, BD-RGA, or BD-GAI with the AD-ATML1 construct, but not those with the empty plasmid, were able to grow on the selection medium in the presence of 3-aminotriazole, indicating that ATML1 is able to interact with the DELLA proteins.
Figure 6.
ATML1 Interacts with DELLA Proteins and PDF2 Interacts with ATML1 and Binds to the LIP1 L1 Box.
(A) Yeast strains containing the BD-RGL2, BD-RGA, or BD-GAI construct were mated to strains containing the AD-ATML1 or AD-PDF2 construct or the corresponding empty plasmid (control).
(B) Yeast strains containing either the 50-bp LIP1 element carrying the wild-type LIP1 L1 box sequence (wild type) or a 2-bp mutation in the L1 box (mut; Castrillo et al., 2011) were mated to strains containing the AD-ATML1 or AD-PDF2 construct or the corresponding empty plasmid (control).
(C) Yeast strains containing the BD-ATML1 construct were mated to strains containing the AD-PDF2 construct or the corresponding empty plasmid (control).
For all the experiments, diploid cells were grown in minimal medium without Leu and Trp (-L-W), cell densities were measured, and equivalent amounts of cells were grown on minimal medium without Leu, Trp, and His (-L-W-H) with increasing concentrations of 3-aminotriazole (3-AT).
[See online article for color version of this figure.]
Abe et al. (2003) described the redundant roles of ATML1 and PDF2, a paralogue of ATML1, in the regulation of shoot epidermal cell differentiation. Using yeast one-hybrid and yeast two-hybrid assays, we observed that PDF2 interacted with the DELLA proteins (Figure 6A) and was able to bind to the L1 box present in the LIP1 element (Figure 6B) and to interact with ATML1 (Figure 6C), suggesting a redundant function in the regulation of DELLA-dependent GA-mediated L1 box gene expression. These results provide molecular evidence of previous genetic observations indicating that both genes participate in the same regulatory programs and are functionally interchangeable.
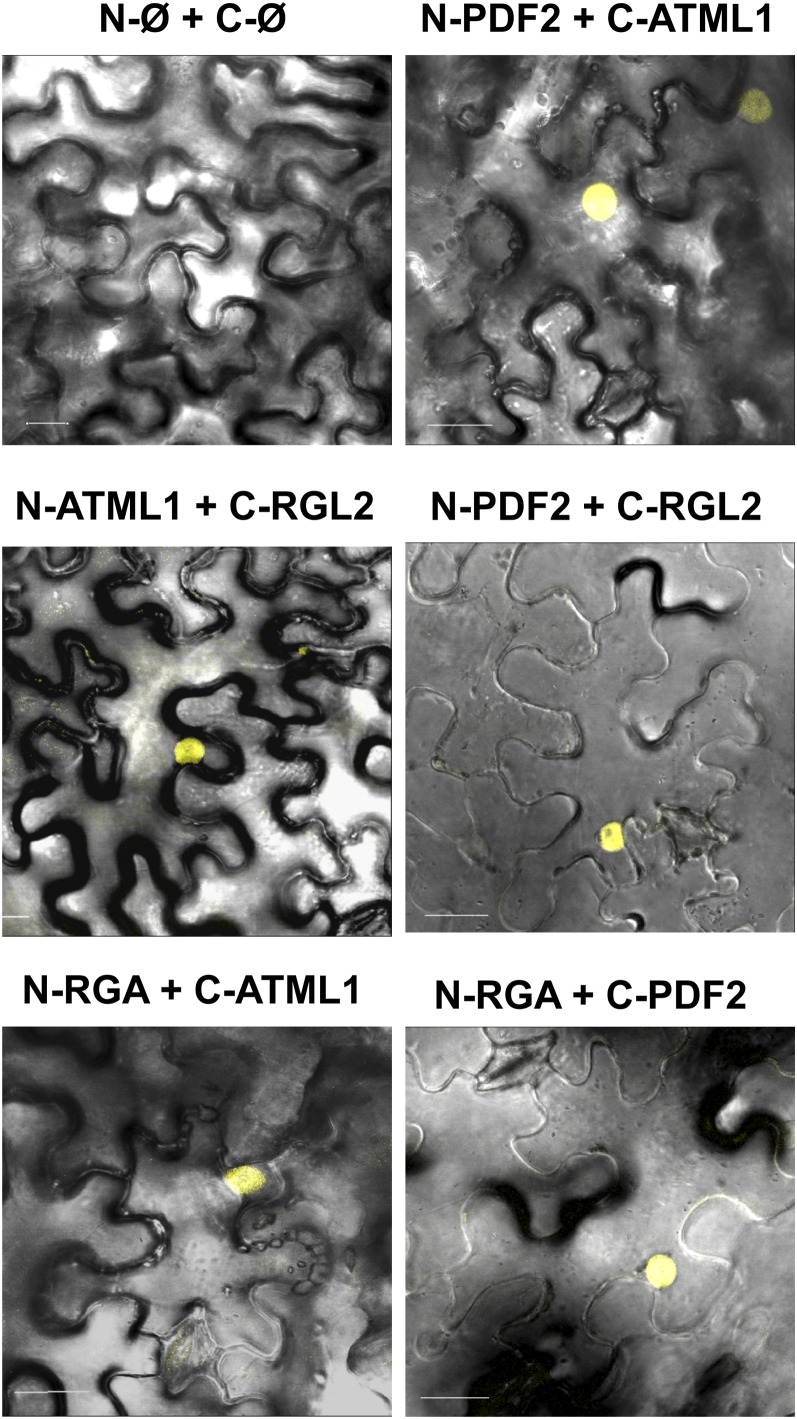
To validate the protein interactions in planta, bimolecular fluorescence complementation (BiFC) experiments were performed by agroinfiltration of Nicotiana benthamiana leaves. ATML1, PDF2, RGL2, RGA, and GAI CDSs were translationally fused to the N-terminal half of the yellow fluorescent protein (N-YFP) or the C-terminal half of the cyan fluorescent protein (C-CFP) as indicated in Figure 7. A strong fluorescent signal was observed in nuclei of N. benthamiana cells, with coexpression of N-YFP:PDF2 with C-CFP:ATML1, C-CFP:RGL2 with N-YFP:ATML1 or N-YFP:PDF2, and N-YFP:RGA with C-CFP:ATML1 or C-CFP:PDF2 (Figure 7), indicating that both HD-ZIP TFs are able to form heterodimers and to interact with the RGL2 and RGA proteins in planta. We did not detect fluorescence when the ATML1 or PDF2 protein was coexpressed with the GAI protein (Supplemental Figure 4), suggesting that the interaction observed in yeast may not be relevant in planta (i.e., posttranslational modifications of the GAI protein might hamper the interaction). No fluorescent signal was observed when different combinations of TFs and/or empty vectors were coexpressed (Figure 7; Supplemental Figure 4).
Figure 7.
ATML1 and PDF2 Interact with DELLA Proteins in Planta.
The ATML1, PDF2, RGA, and RGL2 CDSs were fused to the N-YFP (N) or C-CFP (C) CDS and coexpressed in N. benthamiana cells as indicated for each panel. Ø, empty vector. Overlays of the fluorescence Z projections and differential interference contrast microscopy images are shown.
ATML1 and PDF2 Play Redundant Roles during Seed Imbibition in the Regulation of L1 Box/GA–Mediated Gene Expression in the Epidermis
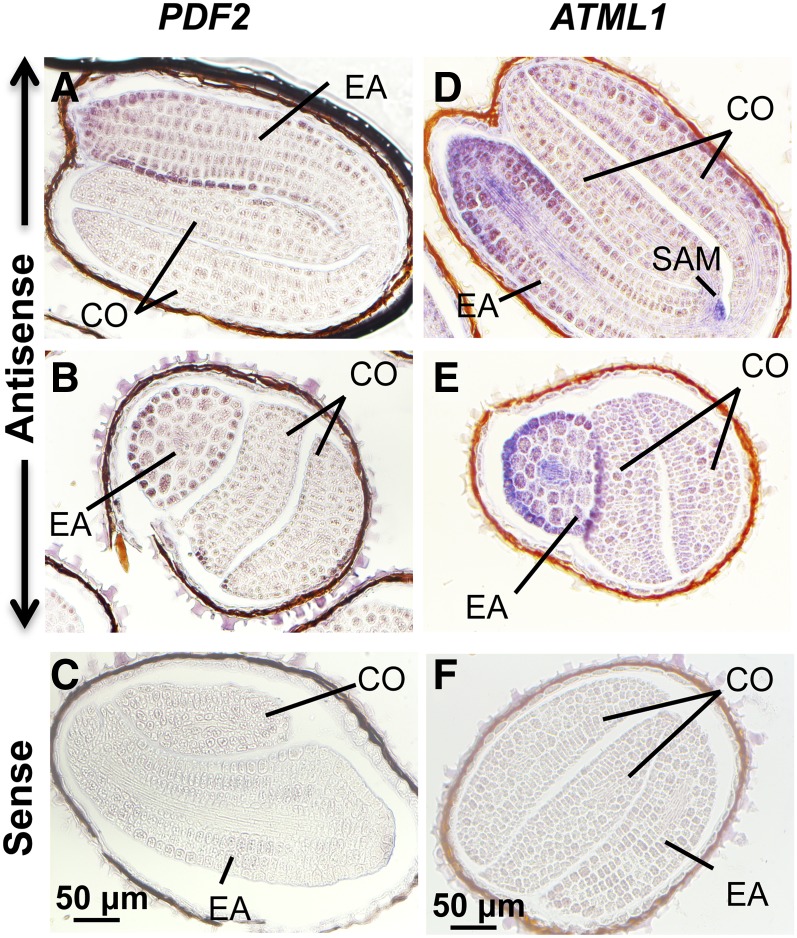
Although ATML1 and PDF2 are expressed specifically in the epidermis of all tissues and developmental stages analyzed to date (Lu et al., 1996; Sessions et al., 1999; Abe et al., 2003; Takada and Jürgens, 2007; Gallego-Bartolomé et al., 2011b; Kamata et al., 2013; Takada et al., 2013), no data have been reported on their expression patterns in imbibed seeds. To test if ATML1 and PDF2 are also specifically expressed during seed germination in the epidermis of the embryo, a location compatible with being regulators of epidermal gene expression during this developmental stage, we performed mRNA in situ hybridizations of imbibed seeds with gene-specific probes. As observed in Figure 8, mRNAs for both genes were detected with antisense probes in longitudinal and transverse sections mainly in the epidermis of the embryonic axis. Moreover, ATML1 mRNAs were also detected in the shoot apical meristem of the embryo, while no hybridization signal was detected when the sense probe was used (Figure 8).
Figure 8.
ATML1 and PDF2 Tissue-Specific Expression during Seed Imbibition.
mRNA in situ hybridization of PDF2 ([A] to [C]) and ATML1 ([D] to [F]) transcripts in Col-0 seed sections. Seeds were imbibed for 24 h and hybridized using antisense ([A], [B], [D], and [E]) or sense ([C] and [F]) probes. CO, cotyledons; EA, embryonic axis; SAM, shoot apical meristem. Bars = 50 μm.
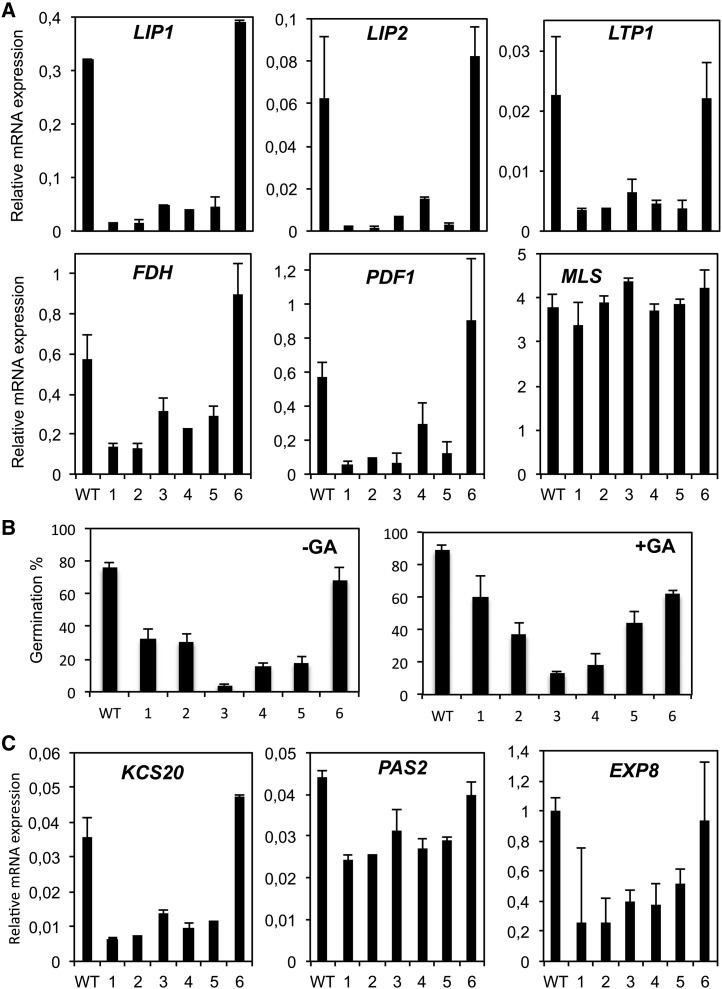
To analyze the functional relevance of ATML1 and PDF2 in planta, we assessed the consequences of the posttranscriptional repression of these genes, since plants harboring individual knockout alleles of both genes display normal growth and morphology, while double mutants show pleiotropic effects and fail to survive (Abe et al., 2003). A transgene designed to express amiRTPL-353, an artificial microRNA (amiRNA) targeting the PDF2 and ATML1 genes (Supplemental Figure 5A), was generated and transferred into Arabidopsis plants as described by Jover-Gil et al. (2014). Several T3 homozygous lines with reduced levels of the ATML1/PDF2 transcripts (Supplemental Figure 5B) were selected. HDG2, the closest HD-ZIP homologue to ATML1 and PDF2 and expressed also in the epidermis (Kamata et al., 2013), did not show a similar reduction in its mRNA levels (Supplemental Figure 5C), indicating that the amiRNA was specific for ATML1/PDF2 silencing. Seeds of the silenced lines showed a clear reduction in the transcript levels for their L1 box/GA–regulated putative targets (Figure 9A), even in the presence of exogenous GA (Supplemental Figure 6). These seeds also had delayed germination kinetics when imbibed in the absence or presence of exogenous GA (Figure 9B) and compared with the wild type or an amiRNA line (line 6) having wild-type levels of the ATML1 and PDF2 mRNAs (Supplemental Figure 5B). These results indicate that ATML1 and PDF2 are positive regulators of GA-regulated epidermal gene expression and that a reduction in their expression levels negatively affects germination by reducing epidermal GA-regulated gene expression.
Figure 9.
Silencing of ATML1 and PDF2 Reduces Epidermal Gene Expression and Delays Germination.
(A) mRNA expression levels in amiRTPL-353 seeds at 24 hai of several epidermal and control genes determined by qRT-PCR. Values are relative to ACT8 expression levels, and averages and se of two replicates are shown.
(B) Germination of amiRTPL-353 seeds in the absence or presence of GA was scored at 40 hai. Averages and se of four replicates are shown.
(C) mRNA expression levels in amiRTPL-353 seeds at 24 hai of additional KCS genes and the EXP8 gene determined by qRT-PCR. Values are relative to ACT8 expression levels, and averages and se of two replicates are shown.
DISCUSSION
The L1 Box Sequence Is an Epidermal GA-Responsive Element
The only specific Arabidopsis GA-responsive element described so far was found in the LEAFY gene (CAACTGTC) using a promoter deletion strategy (Blázquez and Weigel, 2000). Since then, several microarray experiments have examined the effects of GA and/or DELLA on global transcription profiles in germinating seeds, seedlings, and flowers in Arabidopsis (Locascio et al., 2013b), but common specific cis-elements associated with these responses could not be assigned. We have studied gene regulatory networks involved in GA responses by focusing on LIP1, which is highly induced by GA (in terms of mRNA levels and transcription) and repressed by DELLAs during seed imbibition. A 2-bp mutation in the L1 box sequence within the LIP1 promoter abolished its GA inducibility, suggesting that there are no additional GA-responsive elements in this promoter or, if they exist, they require the presence of a functional L1 box. In accordance with having a functional L1 box, LIP1 is also specifically expressed in the epidermis (Figure 3C), and other epidermal genes are also induced by GA (Figure 3B). One of those genes, PDF1, was also shown to be expressed in the epidermis of imbibed ga1-3 embryos only when GAs were added exogenously (Ogawa et al., 2003). In monocots, the GARE (TAACAAA/G) is a major cis-element for GA-induced expression in seeds, but this element is not found in Arabidopsis (Ogawa et al., 2003; Supplemental Figure 3A). It cannot be excluded that anatomical differences between both types of seeds require different regulatory programs in the dry seed and during imbibition. Also, unlike in monocots, genes encoding DELLA proteins in dicots are frequently duplicated, and in some cases, as occurs in the Brassicaceae, the genomes contain up to five DELLA genes (Gallego-Bartolomé et al., 2010), thereby increasing the complexity of their regulation. Since the epidermal genes, as well as LEAFY, have cell type–specific expression, our data support that GA responses may be compartmentalized by having different sets of regulatory cis-elements and interacting TFs. In fact, cis-elements identified as overrepresented among DELLA-regulated promoters in dark-grown seedlings were found to be different conserved elements recognized by several families of TFs (Gallego-Bartolomé et al., 2011a). Also, although GA biosynthetic and catabolic genes are expressed in particular cell layers of the embryo, GA-induced expression is not restricted to GA biosynthetic tissues (Yamaguchi et al., 2001; Ogawa et al., 2003; Curaba et al., 2004; Yamauchi et al., 2004; Frigerio et al., 2006; Piskurewicz et al., 2008). These findings are consistent with the hypothesis that DELLAs regulate the activity of different members of a TF family to drive similar biochemical pathways in different organs but also regulate functionally different genes to drive organ-specific pathways (Cao et al., 2006). In agreement with this model, DELLAs bind DNA through interaction with different families of TFs and function as key plant growth regulators by integrating multiple hormonal signals (Locascio et al., 2013b).
Role of Epidermal HD-ZIP TFs in GA Signaling and Growth
We have found that the LIP1 L1 box sequence is bound not only by ATML1 but also by the ATML1 paralogue PDF2 and that both proteins are able to interact, likely by heterodimerization, via their leucine zipper protein–protein interaction motif. However, the function of these factors appears to be redundant, as indicated by the absence of phenotype in single mutants. Both proteins are key players in the regulation of epidermal gene expression, and their overlapping functions cannot be substituted for by other proteins, since double mutants have severe defects in epidermal cell differentiation and die before producing flowers (Abe et al., 2003). Notably, within the HD-ZIP IV subfamily (16 members) there are additional members besides ATML1 and PDF2 with roles in epidermis-related functions such as trichome differentiation (Rerie et al., 1994; Nakamura et al., 2006; Marks et al., 2009). Double mutants of ATML1 or PDF2 and other HD-ZIP IV TFs show slight defects in cotyledon development (Nakamura et al., 2006) or produce abnormal flowers (Kamata et al., 2013). In the latter case, reduced expression of AP3 was observed in internal cell layers, altogether suggesting a non-cell-autonomous effect of these genes in the regulation of different aspects of plant development. Recently, it was shown that GAs play differential and tissue-specific roles in flowering induction and floral development (Porri et al., 2012). Since Arabidopsis DELLAs have different and overlapping functions, it is possible that different DELLAs interact specifically with different HD-ZIP proteins depending on the plant developmental stage and organ. Whether HD-ZIP IV–mediated regulation of flower development and other plant developmental processes requires GA remains to be determined.
Cells in the epidermis interact with the environment and can restrict or promote growth. In addition to a role as an external barrier, the epidermis was also shown to restrict inner cell layer growth, since increasing brassinosteroid perception in the epidermis affects both shoot growth and root meristem size (Savaldi-Goldstein et al., 2007; Savaldi-Goldstein and Chory, 2008; Hacham et al., 2011). Biosynthesis of very-long-chain fatty acids (VLCFAs) in the epidermis influences organ growth by suppressing cytokinin biosynthesis and restricting cell proliferation of inner tissues (Nobusawa et al., 2013). Genes for enzymes that catalyze the first step of VLCFA elongation (ketoacyl-CoA synthases [KCSs]) are predominantly expressed in the epidermis and have L1 boxes in their promoters (Yephremov et al., 1999; Joubès et al., 2008). We have observed that several of these genes (FDH in Figure 9A and KCS20 and PAS2 in Figure 9C) show reduced expression in our amiRTPL-353 lines, suggesting a role for ATML1/PDF2 in inner tissue growth coordination by influencing VLCFA biosynthesis. Recent studies have also highlighted the importance of GA signaling in the endodermis for several physiological responses. GA signaling in the endodermis but not in other root cell layers is required to regulate root growth, meristem size, and apical hook development (Ubeda-Tomás et al., 2008, 2009; Gallego-Bartolomé et al., 2011b). It is interesting that these previous studies did not find any effect when GA signaling was blocked in the epidermis. However, we have uncovered a role for GA signaling in the epidermis in the context of seed germination with no obvious effects at postgerminative stages. Altogether, these findings suggest that there is a hierarchy in GA responses depending on the cell layer and developmental phase.
A Possible Mechanism Contributing to Seed Germination
Germination of Arabidopsis seeds is not completed as a result of elongation of the radicle but rather of the adjacent transition zone and lower hypocotyl of the embryonic axis (Sliwinska et al., 2009). A promoter trap line wherein the GUS reporter is translationally fused to RGL2 showed GUS activity in the embryonic axis during seed germination, suggesting that this DELLA protein may be preventing its elongation (Lee et al., 2002; Piskurewicz et al., 2008). Thus, the RGL2 expression pattern during seed imbibition overlaps with that observed for LIP1 (Figure 3C) and PDF1 (Ogawa et al., 2003) as well as that of their regulators ATML1 and PDF2 (Figure 8). In addition, loss-of-function mutants of four DELLA genes (GAI, RGA, RGL1, and RGL2) in the ga1-3 background result in embryos with longer embryo axes due to elongated hypocotyl epidermal cells, suggesting that although RGL2 is the predominant DELLA repressor of seed germination, other DELLA proteins have relevant functions in the epidermis (Cao et al., 2005). We have found that LIP1 expression is under the control of more than one DELLA protein (RGL2 and RGA and/or GAI; Figure 1B) and that ATML1 and PDF2 interact in yeast and in planta with the RGL2 and RGA DELLA proteins, revealing an interaction between members of these TF families. Together, these data indicate that the regulation described here is involved in controlling the elongation of the embryo axis that precedes germination.
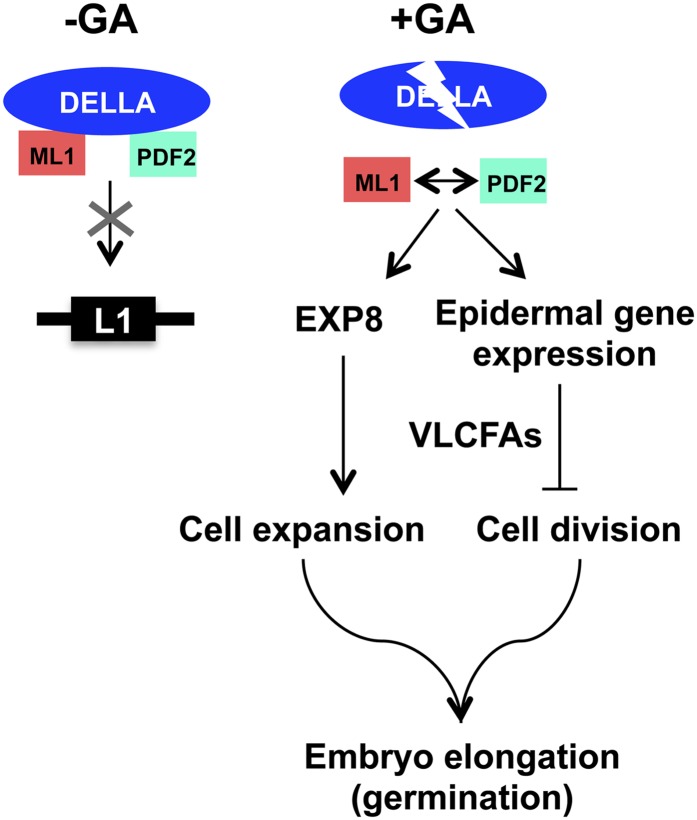
GDSL lipases represent a subfamily of lipolytic enzymes that show very broad substrate specificity (Chepyshko et al., 2012). In an oilseed such as Arabidopsis, which stores lipids predominantly in the embryo, lipolytic activities coupled to gluconeogenesis are vital to germinating seeds and to fuel seedling growth until photosynthetic capacity is acquired (Penfield et al., 2004). Loosening cell walls is also required for growth, and expansins play a major role in this process (Cosgrove, 2000). EXPANSIN8 (EXP8) has been reported to be a direct target of several TFs influencing growth, indicating that it is an important player for cell wall remodeling and a convergence point for several signaling pathways (de Lucas et al., 2008; Feng et al., 2008; Stavang et al., 2009; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Park et al., 2013). In seeds, EXP8 expression is induced by GA during seed imbibition and repressed by DELLAs (Cao et al., 2006; Park et al., 2013), being a direct target of RGL2 (Stamm et al., 2012). PIL5 mediates the phytochrome regulation of seed germination by coordinating hormone signals and modulating cell wall properties by repressing EXP8 expression, among other genes, through direct interaction with its promoter (Oh et al., 2007, 2009). Our finding that EXP8 expression is reduced in the amiRTPL-353 lines (Figure 9C) supports the idea that ATML1/PDF2 are required for normal embryo growth at least during germination. PIL5 regulates GA responsiveness by increasing the expression of RGA and GAI, but not that of RGL2, by binding directly to their promoters (Oh et al., 2007). It was shown that at least RGL2 and RGA are able to interact with PIL5 in vivo (Gallego-Bartolomé et al., 2010), and we have observed that several ATML1/PDF2 targets are downregulated by RGL2 and PIL5 (Oh et al., 2009; Stamm et al., 2012). Whether PIL5 participates in a physical interaction with the DELLAs to block ATML1/PDF2 function remains to be determined. Our results suggest a regulatory model wherein DELLAs block GA signaling in the epidermis by sequestering ATML1/PDF2. Upon imbibition, GA biosynthesis destabilizes the DELLAs and releases ATML1/PDF2 to activate L1 box gene expression, having an effect on epidermis elongation and the coordination of growth in this cell layer with that of inner tissues (Figure 10).
Figure 10.
A Model for ATML1/PDF2 Function.
We propose the following regulatory model based on our results. In the absence of GA, DELLAs sequester ATML1/PDF2, thereby blocking GA signaling in the epidermis. The GA produced upon imbibition destabilizes the interaction between the DELLAs and ATML1/PDF2, leading to the activation of L1 box gene expression and downstream events involved in the elongation of epidermal cells and the coordination of epidermal growth with inner tissues.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Col-0 and Landsberg erecta (Ler) ecotypes were used in this study. The rgl2-13 (Col-0 background) and ProML1:GFP-gai-1 (Ler background) seeds were obtained from Tai Pin Sun (Tyler et al., 2004) and M.A. Blázquez and D. Alabadi (Gallego-Bartolomé et al., 2011b), respectively. The ga1-3 (Koornneef and van der Veen, 1980; Sun and Kamiya, 1994), ga1-3 rgl2-1 (Lee et al., 2002), and ga1-3 rgl2-1 gai-t6 rga-t2 and ga1-3 rgl2-1 gai-t6 rga-t2 rgl1-1 (Yu et al., 2004) mutants were provided by S. Prat. Plants were grown either on Petri dishes containing half-strength Murashige and Skoog (MS) medium buffered with 2 mM MES, pH 5.7, and 0.7% (w/v) agar or in soil and grown to maturity at 16 h of light at 22°C/8 h of dark at 20°C and 60% relative humidity. Seeds were harvested when plants had ceased flowering and siliques were starting to dehisce.
RNA Isolation and qRT-PCR
Total RNA was isolated from seeds and other tissues, as described by Oñate-Sánchez and Vicente-Carbajosa (2008). Seeds (15 mg) from each genotype were germinated on moistened filter paper, and samples were taken at the indicated times (hours) after imbibition. First-strand cDNA was synthesized as described by Rueda-Romero et al. (2012). For quantitative PCR, 8 to 16 ng of cDNA was used as template together with 0.5 µM each forward and reverse specific oligonucleotides (Supplemental Table 1) and Fast Start Universal SYBR Green Master mix (Roche Diagnostics). Cycling conditions (Eco Real-Time PCR System; Illumina) were as follows: 10 min at 95°C and 50 cycles of 15 s at 95°C and 60 s at 60°C, linked to a default dissociation stage program to detect nonspecific amplification. Expression levels were determined as the number of cycles needed for the amplification to reach a threshold fixed in the exponential phase of the PCR (CT; Pfaffl, 2001). To compare data from different cDNA samples, expression values for all selected genes were normalized with the corresponding expression values of ACT8 (Graeber et al., 2011). At least two replicates were used for every experiment.
Seed Germination Assays
Wild-type and mutant seeds were collected at the same time and obtained from plants grown in the same conditions. For each genotype, ∼50 seeds were placed on 0.6% agarose Petri dishes. Plates were sealed with Micropore tape (Micropore 3M) and incubated at 22°C under 16/8-h light/dark conditions. Germination was scored as radicle emergence through the endosperm and testa every 24 h. All germination assays were performed in triplicate with at least two independent seed batches.
In Vivo Imaging of Bioluminescence
In vivo imaging and quantification of luciferase activity were performed at 24 and 48 hai using a cooled CCD camera (NightOwl II LB 983 NC-100; Berthold Technologies) and the provided software. Seeds from representative 4xLIP and -58F8-pYRO lines (Castrillo et al., 2011) were sown on Murashige and Skoog agar plates containing 50 μM luciferin in the presence or absence of 5 μM PAC or 50 μM PAC plus 5 μM GA and stratified for 2 d (4°C in the dark). Plates were transferred to a chamber at a constant temperature of 22°C and a photoperiod of 16 h of light/8 h of dark. Since some of these treatments accelerate or delay testa rupture, an event required to detect light coming from inner tissues, we used only seeds producing detectable amounts of light for quantification.
Promoter Constructs
The 4xLIP:LUC and min:LUC constructs are described by Castrillo et al. (2011) and the ProML1:GFP-gai-1 construct by Gallego-Bartolomé et al. (2011b). To produce promoter:GUS fusions for transient expression assays in Arabidopsis mesophyll protoplasts, an ∼1000-bp LIP1 promoter (from −845 to +124 related to the TSS) was amplified from Arabidopsis Col-0 genomic DNA with the primers LO1502 and LO1503 (Supplemental Table 1), cloned into pGEM-T Easy, digested with XbaI and NcoI restriction enzymes, and ligated into pBT10 (Sprenger-Haussels and Weisshaar, 2000). The mutated version (pLIPm) was generated by PCR using two primer combinations (LO1502+1500 and LO1501+1503) that produced two fragments with overlapping ends containing the mutated sequence. An equimolecular mixture of both fragments was used as a template for a final PCR with primers LO1502+1503, and the final product was cloned into pBT10.
Transient Expression in Arabidopsis Mesophyll Protoplasts
Arabidopsis mesophyll protoplasts were produced as described by Yoo et al. (2007). Five micrograms of the appropriate GUS reporter plasmid and 1 μg of 35S:NAN plasmid for normalization were mixed in a solution of 4 mM MES, pH 5.7, 0.5 M mannitol, and 20 mM KCl and used to transform Arabidopsis mesophyll protoplasts. After 16 h of incubation in a growth cabinet at 22°C, protoplasts were collected and GUS and NAN activities were quantified according to Jefferson et al. (1987) and Kirby and Kavanagh (2002), respectively.
Yeast Constructs and Assays
A Gateway cassette flanked by attR recombination sequences was introduced into the NdeI/BamHI restriction sites of the pGBKT7 and pGADT7 plasmids (Clontech) to produce their corresponding Gateway-compatible versions (R. Solano, Spanish National Biotechnology Centre). CDSs for the truncated DELLA versions (de Lucas et al., 2008; Gallego-Bartolomé et al., 2010) fused to the GAL4 DNA binding domain (BD-DELLA) in the Gateway plasmid pGBKT7-GW, as well as the empty pGADT7-GW plasmid, were provided by S. Prat. We used a truncated version of DELLAs, since they have reduced autoactivation (de Lucas et al., 2008; Gallego-Bartolomé et al., 2010; Hou et al., 2010). CDSs of ATML1 and PDF2 fused to the GAL4 activation domain were obtained as follows. The ATML1 CDS was transferred from the pDEST22 plasmid (Invitrogen; Castrillo et al., 2011) into the pDONR221 (Invitrogen) and pGADT7-GW or pDEST32 plasmids by sequential BP and LR reactions (Invitrogen), respectively. The PDF2 CDS was amplified with primers LO1342 and LO1343 (Supplemental Table 1) from the SSP Gateway clone U16775 (Yamada et al., 2003) and cloned into the pDONR221 and pGADT7-GW or pDEST22 plasmids by sequential BP and LR reactions, respectively. The construction of plasmids carrying the wild-type or mutated LIP1 element, and growth and mating of yeast cells, were as described previously (Castrillo et al., 2011; Rueda-Romero et al., 2012). The Saccharomyces cerevisiae Y187α and YM4271 strains carrying bait and prey constructs, respectively, were used for DNA–protein interactions, and the pJ694α and YM4271 yeast strains carrying bait and prey constructs, respectively, were used for protein–protein interactions.
BiFC Constructs, Assays, and Confocal Imaging
For the BiFC constructs, CDSs of the studied proteins were initially cloned in entry Gateway-compatible plasmids as follows: ATML1 and PDF2 were described in the previous section, the RGL2 and GAI CDSs were obtained from an arrayed library (Castrillo et al., 2011) and transferred from the pDEST22 plasmid (Invitrogen) into the pDONR221 plasmid (Invitrogen) by BP reactions (Invitrogen), and the RGA CDS cloned into the pDONR/SD/D-TOPO plasmid (Invitrogen) was provided by S. Prat. Then, the entry plasmids were recombined into the Gateway binary destination vectors pXNGW, pNXGW, pCXGW, and pXCGW (W. Frommer, Carnegie Institution for Science; provided by R. Bustos) using LR Clonase (Invitrogen). Thus, each protein was independently tagged with cCFP or nYFP at either the N or C terminus. The binary vector backbone is derived from pPZP312, which contains a single 35S cauliflower mosaic virus promoter and terminator derived from pRT100.
The BiFC constructs were introduced into Agrobacterium tumefaciens C58 GV3101::pMP90, and the resulting strains were used for agroinfiltration of Nicotiana benthamiana leaves. A pBIN61-35S:P19 plasmid was always coinfiltrated to avoid gene silencing (Voinnet et al., 2003).
BiFC images were taken by laser scanning microscopy using a Leica TCS SP8 confocal microscope.
Bioinformatic (in Silico) Analyses
The total number of occurrences of a given cis-element (X) in all 500-bp Arabidopsis gene promoters (Y) was obtained using the patmatch tool on the TAIR web page (http://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl). The total number of occurrences of a given cis-element (A) in all 500-bp gene promoters (Z) of specific transcriptomes was obtained by using the DNA-pattern tool of the RSAT web page (http://rsat.ulb.ac.be/). The average occurrences were calculated by dividing X/Y and A/Z. The average occurrence in the Arabidopsis genes represented on the Affymetrix chip used in transcriptomic analyses was taken arbitrarily as 1. The χ2 test was used for statistical analysis.
LIP2 coregulation data were obtained by querying the Botany Array Resource data set with the ATML1 locus (At4g21750) using the expression angler tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi; Toufighi et al., 2005).
mRNA in Situ Hybridization
In situ hybridization was performed as described by Iglesias-Fernández et al. (2011). Oligonucleotides used to amplify gene-specific probes for LIP1 (LO1496 and LO1497), ATML1 (LO1827+LO1828), and PDF2 (LO1829+LO1830) are described in Supplemental Table 1.
amiRNA Construction
Arabidopsis transgenic lines expressing the amiRTPL-353 amiRNA (5′-UUUUAUACAUGUUGUGAGCCG-3′) were obtained in the laboratory of M.R. Ponce as described by Jover-Gil et al. (2014). In brief, the transgene designed to express amiRTPL-353 was constructed in the backbone of the gene encoding miR319a, an endogenous Arabidopsis microRNA. The sequence of the mature miR319a was replaced by that of amiRTPL-353, as described by Schwab et al. (2006) and at http://wmd3.weigelworld.org/cgi-bin/webapp.cgi. The amiRTPL-353 construct was flanked with attB1 and attB2 sites to enable use of the Gateway technology and inserted by means of a BP reaction into pGEM-T Easy221. The plasmid obtained in this way was used to transform Escherichia coli DH5α cells. Plasmid DNA was isolated from transformants, and the insert was transferred into the pMDC32 destination vector (Curtis and Grossniklaus, 2003) and then mobilized into Agrobacterium C58C1 cells. Arabidopsis plants were transformed by infection using the floral dip method (Clough and Bent, 1998).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATML1 (At4g21750), PDF1 (At2g42840), FDH (At2g26250), LTP1 (At2g38540), LIP2 (At4g18970), LIP1 (At5g45670), PDF2 (At4g04890), MAN7 (At5g66460), MLS (At5g03860), ACT8 (At1g49240), RGL2 (At3g03450), RGA (At2g01570), GAI (At1g14920), HDG2 (At1g05230), ACR4 (At3g59420), KCS20 (At5g43760), PAS2 (At5g10480), EXP8 (At2g40610), and GA20ox3 (At5g07200).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Kinetics of Seed Germination and AtGA20ox3 Expression in Samples Used in Figure 1A.
Supplemental Figure 2. Expression of Minimal and 4xLIP1 Promoter Constructs in Response to GA.
Supplemental Figure 3. GARE Occurrences in GA-Related Arabidopsis Seed Transcriptomes and L1 Box Sequences in the LIP1 and LIP2 Promoters.
Supplemental Figure 4. BiFC Negative Assays.
Supplemental Figure 5. Sequence of amiRTPL-353 and mRNA Levels of Target Genes.
Supplemental Figure 6. GA Cannot Restore Epidermal Gene Expression in ATML1/PDF2-Silenced Lines.
Supplemental Table 1. List of Oligonucleotides Used.
Supplementary Material
Acknowledgments
We thank M.R. Ponce and J.L. Micol (Instituto de Bioingeniería, Universidad Miguel Hernández) for the design, construction, cloning, and transfer into Arabidopsis plants of amiRTPL-353; Tai Pin Sun (Duke University) for rgl2-13 seeds; M.A. Blázquez and D. Alabadi (Institute for Plant Molecular and Cell Biology, Spain) for ProML1:GFP-gai-1 seeds; S. Prat (Spanish National Biotechnology Centre) for providing the BD-DELLA and pDONR/SD/D-TOPO:RGA constructs, the pGADT7-GW plasmid, and the ga1-3, ga1-3 rgl2-1, ga1-3 rgl2-1 gai-t6 rga-t2, and ga1-3 rgl2-1 gai-t6 rga-t2 rgl1-1 seeds; and R. Bustos (Centre for Plant Biotechnology and Genomics, Spain) for advice regarding statistical analyses and BiFC assays, providing the BiFC plasmids, and critical reading of the article. This work was supported by the Spanish Ministry of Science and Innovation (Grants BIO2010-17334 and CONSOLIDER CSD2007-00057 and a predoctoral fellowship to P.R.-R.). This article is dedicated to the memory of Gabriel Salcedo-Durán.
AUTHOR CONTRIBUTIONS
B.R.-C., P.R.-R., R.I.-F., and L.O.-S. performed research. P.C. and L.O.-S. designed the research and analyzed data. L.O.-S. wrote the article.
Glossary
- GA
gibberellin
- TF
transcription factor
- qRT-PCR
quantitative RT-PCR
- PAC
paclobutrazol
- hai
hours after imbibition
- Col-0
Columbia-0
- GARE
gibberellin-responsive cis-element
- CDS
coding sequence
- BiFC
bimolecular fluorescence complementation
- amiRNA
artificial microRNA
- VLCFA
very-long-chain fatty acid
- Ler
Landsberg erecta
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abe M., Katsumata H., Komeda Y., Takahashi T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643. [DOI] [PubMed] [Google Scholar]
- Abe M., Takahashi T., Komeda Y. (1999). Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 40: 571–580. [DOI] [PubMed] [Google Scholar]
- Abe M., Takahashi T., Komeda Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26: 487–494. [DOI] [PubMed] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T.A., Lawrenson T., Sablowski R., Østergaard L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24: 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Weigel D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892. [DOI] [PubMed] [Google Scholar]
- Cao D., Cheng H., Wu W., Soo H.M., Peng J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142: 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Hussain A., Cheng H., Peng J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113. [DOI] [PubMed] [Google Scholar]
- Castrillo G., Turck F., Leveugle M., Lecharny A., Carbonero P., Coupland G., Paz-Ares J., Oñate-Sánchez L. (2011). Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6: e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Singh K.B. (1999). The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 19: 667–677. [DOI] [PubMed] [Google Scholar]
- Chepyshko H., Lai C.P., Huang L.M., Liu J.H., Shaw J.F. (2012). Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: New insights from bioinformatics analysis. BMC Genomics 13: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cornah J.E., Germain V., Ward J.L., Beale M.H., Smith S.M. (2004). Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279: 42916–42923. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2000). Loosening of plant cell walls by expansins. Nature 407: 321–326. [DOI] [PubMed] [Google Scholar]
- Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., Vachon G. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136: 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.M., Achard P. (2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Davière J.M., de Lucas M., Prat S. (2008). Transcriptional factor interaction: A central step in DELLA function. Curr. Opin. Genet. Dev. 18: 295–303. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Duan L., Dietrich D., Ng C.H., Chan P.M., Bhalerao R., Bennett M.J., Dinneny J.R. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado J.A., Huang D., Wicki-Stordeur L., Hemstock L.E., Potentier M.S., Tsang E.W., Cutler A.J. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23: 1772–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio M., Alabadí D., Pérez-Gómez J., García-Cárcel L., Phillips A.L., Hedden P., Blázquez M.A. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142: 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Alabadí D., Blázquez M.A. (2011a). DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Arana M.V., Vandenbussche F., Zádníková P., Minguet E.G., Guardiola V., Van Der Straeten D., Benkova E., Alabadí D., Blázquez M.A. (2011b). Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 67: 622–634. [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Marín J.A., Prat S., Blázquez M.A., Alabadí D. (2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27: 1247–1256. [DOI] [PubMed] [Google Scholar]
- Geng Y., Wu R., Wee C.W., Xie F., Wei X., Chan P.M., Tham C., Duan L., Dinneny J.R. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25: 2132–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K., Linkies A., Wood A.T., Leubner-Metzger G. (2011). A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 23: 2045–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Jacobsen J.V. (1992). Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell 4: 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M.J., Chory J., Savaldi-Goldstein S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale A.L., Ariizumi T., Steber C.M. (2012). Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol. 160: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J.O., Chang K.S., Kim I.A., Lee M.H., Lee S.A., Song S.K., Lee M.M., Lim J. (2011). Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 108: 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54. [DOI] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R., Barrero-Sicilia C., Carrillo-Barral N., Oñate-Sánchez L., Carbonero P. (2013). Arabidopsis thaliana bZIP44: A transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J. 74: 767–780. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R., Rodríguez-Gacio M.C., Barrero-Sicilia C., Carbonero P., Matilla A. (2011). Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233: 25–36. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse E.M., Gan Y., Bou-Torrent J., Stewart K.L., Gilday A.D., Jeffree C.E., Vaistij F.E., Martínez-García J.F., Nagy F., Graham I.A., Halliday K.J. (2011). A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell 23: 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J., Raffaele S., Bourdenx B., Garcia C., Laroche-Traineau J., Moreau P., Domergue F., Lessire R. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67: 547–566. [DOI] [PubMed] [Google Scholar]
- Jover-Gil S., Paz-Ares J., Micol J.L., Ponce M.R. (2014). Multi-gene silencing in Arabidopsis: A collection of artificial microRNAs targeting groups of paralogs encoding transcription factors. Plant J., in press. [DOI] [PubMed]
- Kamata N., Okada H., Komeda Y., Takahashi T. (2013). Mutations in epidermis-specific HD-ZIP IV genes affect floral organ identity in Arabidopsis thaliana. Plant J. 75: 430–440. [DOI] [PubMed] [Google Scholar]
- Kirby J., Kavanagh T.A. (2002). NAN fusions: A synthetic sialidase reporter gene as a sensitive and versatile partner for GUS. Plant J. 32: 391–400. [DOI] [PubMed] [Google Scholar]
- Koornneef M., van der Veen J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58: 257–263. [DOI] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turecková V., Strnad M., Lopez-Molina L. (2010). A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. USA 107: 19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16: 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A., Blázquez M.A., Alabadí D. (2013a). Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Curr. Biol. 23: 804–809. [DOI] [PubMed] [Google Scholar]
- Locascio A., Blázquez M.A., Alabadí D. (2013b). Genomic analysis of DELLA protein activity. Plant Cell Physiol. 54: 1229–1237. [DOI] [PubMed] [Google Scholar]
- Löfke C., Zwiewka M., Heilmann I., Van Montagu M.C., Teichmann T., Friml J. (2013). Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc. Natl. Acad. Sci. USA 110: 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Porat R., Nadeau J.A., O’Neill S.D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8: 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M.D., Wenger J.P., Gilding E., Jilk R., Dixon R.A. (2009). Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant 2: 803–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K.T., Takahashi T. (2006). Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 141: 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa T., Okushima Y., Nagata N., Kojima M., Sakakibara H., Umeda M. (2013). Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 11: e1001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Vicente-Carbajosa J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Nguyen K.T., Park E., Jeon J.S., Choi G. (2013). DELLA proteins and their interacting RING finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006. [DOI] [PubMed] [Google Scholar]
- Penfield S., Li Y., Gilday A.D., Graham S., Graham I.A. (2006). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Rylott E.L., Gilday A.D., Graham S., Larson T.R., Graham I.A. (2004). Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28: 2259–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209. [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P., Barrero-Sicilia C., Gómez-Cadenas A., Carbonero P., Oñate-Sánchez L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63: 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Chory J. (2008). Growth coordination and the shoot epidermis. Curr. Opin. Plant Biol. 11: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202. [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263. [DOI] [PubMed] [Google Scholar]
- Shani E., Weinstain R., Zhang Y., Castillejo C., Kaiserli E., Chory J., Tsien R.Y., Estelle M. (2013). Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. USA 110: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K., Olsen F.L., Rogers J.C., Mundy J. (1991). Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88: 7266–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwinska E., Bassel G.W., Bewley J.D. (2009). Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J. Exp. Bot. 60: 3587–3594. [DOI] [PubMed] [Google Scholar]
- Sprenger-Haussels M., Weisshaar B. (2000). Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 22: 1–8. [DOI] [PubMed] [Google Scholar]
- Stamm P., Ravindran P., Mohanty B., Tan E.L., Yu H., Kumar P.P. (2012). Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol. 12: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang J.A., Gallego-Bartolomé J., Gómez M.D., Yoshida S., Asami T., Olsen J.E., García-Martínez J.L., Alabadí D., Blázquez M.A. (2009). Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60: 589–601. [DOI] [PubMed] [Google Scholar]
- Sun T.P., Kamiya Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Jürgens G. (2007). Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134: 1141–1150. [DOI] [PubMed] [Google Scholar]
- Takada S., Takada N., Yoshida A. (2013). ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919–1923. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Watanabe M., Watanabe D., Tanaka T., Machida C., Machida Y. (2002). ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 43: 419–428. [DOI] [PubMed] [Google Scholar]
- Thoma S., Hecht U., Kippers A., Botella J., De Vries S., Somerville C. (1994). Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 105: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: E-northerns, expression angling, and promoter analyses. Plant J. 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135: 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199. [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Swarup R., Coates J., Swarup K., Laplaze L., Beemster G.T., Hedden P., Bhalerao R., Bennett M.J. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10: 625–628. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wild M., Davière J.M., Cheminant S., Regnault T., Baumberger N., Heintz D., Baltz R., Genschik P., Achard P. (2012). The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y., Sun T. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28: 443–453. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A., Wisman E., Huijser P., Huijser C., Wellesen K., Saedler H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu H., Ito T., Zhao Y., Peng J., Kumar P., Meyerowitz E.M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 101: 7827–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Galvão V.C., Zhang Y.C., Horrer D., Zhang T.Q., Hao Y.H., Feng Y.Q., Wang S., Schmid M., Wang J.W. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. (2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.