This work reports the discovery of the DELLA-binding transcription factor GAF1 and shows that DELLAs and TPR act as coactivators and a corepressor with GAF1, respectively. GA converts the GAF1 complex from transcriptional activator to repressor via degradation of DELLAs. Accordingly, DELLAs turn on or off two sets of GA-regulated genes by dual functions, namely titration and coactivation.
Abstract
Gibberellins (GAs) are essential regulators of plant development, and DELLAs are negative regulators of GA signaling. The mechanism of GA-dependent transcription has been explained by DELLA-mediated titration of transcriptional activators and their release through the degradation of DELLAs in response to GA. However, the effect of GA on genome-wide expression is predominantly repression, suggesting the existence of unknown mechanisms of GA function. In this study, we identified an Arabidopsis thaliana DELLA binding transcription factor, GAI-ASSOCIATED FACTOR1 (GAF1). GAF1 shows high homology to INDETERMINATE DOMAIN1 (IDD1)/ENHYDROUS. GA responsiveness was decreased in the double mutant gaf1 idd1, whereas it was enhanced in a GAF1 overexpressor. GAF1 binds to genes that are subject to GA feedback regulation. Furthermore, we found that GAF1 interacts with the corepressor TOPLESS RELATED (TPR) and that DELLAs and TPR act as coactivators and a corepressor of GAF1, respectively. GA converts the GAF1 complex from transcriptional activator to repressor via the degradation of DELLAs. These results indicate that DELLAs turn on or off two sets of GA-regulated genes via dual functions, namely titration and coactivation, providing a mechanism for the integrative regulation of plant growth and GA homeostasis.
INTRODUCTION
Hormonal regulation of transcription is a key control point for plant growth and development. Gibberellins (GAs), which are tetracyclic diterpenoid growth factors, are essential regulators of many aspects of plant development, including seed germination, stem elongation, flower induction, and anther development. The endogenous levels of GAs are fine-tuned by feedback control at several steps in their metabolic pathway, including GA 20-oxidase and GA 3-oxidase (Yamaguchi, 2008). GA feedback regulation has been shown to depend on GA signaling components (Sun, 2011), but its molecular mechanisms are still largely unknown.
DELLA proteins are major plant growth repressors. Upon binding to a soluble GA receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1), GA triggers the degradation of DELLAs through the ubiquitin-proteasome pathway, thereby promoting plant growth (Ueguchi-Tanaka et al., 2007b; Sun, 2011). Arabidopsis thaliana contains five DELLAs, GIBBERELLIN-INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3, which display partially overlapping but also distinct functions in repressing GA responses (Sun and Gubler, 2004). Among the DELLAs, GAI and RGA are the major GA repressors during vegetative growth and floral induction (Mutasa-Göttgens and Hedden, 2009). Because DELLAs localize in the nucleus and show structural similarities to mammalian STAT (for signal transducers and activators of transcription) proteins (Richards et al., 2000), they are thought to be involved in transcription. Several DELLA binding transcription factors have been identified to date. For example, DELLAs regulate hypocotyl elongation by interacting with PHYTOCHROME INTERACTING FACTORS (PIFs) (de Lucas et al., 2008; Feng et al., 2008) and BRASSINAZOLE RESISTANT1 (BZR1) (Bai et al., 2012; Gallego-Bartolomé et al., 2012) and also play a role in plant defense by interacting with JASMONATE ZIM-DOMAIN (JAZ) proteins (Hou et al., 2010). Through these interactions, DELLAs inhibit the activity of these proteins (Hauvermale et al., 2012). Thus, DELLAs function as signaling nodes that mediate the crosstalk of endogenous programs and various environmental stimuli.
Among these transcription factors, PIFs are the most studied. PIFs promote hypocotyl elongation and are negatively regulated by the photoreceptor PHYTOCHROME B. The interaction between DELLAs and PIFs inhibits PIF-induced hypocotyl elongation by blocking the DNA binding activities of PIFs (de Lucas et al., 2008; Feng et al., 2008). GA triggers the degradation of DELLAs, which release PIFs to activate the target genes, including LIPID TRANSFER PROTEIN3, β-EXPANSIN, and PACLOBUTRAZOL RESISTANCE, and thus promotes hypocotyl elongation (de Lucas et al., 2008). The titration of transcriptional activators by DELLAs partly explains GA-dependent transcriptional activation and how plants integrate environmental stimuli and GA signals to optimize growth and development.
However, genes encoding GA 20-oxidase and GA 3-oxidase are downregulated by GA via feedback regulation. Genome-wide analysis revealed that the effect of GA on gene expression is predominantly through repression, whereas that of DELLAs is through activation (Zentella et al., 2007; Hirano et al., 2012). Furthermore, a chromatin immunoprecipitation (ChIP) assay showed an in vivo association of DELLAs with promoters of several genes, although DELLAs lack known DNA binding motifs (Zentella et al., 2007; Lim et al., 2013; Park et al., 2013). All of the DELLA-regulated genes, including GA biosynthetic enzyme genes and GA receptor genes, are repressed by GA and activated by DELLAs (Zentella et al., 2007). These observations cannot be explained by the conventional titration model. Thus, other molecular mechanisms underlying GA-dependent transcriptional regulation must exist.
Here, we describe a previously unknown DELLA binding transcription factor, designated GAI-ASSOCIATED FACTOR1 (GAF1). DELLAs and TOPLESS-RELATED (TPR) act as coactivators and a corepressor of GAF1, respectively, in GA-mediated transcriptional regulation. GA converts the GAF1 complex from transcriptional activator to repressor. DELLAs simultaneously turn on or off two sets of GA-regulated genes via two mechanisms, namely titration and coactivation. Our results thus provide insight into the mechanism of regulation of GA homeostasis and plant growth by DELLA proteins.
RESULTS
Isolation of GAF1, a DELLA-Interacting Protein
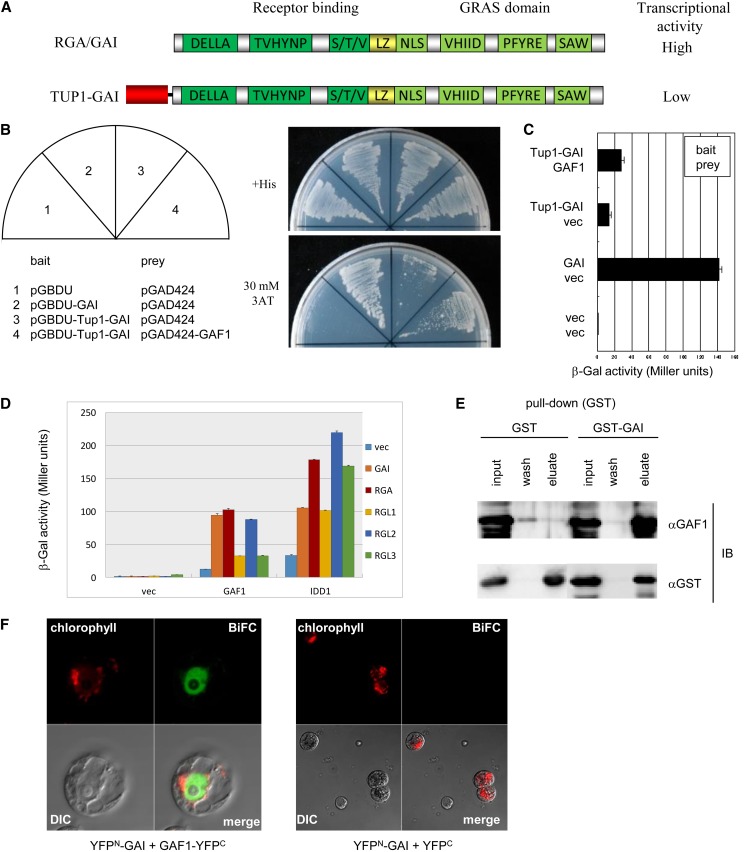
Because DELLAs exhibit strong transcriptional activity in yeast, it is difficult to screen for DELLA-interacting proteins using the conventional yeast two-hybrid (Y2H) system with a full-length DELLA as bait (Figures 1A to 1C). To overcome this problem, we developed a modified Y2H system in which a bait protein, the Arabidopsis DELLA GAI, was fused to Tup1, a general repressor from yeast. The N-terminal domain of Tup1 (1 to 200 bp), which was sufficient for repression (Jabet et al., 2000; Hirst et al., 2001), reduced the transcriptional activity of GAI in the Tup1-GAI fusion protein (Figures 1B and 1C). We performed a Y2H screen with Tup1-GAI as bait using an Arabidopsis cDNA library. The GAI-interacting protein GAF1 was isolated from 1.6 × 106 transformants. Y2H assays showed that GAF1 interacted with all Arabidopsis DELLAs, namely, GAI, RGA, and RGL1 to RGL3 (Figure 1D), and pull-down assays showed direct interaction between GAI and GAF1 (Figure 1E).
Figure 1.
Identification of a DELLA Interactor Using a Modified Y2H System.
(A) Schematic representation of DELLA proteins. The fusion of the repression domain of Tup1 repressed the strong transcriptional activity of GAI.
(B) Y2H assay. Tup1-GAI interacts with GAF1 in Y2H assays. Transformed yeast cells were streaked on a plate with His (+His) or without His but with 30 mM aminotriazole (3AT).
(C) β-Galactosidase activity for the Tup1 two-hybrid system. Data are means ± sd; n = 3.
(D) GAF1 and IDD1 interact with five DELLA proteins in yeast β-galactosidase assays. Data are means ± sd; n = 3. vec indicates empty vector used as a negative control.
(E) In vitro pull-down assays with GST-GAI protein. GST and GST-GAI proteins were incubated with recombinant 6×His-GAF1 protein bound to Glutathione Sepharose 4B and then eluted and analyzed by immunoblotting (IB) with anti-GAF1 antibody (top) and anti-GST antibody (bottom).
(F) BiFC analysis showing interaction between GAF1 and GAI. CaMV35S:GAF1-YFPC and CaMV35S:YFPN-GAI plasmids were introduced and transiently expressed in protoplasts of T87 Arabidopsis cultured cells (left). CaMV35S:YFPC and CaMV35S:YFPN-GAI plasmids were introduced into protoplasts of T87 Arabidopsis cultured cells as a negative control (right). DIC, differential interference contrast.
To investigate protein–protein interaction between GAI and GAF1 in plant cells, we performed bimolecular fluorescence complementation (BiFC) analysis using Arabidopsis T87 cultured cells. The reconstituted yellow fluorescent protein (YFP) signal, caused by interaction between YFPN-GAI and GAF1-YFPC, was observed in the nucleus of the protoplasts of T87 cells; no YFP signal was observed when YFPN-GAI was cotransfected with YFPC (Figures 1F and 1G). These results suggest that GAF1 binds to GAI in the nucleus of plant cells.
GAF1 Belongs to the IDD Transcription Factor Family
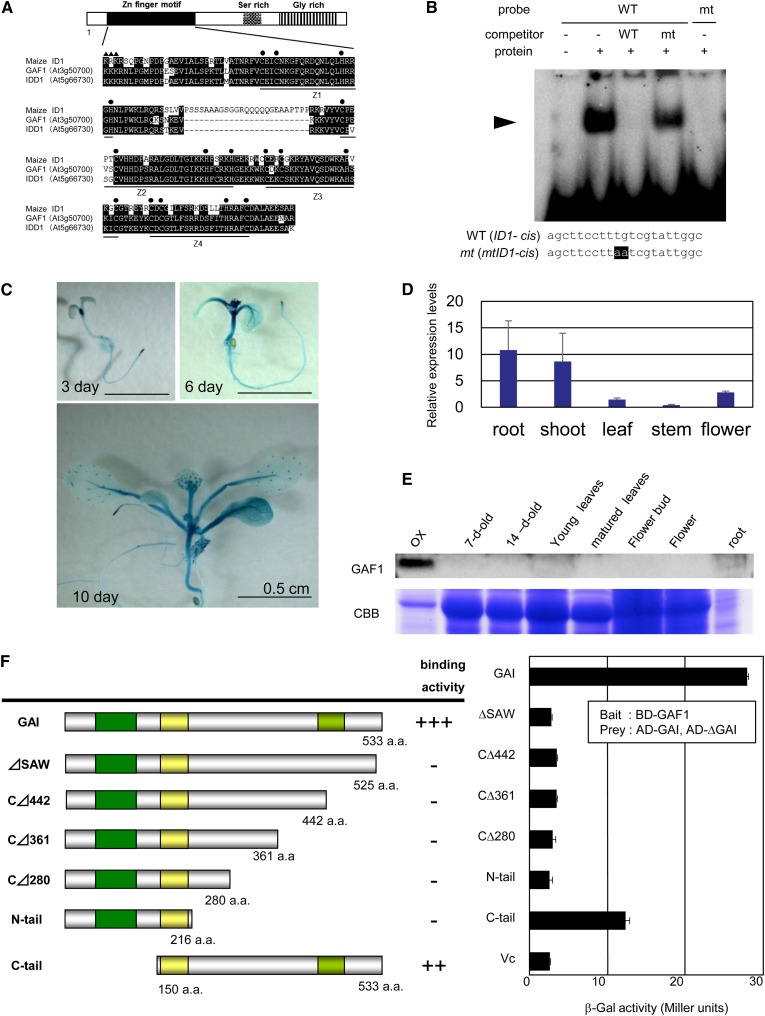
GAF1 encodes a transcription factor with zinc finger motifs that shows similarity to maize (Zea mays) INDETERMINATE1 (ID1) (Colasanti et al., 1998, 2006). Mutations in maize ID1 have severe effects on floral transition (Singleton, 1946; Colasanti et al., 1998), resulting in late flowering, demonstrating that ID1 is essential for normal floral transition in monocots. ID1 appears to be specific to monocots, and functional orthologs are not found in Arabidopsis (Colasanti et al., 2006). Although the Arabidopsis genome contains 16 ID1-related proteins (IDD, for ID1 domain protein), their amino acid sequence similarity to ID1 is limited to the zinc finger motif that is important for DNA binding (Figure 2A). Maize ID1 binds to the consensus sequence TTTTGTCG (Kozaki et al., 2004). To determine whether GAF1 binds to this sequence, we performed a gel retardation assay. Recombinant GAF1 protein specifically bound to the ID1 binding sequence (Figure 2B), suggesting that GAF1 is a sequence-specific DNA binding protein.
Figure 2.
Characterization of GAF1.
(A) Alignments of the zinc finger domains of GAF1, IDD1, and maize ID1 proteins. Identical residues are highlighted in black. The solid circles indicate the Cys and His residues in the C2H2-type zinc finger motifs. The solid triangles indicate the nuclear localization signal.
(B) Gel retardation assay using recombinant GAF1 protein. Oligonucleotides containing ID1-cis (WT; lanes 1 to 4) or mtID1-cis (mt; lane 5) were used as probes. The mutated bases are highlighted. WT and mt, competition with a 1000-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1-DNA complexes are indicated by the arrowhead. +, addition to the reaction mixtures; –, omission from the reaction mixtures.
(C) GUS expression pattern in transgenic plants expressing GAF1 promoter:GUS. Analysis was performed in 3-, 6-, and 10-d-old seedlings. Bars = 5 mm.
(D) Quantitative real-time PCR analysis of GAF1 expression in different tissues. UBQ11 was used as an internal control. Data are means ± sd; n = 3.
(E) Immunoblot analysis of GAF1 protein expression in different tissues of Col-0 and 35S:GAF1-overexpressing transgenic Arabidopsis. Coomassie blue (CBB) staining was used as a loading control.
(F) Identification by Y2H assay of the domain of GAI that binds to GAF1. Schematic representations of GAI-truncated proteins used for Y2H assays. Yeast was transformed with a combination of the indicated plasmids, and subsequently, β-galactosidase activity was determined. Data are means ± sd; n = 3.
To investigate the expression pattern of GAF1 in plants, we monitored the activity of β-glucuronidase (GUS) driven by the GAF1 promoter (Figure 2C). Histochemical analysis indicated that the GAF1 promoter is mainly expressed in hypocotyls, petioles, shoot apices, root tips, and trichomes (Figure 2C). This observation is consistent with the microarray data derived from different stages of Arabidopsis development (AtGenExpress). Expression of GAF1 mRNAs in various organs was confirmed by quantitative real-time PCR (Figure 2D). We further investigated the expression of GAF1 protein by immunoblot analysis using antibody raised against recombinant GAF1. Our analysis detected GAF1 in transgenic plants expressing GAF1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter but not in wild-type plants (Figure 2E). These results indicate that GAF1 is mainly expressed in vegetative tissues at low levels.
The SAW DOMAIN of GAI Is Important for GAF1 Binding
DELLAs belong to a subclass of the plant-specific GRAS family, a family of transcriptional regulators. Like all GRAS proteins, DELLAs share a conserved C-terminal domain that is involved in transcriptional regulation and is characterized by two Leu heptad repeats (LHRI and LHRII) and three conserved motifs, VHIID, PFYRE, and SAW (Itoh et al., 2002; Ueguchi-Tanaka et al., 2007a) (Figure 1A). DELLAs are distinguished from the rest of the GRAS family by a specific N-terminal sequence containing two conserved domains: the DELLA domain (which gives them their name) and the TVHYNP domain. To determine which region of GAI is important for interaction with GAF1, we generated various deletion mutants of GAI (Figure 2F) and performed Y2H assays. As shown in Figure 2F, deletion of the N-terminal region of GAI did not affect GAF1 binding. By contrast, deletion of the C-terminal eight amino acids containing the core sequence of the SAW domain abolished GAF1 binding (ΔSAW). Because the mutation in the SAW domain of SLENDER RICE1 (SLR1), a rice (Oryza sativa) DELLA, caused slender phenotypes in rice (Ikeda et al., 2001), the SAW domain of DELLA is thought to be necessary for the repression of GA responses (Itoh et al., 2002; Ueguchi-Tanaka et al., 2007a). These results suggest that GAF1 might be involved in GA responses through binding to the SAW domain of DELLA.
The gaf1 idd1 Double Mutant and GAF1 Overexpressor Lines Show GA-Related Phenotypes
The most closely related protein to GAF1 among the Arabidopsis IDD family is IDD1. The two proteins share 68.5% amino acid sequence similarity. IDD1 is also known as ENHYDROUS (ENY) and promotes the transition of seeds to germination by regulating light and hormonal signaling during their maturation (Feurtado et al., 2011). IDD1/ENY also interacts with DELLAs (Feurtado et al., 2011; Figure 1D); however, the functional significance of this remains unclear. GAF1 and IDD1 show distinct but overlapping expression patterns (i.e., GAF1 is expressed mainly in vegetative tissues) (Figure 2C), while IDD1/ENY is expressed mainly in seeds and at lower levels in vegetative tissues (Feurtado et al., 2011). These observations suggest that GAF1 and IDD1 could play partially redundant roles in plants.
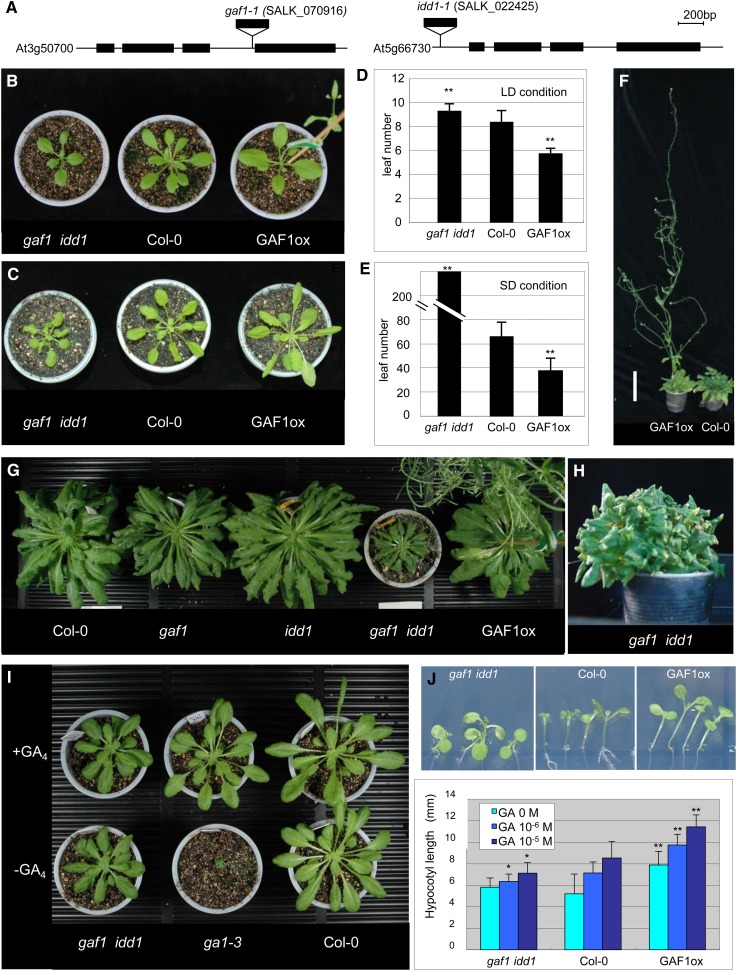
To investigate the function of GAF1, we compared the gaf1 and idd1 mutants with GAF1 overexpressors (Supplemental Figure 1). A T-DNA insertional mutant line for GAF1 (SALK_070916) was obtained and confirmed as a transcriptional knockout (Figure 3A; Supplemental Figure 2A). However, the only available mutant line for IDD1 (SALK_022425) has a T-DNA insertion in the promoter region, leading to reduced expression of IDD1 as compared with that of the Columbia-0 (Col-0) wild type (Figure 3A; Supplemental Figure 2A).
Figure 3.
Phenotypes of GAF1 Overexpressor and the gaf1 idd1 Mutant.
(A) Positions of the T-DNA insertions within At3g50700 in line SALK_070916 (gaf1) and At5g66730 in line SALK_022425 (idd1). Bar = 200 bp.
(B) A 25-d-old of gaf1 idd1 double mutant plant exhibiting a semidwarf phenotype under long-day (LD) conditions.
(C) A 40-d-old of gaf1 idd1 double mutant plant exhibiting a semidwarf phenotype under SD conditions.
(D) and (E) Flowering time analysis (rosette leaf number) under LD and SD conditions. The GAF1 overexpressor exhibits early flowering under LD and SD conditions. The gaf1 idd1 mutant exhibits slightly delayed flowering in LD conditions and extremely delayed flowering under SD conditions (n > 8). Asterisks represent significance by Student’s t test compared with Col-0 (**P < 0.01).
(F) One hundred-day-old GAF1-overexpressing plants exhibiting early flowering and taller phenotypes. Bar = 10 cm.
(G) Phenotypes of Col-0, gaf1, idd1, gaf1 idd1, and GAF1 overexpressor plants grown for 90 d under SD conditions.
(H) Extremely delayed flowering phenotype of the gaf1 idd1 mutant (8 months old) under SD conditions.
(I) The gaf1 idd1 mutant is GA insensitive. Plants were treated or not with bioactive GA4 (1 μM). GA4 was applied once per week for 2 months.
(J) Hypocotyl lengths of 11-d-old gaf1 idd1, Col-0, and GAF1 overexpressor plants grown on GA3-containing medium. Data are means ± sd; n = 10. The photographs in the top panel show hypocotyl lengths of 11-d-old gaf1 idd1, Col-0, and GAF1 overexpressor plants grown on medium containing 0, 1, or 10 μM GA3. Asterisks represent significance by Student’s t test compared with Col-0 (**P < 0.01 and *P < 0.05).
Because gaf1 showed slightly late flowering under short-day (SD) conditions, which is a GA-related phenotype, we generated and characterized a gaf1 idd1 double mutant (Figure 3). GAs affect flowering: GA-deficient mutants are late flowering and a DELLA quadruple mutant was shown to be early flowering under SD conditions (Cheng et al., 2004). We examined the effects of mutations and the overexpression of GAF1 on flowering. The gaf1 idd1 double mutant flowered later than the wild type, especially under SD conditions (Figures 3B to 3E), while the GAF1 overexpressor flowered earlier (Figures 3D to 3F). In addition to this, the gaf1 idd1 double mutant showed a semidwarf phenotype (Figures 3G and 3H). To confirm that the late flowering of gaf1 idd1 under SD conditions is due to the mutations in GAF1 and IDD1, we performed a complementation experiment. The late-flowering phenotype of gaf1 idd1 was rescued by the expression of a GAF1-GFP fusion gene under the control of the GAF1 promoter (Supplemental Figure 3). The GAF1 overexpressor plants were also taller (reaching 100 cm) than the wild type under SD conditions (Figure 3F). Furthermore, treatment with GA4, which restores the growth of ga1-3, a GA-deficient mutant, did not affect the semidwarf phenotype of gaf1 idd1 (Figure 3I), suggesting that GA responsiveness is reduced in gaf1 idd1.
As GAs promote the elongation of the hypocotyl, we examined the effects of bioactive GA application on the hypocotyl length of gaf1 idd1 and GAF1 overexpressor. The hypocotyl of the GAF1 overexpressor was longer than that of the wild type either in the presence or absence of exogenous GA, whereas that of gaf1 idd1 was shorter even in the presence of GA (Figure 3J). Another mutant allele of IDD1 (TL-7) exhibited similar phenotypes in the gaf1 background (Supplemental Figure 4). These results suggest that the phenotype of gaf1 idd1 results from reduced responsiveness to GA and that both GAF1 and IDD1 are involved in GA signaling.
Binding Domain of GAF1 to GAI
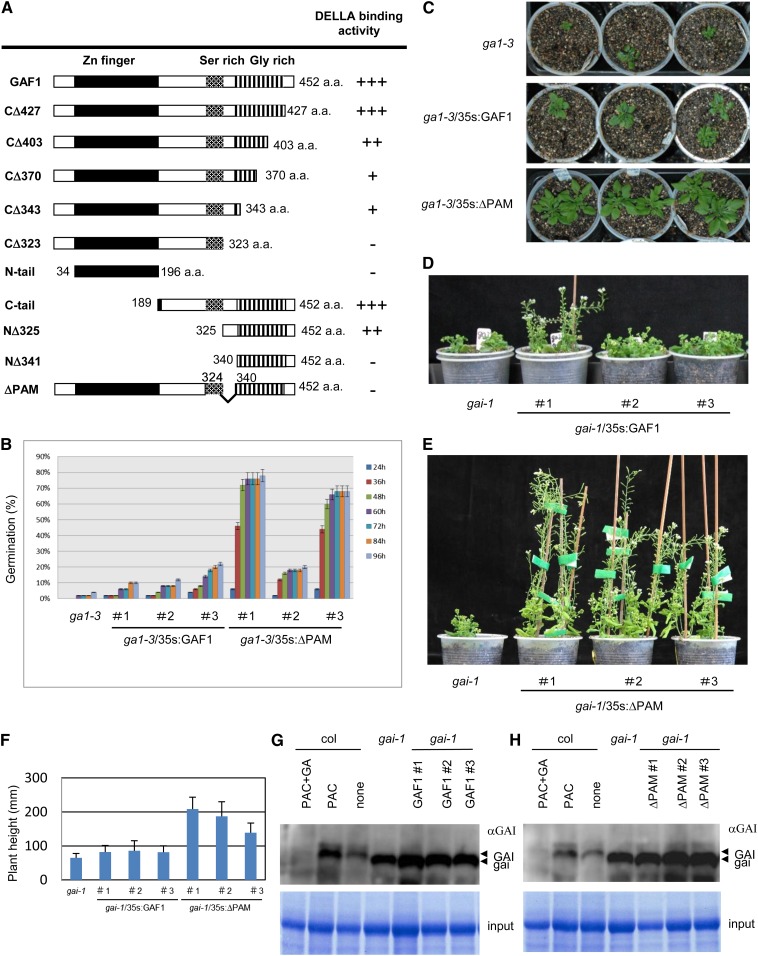
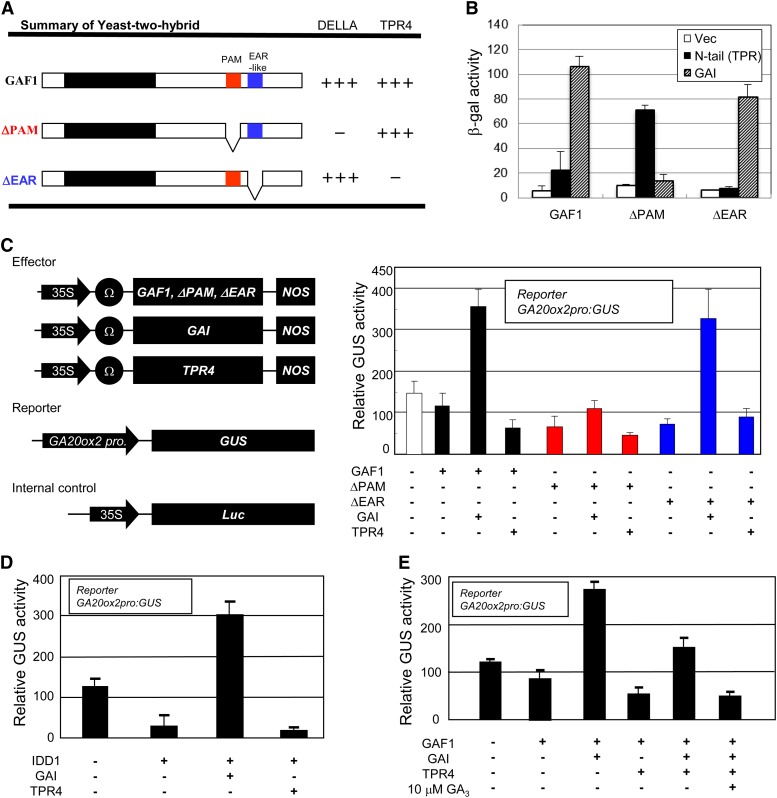
To identify the domain of GAF1 that interacts with GAI, we generated various deletion mutants of GAF1 (Figure 4A). GAI bound to the C-terminal half of GAF1 (189 to 452) but not to the zinc finger domain of GAF1 (34 to 196) in the Y2H assay (Figure 4A). Internal deletion of 16 amino acids from 325 to 340 of GAF1, which we refer to hereafter as the PAM domain, abolished GAI binding (Figure 4A; Supplemental Figure 5A); however, this deletion did not affect binding to another protein, TPR (see below). These results suggest that the PAM domain of GAF1 is necessary for GAI binding.
Figure 4.
Effect of Overexpression of GAF1 or ΔPAM in the ga1-3 or gai-1 Mutant.
(A) Schematic representations of GAF1 proteins used for Y2H assays. Levels of DELLA binding activity are indicated by + or –. The + and – signs indicate interaction and no interaction between truncated GAF1 and GAI, respectively.
(B) Germination percentage of seeds of the ga1-3 mutant compared with that of ga1-3/35S:GAF1 or ga1-3/35S:ΔPAM. Two days after stratification, seeds were transferred to medium without GA. Germination was monitored every 12 h between 24 and 96 h after imbibition.
(C) Representative plants of ga1-3, ga1-3/35S:GAF1, and ga1-3/35S:ΔPAM grown in soil for 40 d.
(D) Representative plants of gai-1 and gai-1/35S:GAF1 grown in soil for 70 d.
(E) Representative plants of gai-1 and gai-1/35S:ΔPAM grown in soil for 70 d.
(F) Final height of gai-1, gai-1/35S:GAF1, and gai-1/35S:ΔPAM transgenic plants. Error bars represent sd.
(G) and (H) Immunoblot analysis of gai-1, gai-1/35S:GAF1, and gai-1/35S:ΔPAM transgenic plants with anti-GAI antibody. Seven-day-old Col-0 seedlings were transferred to half-strength MS plates with or without 1 μM PAC or 1 μM PAC and 10 μM GA3 and incubated for 1 week. gai-1, gai-1/35S:GAF1, and gai-1/35S:ΔPAM transgenic plants were grown without PAC and GA3 for 2 weeks. Total proteins were extracted from whole plants and analyzed by immunoblotting with affinity-purified anti-GAI antibody. Coomassie blue staining was used to confirm equal loading. Arrowheads indicate the positions of the GAI protein (top) and the gai-1 protein (bottom). Immunoblot analysis used overexpression of GAF1 in gai-1 (G) and overexpression of ΔPAM in gai-1 (H).
Suppression of GA-Deficient and GA-Insensitive Mutants by GAF1 and ΔPAM
GA-deficient mutants display characteristic phenotypes, including reduced germination, dark green leaves, and a dwarf growth habit attributable to reduced stem elongation. A GA-insensitive semidominant mutant, gai-1, in which GAI is resistant to GA-dependent proteolysis because of a lack of the N-terminal DELLA domain, shows phenotypes similar to those of GA-deficient mutants except for reduced germination. To investigate the biological role of GAI binding to GAF1, we generated transgenic plants expressing GAF1 or the mutant version of GAF1 that cannot bind GAI (ΔPAM) under the control of the CaMV 35S promoter in the ga1-3 or gai-1 background (Supplemental Figures 2B to 2D). The expression of ΔPAM rescued the reduced germination phenotype of ga1-3 more effectively than did that of wild-type GAF1 (Figure 4B). Although both wild-type GAF1 and the GAF1 mutant ΔPAM could partially rescue the dwarf phenotypes of ga1-3 and gai-1, ΔPAM was found to be more effective (Figures 4B to 4F). These results suggest that the GAF1 mutant ΔPAM is more effective in suppressing the GA-deficient mutant ga1-3 and the GA-insensitive mutant gai-1. As accumulation and reduction of DELLAs inhibits and promotes plant growth, respectively, we examined whether the overexpression of GAF1 promotes plant growth by repressing the expression of GAI. Immunoblot analysis showed that GAI protein levels were not reduced by the expression of GAF1 or the GAF1 mutant ΔPAM (Figures 4G and 4H), suggesting that overexpression of GAF1s can rescue plant growth in ga1-3 and gai-1 without affecting the expression of GAI.
TOPLESS Is a GAF1 Binding Protein
The fact that the mutant version of GAF1 (ΔPAM), which cannot bind to GAI, suppresses the dwarf phenotypes of ga1-3 or gai-1 more effectively than GAF1 (Figure 4) raised the possibility that GAF1 plays a role in promoting plant growth after DELLAs are degraded in response to GAs. Transcription factors often form multiprotein complexes with other proteins, including general transcription factors, cofactors, or enzymes. To investigate the non-DELLA-dependent function of GAF1, we searched for GAF1 binding proteins other than DELLAs using a conventional Y2H system. From 1.5 × 105 transformants, we identified TPR1 and TPR4 as GAF1 binding proteins in addition to DELLAs. In Arabidopsis, TOPLESS (TPL) and TPRs form a family of transcriptional corepressors (Long et al., 2006; Szemenyei et al., 2008). The TPL/TPR family belongs to a larger Groucho/Tup1 family, first identified in Drosophila and yeasts, respectively. This family represents an ancient class of corepressors recruited by a range of DNA binding transcription factors to elicit a repressed chromosomal state (Liu and Karmarkar, 2008; Krogan et al., 2012; Wang et al., 2013). Recent studies showed that TPL/TPR corepressors interact with transcription complexes involved in auxin and jasmonate signal transduction, meristem maintenance, and defense responses (Long et al., 2006; Szemenyei et al., 2008; Pauwels et al., 2010).
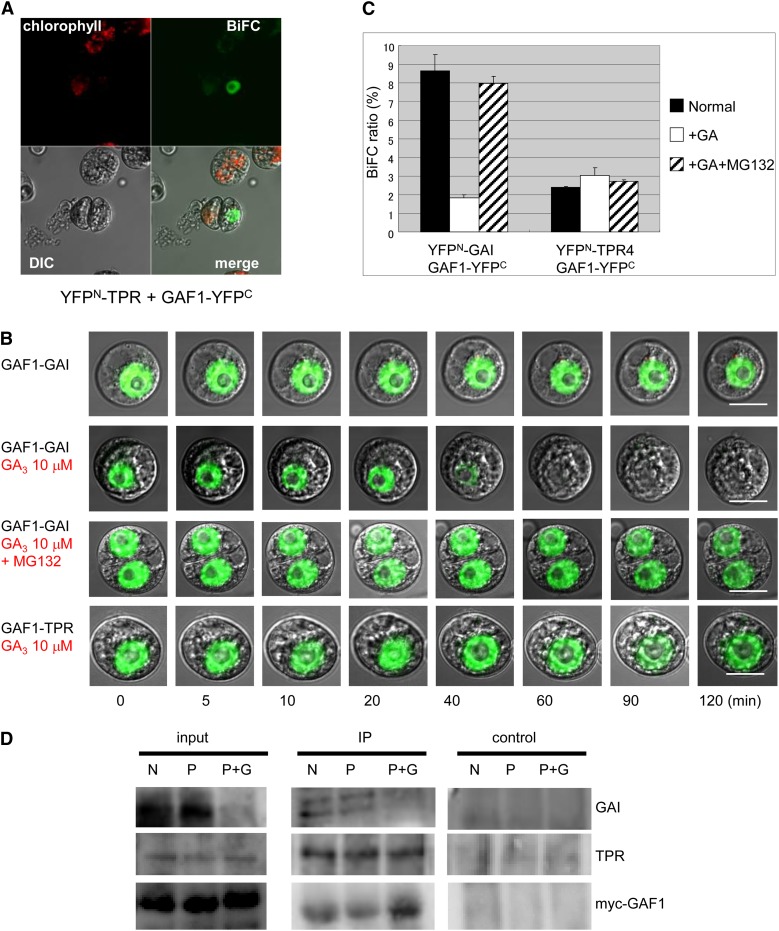
Using BiFC analysis, we confirmed the in vivo interaction between GAF1 and TPR4, and GAF1 and GAI in the nucleus of Arabidopsis T87 cultured protoplast cells (Figure 5A). GAs release the growth inhibition imposed by DELLAs via the degradation of DELLAs through the ubiquitin-26S proteasome pathway (Sun, 2010). We examined whether the interactions between GAF1 and GAI and between GAF1 and TPR4 are regulated by GAs. Treatment with GA induced the disappearance of the interaction between GAF1 and GAI within 60 min, while it did not affect the interaction between GAF1 and TPR4 (Figures 5B and 5C). However, treatment with MG132, a proteasome inhibitor, blocked the GA-induced disappearance of BiFC fluorescence. The subcellular localization and stability of GAF1-GFP were not affected by treatment with GAs (Supplemental Figures 5B to 5D). These observations were confirmed by coimmunoprecipitation experiments using transgenic plants expressing myc-tagged GAF1 under the control of the CaMV 35S promoter. The myc-tagged GAF1 protein was immunoprecipitated with anti-myc antibodies, and GAF1-bound materials were blotted and probed with anti-GAI antibodies or anti-TPR4 antibodies. As shown in Figure 5D, GAI and TPR4 coimmunoprecipitated with myc-tagged GAF1. GAs promoted the loss of interaction between GAI and GAF1 but did not affect the interaction between TPR4 and GAF1 (Figure 5B). These results suggest that the binding partners of GAF1 in plant cells are regulated by GA levels.
Figure 5.
In Vivo Interaction between GAF1 and GAI or TPR4 with or without GA.
(A) BiFC analysis of the interaction between GAF1 and TPR4 in T87 Arabidopsis cultured cells. CaMV35S:GAF1-YFPC and CaMV35S:YFPN-TPR4 plasmids were introduced and transiently expressed in protoplasts of T87 Arabidopsis cultured cells. DIC, differential interference contrast.
(B) BiFC analysis of the interaction between GAF1 and GAI or TPR4 in T87 Arabidopsis cultured cells. GA treatment disrupts the interaction between GAF1 and GAI. The top three rows show the interaction between GAF1 and GAI, while the bottom row shows the interaction between GAF1 and TPR4. The cells in the second and bottom rows were treated with 10 μM GA3.The cells in third row were treated with 10 μM GA3 and 10 μM MG132. Each cell was observed at 0, 5, 10, 20, 40, 60, 90, and 120 min after treatment with or without GA3 or GA3 + MG132. Bars = 10 μm.
(C) Measure of the interaction between GAF1 and GAI or TPR4 in the presence or absence (Normal) of GA and GA + MG132 using BiFC. Error bars represent sd (n = 3).
(D) In vivo interaction between GAF1 and GAI or TPR4 in Arabidopsis. Extracts from 35S:4×myc-GAF1 seedlings were immunoprecipitated (IP) using anti-myc antibody. The coimmunoprecipitated proteins were detected by either anti-GAI or anti-TPR4 antibody. Each plant was grown with or without GA and PAC. N, control; P, 1 μM PAC; P+G, 1 μM PAC and 10 μM GA3.
The Domain of GAF1 That Binds TPR4
To identify the interaction domain of GAF1 for TPR4, we used various deletion mutants of GAF1 (Supplemental Figure 6A). We found that internal deletion of 18 amino acids from residues 367 to 384 of GAF1 diminished TPR4 binding in the Y2H assay, but the deletion did not affect GAI binding (Figures 6A and 6B; Supplemental Figure 5A). This region includes the ethylene-responsive element binding factor–associated amphiphilic repression (EAR) motif, which is a target of TPLs (Tiwari et al., 2004; Szemenyei et al., 2008). The EAR motif was found only in GAF1 and IDD1 among Arabidopsis IDD proteins. These results suggest that GAF1 binds to GAI and TPR4 through independent binding motifs.
Figure 6.
A DELLA and TPR Act as a Coactivator and a Corepressor of GAF1, Respectively.
(A) Interaction domain of GAF1 for GAI or TPR4. Positive and no interactions are indicated by + and –, respectively. The PAM domain, which is necessary to bind to GAI, and the EAR-like motif, which is necessary to bind to TPR4, are indicated.
(B) β-Galactosidase activity of Y2H assay showing that ΔPAM or ΔEAR cannot interact with GAI or TPR but can interact with TPR or DELLAs, respectively. Data are means ± sd; n = 3.
(C) Transactivation assay of GAF1, GAI, and TPR4. The reporter, effector, and internal control constructs used in the assay are shown in the left panel. The reporter, effector, and internal control plasmids were cotransfected into Arabidopsis T87 cell protoplasts. The transfected protoplasts were incubated for 20 h, and then the GUS and LUC activities were measured. The results are shown as GUS/LUC activity. Error bars indicate sd (n = 3).
(D) The functions of IDD1 are similar to that of GAF1. The results are shown as GUS/LUC activity. Error bars indicate sd (n = 3).
(E) Transactivation assay showing that GAF1-GAI activates while GAF1-TPR represses the activity of the GA20ox2 promoter. GA represses the transcriptional activation of the GA20ox2 promoter via the GAF1-GAI-TPR complex. Transfected protoplasts were incubated for 20 h with or without 10 μM GA, and then GUS and LUC activities were measured. Error bars indicate sd (n = 3).
GAI and TPR4 Act as Transcriptional Cofactors of GAF1 in Plant Cells
Previous studies showed that DELLAs negatively regulate transcriptional activators, including PIFs, MYC2, and BZR1, by inhibiting DNA binding (Bai et al., 2012; Hong et al., 2012). GAs relieve the activities of transcriptional activators by promoting the degradation of DELLAs via the ubiquitin-26S proteasome pathway. Nevertheless, this model does not explain the feedback regulation of GA biosynthetic genes, which are upregulated in the presence of accumulated DELLA protein under GA-deficient conditions. However, the fact that TPR4 is a corepressor and GAI exhibits transcriptional activity in yeast (Figure 1C) suggests that GAI and TPR4 may function as coactivator and corepressor of GAF1, respectively. This hypothesis would provide a molecular mechanism for GA feedback regulation, namely transcriptional activation of GA biosynthetic genes by GAF1 and DELLAs under GA-deficient conditions.
To test this, we performed transient assays using Arabidopsis T87 protoplasts. The promoter of AtGA20ox2 encoding GA 20-oxidase fused with GUS was used as a reporter because AtGA20ox2 is under GA feedback regulation and activated by the induction of DELLAs within 2 h (Zentella et al., 2007) and contains the putative GAF1 target sequences in its promoter. Although neither GAF1 nor GAI alone affected the expression of AtGA20ox2:GUS, cotransfection of GAI with GAF1 greatly enhanced the expression of the reporter gene (Figure 6C; Supplemental Figure 6B). Cotransfection of GAI with the mutant version of GAF1 (ΔPAM) that cannot bind to GAI failed to activate the reporter gene, indicating that GAI binding to GAF1 is indispensable for transcriptional activation. Contrary to GAI, TPR4 showed corepressor activity with GAF1 in the transient assay. Cotransfection of TPR4 with the mutant version of GAF1 (ΔEAR) that cannot bind TPR4 did not repress the expression of the reporter gene, indicating that TPR4 binding to GAF1 is necessary for transcriptional repression. IDD1 also exhibited similar activity (Figure 6D).
With respect to feedback regulation of GA, the expression of AtGA20ox2 is negatively regulated by GAs. Thus, we examined if transcriptional regulation of AtGA20ox2 by the GAF1 transcription complex is modulated by GAs. As expected, the transcriptional activation of AtGA20ox2 by GAF1 and GAI was completely inhibited by treatment with gibberellic acid (GA3) in the presence of TPR4 (Figure 6E).
The GAF1 Complex Is Involved in GA Feedback Regulation of AtGA20ox2
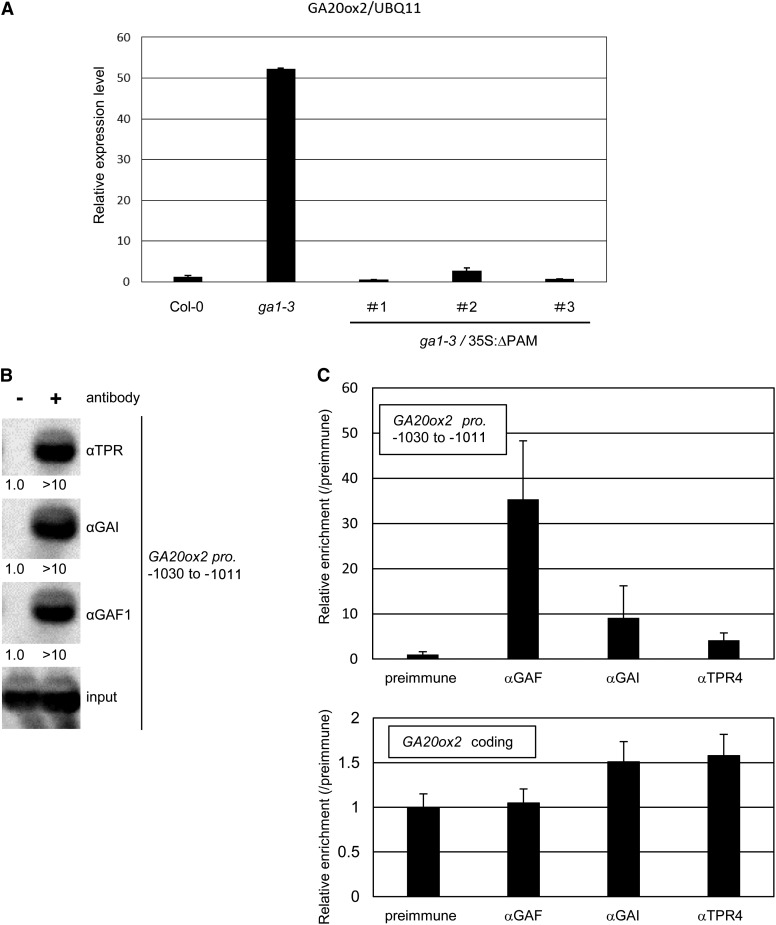
To evaluate the contribution of the GAF1 complex to GA feedback regulation of AtGA20ox2 in plants, we investigated AtGA20ox2 mRNA levels in the wild type (Col-0), the GA-deficient mutant ga1-3, and a transgenic line expressing ΔPAM in the ga1-3 background. The upregulation of AtGA20ox2 in the GA-deficient mutant ga1-3 was repressed by the expression of ΔPAM, showing that the GAF1-DELLA complex is important for GA feedback regulation in plants (Figure 7A). Furthermore, we confirmed that GAF1 associates with DELLAs and TPR on the AtGA20ox2 promoter in plants. ChIP assay using anti-GAF1, anti-GAI, or anti-TPR4 antibody showed that GAF1, GAI, or TPR4 binds to the AtGA20ox2 promoter (−1030 to −1011) in plants, indicating that the coactivator GAI and the corepressor TPR4 bind to the target gene through the DNA binding transcription factor GAF1 (Figures 7B and 7C). These results suggest that GAF1 directly regulates AtGA20ox2 either positively or negatively with the coactivator GAI or the corepressor TPR4, respectively, in response to GA levels.
Figure 7.
The GAF1 Complex Is Involved in GA Feedback Regulation of AtGA20ox2.
(A) The overexpression of ΔPAM in ga1-3 represses the upregulation of GA20ox2 by GA feedback regulation. Relative expression levels of GA20ox2 in Col-0, ga1-3, and 35S:ΔPAM in ga1-3 are shown. Error bars indicate the sd of three biological replicates (n = 3).
(B) ChIP analysis of GAF1, GAI, and TPR4 binding to the GAF1 binding regions of GA20ox2 promoters (−1030 to −1011). ChIP was performed with Col-0 using anti-GAF1, anti-GAI, and anti-TPR4 antibodies (+) or preimmune serum (−). The GA20ox2 promoter was detected by PCR and DNA gel blot hybridization. The values at the bottom of each panel indicate the relative signal strength. The value for preimmune serum was set to 1.0.
(C) ChIP assays performed with preimmune serum, anti-GAF1, anti-GAI, or anti-TPR4 antibody in Col-0. The coprecipitated level of each DNA fragment was quantified by real-time PCR and normalized with respect to the input DNA. The relative coprecipitated level of each DNA fragment using preimmune serum was set to 1, and the relative enrichment of each DNA fragment using anti-GAF1, anti-GAI, or anti-TPR4 is shown. Error bars indicate the sd of three biological replicates (n = 3).
Target Genes of GAF1
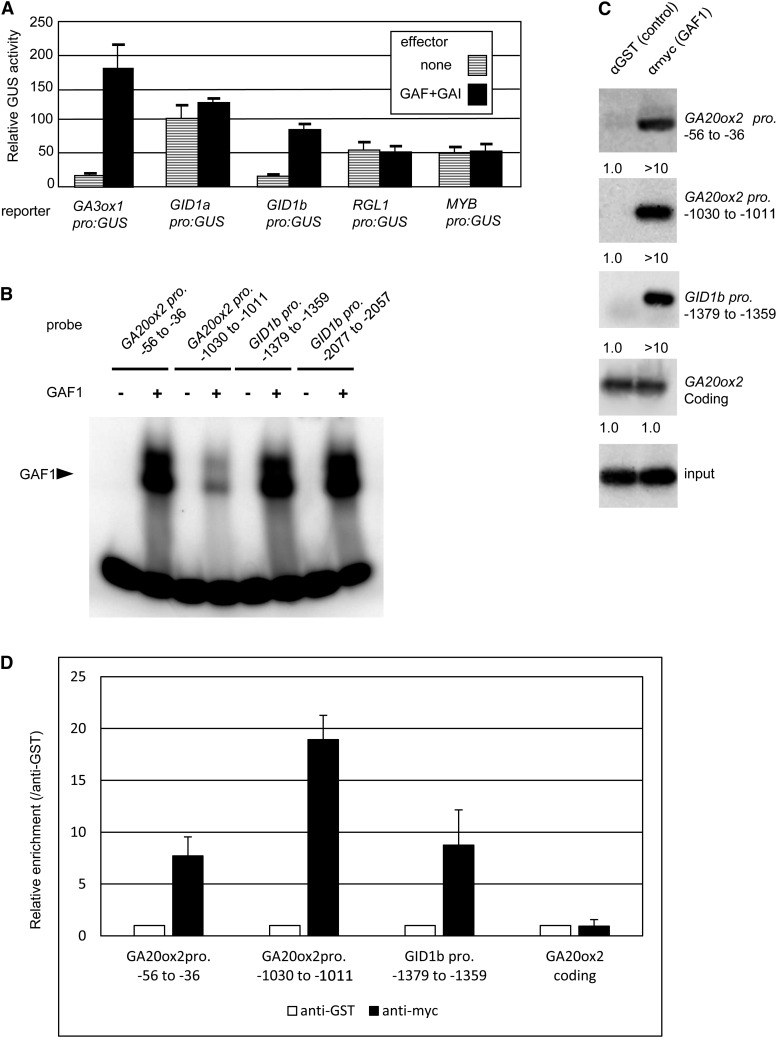
To identify other target genes of GAF1, we focused on DELLA-regulated genes (Zentella et al., 2007) because DELLAs are thought to bind to target genes indirectly through GAF1 or through an unknown DNA binding protein. We isolated the promoter regions of several DELLA-regulated genes and fused them to GUS. In addition to AtGA20ox2, GAF1 and GAI activated the promoters of AtGA3ox1 encoding a GA 3-oxidase and GID1b encoding a GA receptor among the DELLA-regulated genes tested (Figure 8A). Putative GAF1 target sequences were found in the AtGA20ox2 and GID1b promoters. A gel retardation assay indicated that recombinant GAF1 directly binds to these sequences (Figure 8B). Furthermore, ChIP assays using transgenic plants expressing myc-GAF1 under the control of the CaMV 35S promoter showed that GAF1 binds to the AtGA20ox2 and GID1b promoters in vivo (Figures 8C and 8D). These results suggest that GAF1 directly regulates GID1b as well as AtGA20ox2 and is involved in the feedback regulation of GA biosynthesis.
Figure 8.
Identification of GAF1 Target Genes.
(A) Transient transactivation assay showing that the GAF1/GAI complex also activates the GA3ox1 and GID1b promoters. The reporter plasmids consist of a 3-kb promoter region of GA3ox1, GID1a, GID1b, RGL1, and MYB fused with the GUS reporter gene. The results are shown as GUS/LUC activity. Error bars indicate sd (n = 3).
(B) Gel retardation assay showing that recombinant GAF1 binds to promoters of GA20ox2 and GID1b. Oligonucleotides (20 or 21 bp) were used as probes. The numbers adjacent to the gene names indicate base pairs upstream of the initiation ATG of each gene. The specific GAF1-DNA complexes are indicated by the arrowhead. +, addition to the reaction mixtures; –, omission from the reaction mixtures.
(C) ChIP analysis of 4×myc-GAF1 binding to the GAF1 binding regions of GA20ox2 and GID1b promoters described in (B). ChIP was performed with a 35S:4×myc-GAF1 transgenic plant using anti-myc or anti-GST antibody. The GA20ox2 and GID1b promoters and the GA20ox2 coding region were detected by PCR and DNA gel blot hybridization. The values at the bottom of each panel indicate the relative signal strength. The value for anti-GST antibody was set to 1.0.
(D) ChIP assays performed with anti-GST or anti-myc antibody in a 35S:4×myc-GAF1 transgenic plant. The coprecipitated level of each DNA fragment was quantified by real-time PCR and normalized with respect to the input DNA. The relative coprecipitated level of each DNA fragment using anti-GST antibody was set to 1, and the relative enrichment of each DNA fragment using anti-myc antibody compared with anti-GST antibody is shown. Error bars indicate the sd of three biological replicates (n = 3).
DISCUSSION
Defining the precise molecular mechanisms that determine patterns of transcription in response to specific signals is essential for understanding development and homeostasis. Transcriptional cofactors, in addition to DNA binding transcription factors, play key roles in such biological responses. Transcriptional complexes can coordinately turn target genes on or off depending on the functions of coactivators and corepressors.
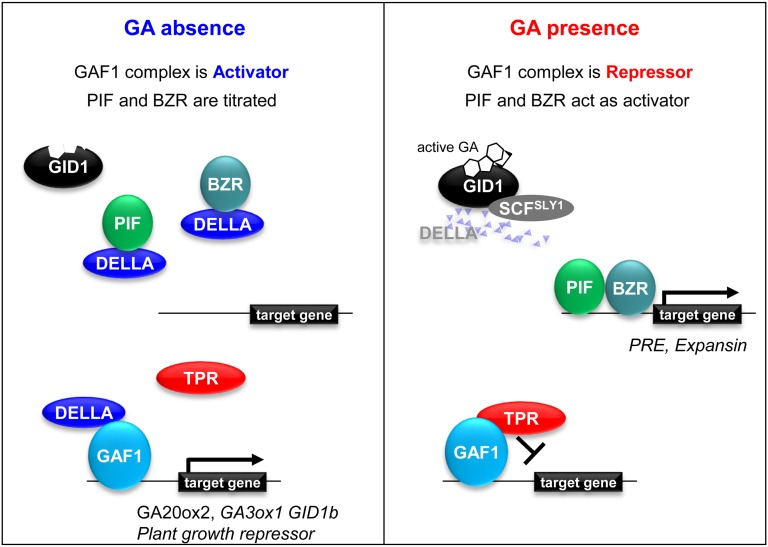
To date, the molecular mechanism of GA-dependent transcriptional regulation has been explained by the titration of transcriptional activators, including PIFs, with DELLAs and their release through the degradation of DELLAs in response to GA (de Lucas et al., 2008; Feng et al., 2008). However, the effect of GA on gene expression is predominantly repression, and DELLAs associate with the promoters of DELLA-induced genes even though DELLA proteins lack DNA binding motifs (Zentella et al., 2007; Hirano et al., 2012; Lim et al., 2013; Park et al., 2013). These observations suggest the existence of hitherto unknown mechanisms of GA-dependent transcriptional regulation. In this study, we found that DELLAs promote the transcription of genes encoding GA biosynthetic enzymes and GA receptors as coactivators of a DNA binding transcription factor, GAF1. Upon GA perception, these genes are actively repressed by a GAF1 corepressor–TPR complex that appears upon the degradation of DELLAs via the ubiquitin-26S proteasome pathway. Thus, the transcriptional state of GAF1 target genes is determined by the balance between coactivator and corepressor. This mechanism clearly accounts for GA feedback regulation and explains why the effect of GA on gene expression is predominantly repression, neither of which can be explained by the conventional titration model (Figure 9).
Figure 9.
A Coactivator and Corepressor Model for GA Signaling.
Under GA-deficient conditions, DELLA proteins are stable and localized in nuclei. DELLAs titrate PIF and BZR transcription factors by inhibiting DNA binding activity while exhibiting high transcriptional activity with GAF1. In the presence of GA, DELLAs are degraded via the 26S proteasome pathway. On the one hand, PIF and BZR activate target genes such as PACLOBUTRAZOL RESISTANCE (PRE) and EXPANSIN. On the other hand, the GAF1-TPR complex exhibits transcriptional repression activity. GA-induced functional conversion of the GAF1 complex in plants depends on the degradation of coactivator DELLA proteins.
Mammalian nuclear receptors are the best investigated examples of proteins with dual transcriptional regulation properties (Rosenfeld et al., 2006). Nuclear receptors for steroids, retinoic acid, and thyroid hormone are potent repressors in the absence of ligand, while they function as activators of transcription when bound to their cognate ligands. Upon ligand binding, the conformational change in the ligand binding domain of these receptors induces corepressors to dislodge and coactivators to bind, allowing transactivation (Rosenfeld et al., 2006). The exchange of coactivators/corepressors is an interesting common molecular framework of the hormonal regulation of transcription in two evolutionally distant organisms (i.e., plants and mammals). However, there is a significant difference: the GA-induced functional conversion of the GAF1 complex depends on the degradation of coactivator DELLAs through the ubiquitin-proteasome pathway, whereas ligand-induced activation of the transcription of nuclear receptors in mammals depends on a conformational change.
Many of the cofactors do not operate in isolation but, rather, are part of large multiprotein complexes. Transcriptional complexes exhibit a diversity of enzymatic activities that can be divided into two generic classes: enzymes capable of remodeling the structure of the nucleosome in an ATP-dependent manner and enzymes capable of covalently modifying histone tails. The latter group includes those with acetylating and deacetylating activities or methylating and demethylating activities, as well as kinases and phosphatases, poly(ADP) ribosylases, and ubiquitin and SUMO ligases (reviewed in Rosenfeld et al., 2006). Serial posttranslational modifications of histones and transcription factors are involved in the specific activation or repression of the genes. DELLAs could have activities for chromatin remodeling or covalent modifications; however, they do not have conserved catalytic motifs. Alternatively, DELLA proteins could recruit these enzymes to the promoters of target genes. In this context, it is worth noting that animal STAT, which shows structural similarity to DELLA proteins, associates with various cofactors, including histone acetyltransferase p300/CBP (CREB binding protein) and PCAF (p300/CBP-associated factor), histone deacetylase HDAC, and chromatin remodeler BRG1 (brm/SWI2-related gene 1) to regulate transcription (Rosenfeld et al., 2006).
In addition to this coactivator function, DELLAs inhibit transcriptional activators by preventing DNA binding. Thus, DELLAs can have opposite transcriptional effects on two sets of GA-regulated genes by acting as a coactivator of GAF1 and by titrating transcriptional activators, including PIFs and BZR1 (Bai et al., 2012; Hong et al., 2012). This mechanism remarkably suits GA signaling. A decrease in GA levels will turn off the expression of genes involved in the promotion of plant growth by titration of transcriptional activators with DELLA proteins and simultaneously turn on the expression of genes subject to GA feedback regulation by a GAF1-DELLA complex. Conversely, an increase in GA will have opposite effects on the two sets of genes. Such dual properties of a cofactor introduce a new paradigm for hormone action through which variations in the levels of GA can coordinately turn on or off two sets of genes, thus enabling the integrative regulation of plant growth and GA homeostasis. Furthermore, because DELLAs mediate the crosstalk of internal and external stimuli through the interaction with key regulators of signaling, including PIFs of light, BZR1 of brassinosteroids, and JAZ of jasmonates (Bai et al., 2012; Hong et al., 2012), they serve as central regulators of plant development.
Our understanding of GA signaling has advanced considerably in recent years (Hauvermale et al., 2012); however, the mechanisms by which DELLAs repress plant growth remain to be elucidated. Sequence-specific transcription factors collectively function as the key interface between genetic information encoded in the DNA sequence and the signal transduction systems in response to internal and external stimuli. Our results suggest that the DNA binding transcription factor GAF1 regulates genes encoding GA biosynthetic enzymes and GA receptors either positively or negatively depending on GA levels. The mutant version of GAF1 that cannot bind GAI (ΔPAM) rescued vegetative growth more effectively than did wild-type GAF1 in DELLA-accumulating plants (Figure 4), suggesting that the GAF1-TPR repressor complex rather than the GAF1-DELLA activator complex plays a role in promoting plant growth. Conversely, we found that GAF1 binds to the DELLA SAW domain (Figure 2F), which is necessary for the repression of GA responses by DELLAs, suggesting that GAF1 is involved in DELLA-mediated growth repression. One possible interpretation of these seemingly conflicting observations is that the GAF1-DELLA complex regulates the transcription of a gene encoding a growth repressor as well as that of genes encoding GA biosynthetic enzymes and a GA receptor. A decrease in GA levels could promote the expression of a growth repressor gene through a GAF1-DELLA complex, resulting in growth arrest, while an increase in GA levels could actively repress the growth repressor gene through the GAF1-TPR complex, promoting plant growth. Expression of SLR1, the rice DELLA protein, fused to the activation domain of the herpes simplex virus protein VP16 has been shown to more severely inhibit plant growth than that of wild-type SLR1 (Hirano et al., 2012), suggesting that DELLAs suppress plant growth through transcriptional activity. This supports the hypothesis that an unknown growth repressor gene is one of the target genes of the GAF1-DELLA complex. Identification of such a gene will provide an important clue for understanding the mechanisms downstream of DELLAs in GA signaling, which influences many aspects of plant growth and development.
METHODS
Plant Material and Growth Conditions
All mutant and transgenic lines in this study were derived from Arabidopsis thaliana ecotype Col-0 (wild type), with the exception of the gai-1 mutant, which was isolated from the Landsberg erecta ecotype. The ga1-3 mutant was introgressed into Col-0 by backcrossing six times (Tyler et al., 2004). The gaf1-1 (SALK_070916), idd1 (SALK_022425), and gai-1 (CS63) mutants were obtained from the ABRC (Ohio State University). gaf1 idd1 double mutants were generated by crossing the gaf1 and idd1 mutants. To generate transgenic plants overexpressing AtGAF1 or ΔPAM, GAF1 or ΔPAM cDNA was cloned into the NotI-XhoI site of the binary pBIJ4 vector, which contains a CaMV 35S promoter with an Ω sequence and a Kozak sequence to enhance translation activity, as described previously (Fukazawa et al., 2010). To generate transgenic plants overexpressing myc-tagged GAF1, the coding sequence for the 4×myc tag was amplified and cloned into the NotI site of pBIJ4-GAF1. To generate GAF1 promoter–driven GAF1-GFP transgenic plants in the gaf1 idd1 background, the GAF1 promoter was cloned into the HindIII-PstI site of the binary vector pBI101, and GAF1-GFP was cloned into XbaI-SacI site of GAF1 promoter–GUS plasmid. Agrobacterium tumefaciens–mediated Arabidopsis transformation was performed according to the floral dip method (Clough and Bent, 1998). The primer sets for cloning are listed in Supplemental Table 1. Plants were grown in a controlled growth chamber at 22°C under long days (16 h of light/8 h of dark) or short days (8 h of light/16 h of dark) with white light illumination.
Y2H Assay
The coding region of GAI was cloned into the BamHI site of pGBT9 vector (Clontech). The Tup1 repression domain was cloned into the EcoRI site of the pGBT9-GAI plasmid construct. The coding region of GAF1 was also cloned into the BamHI site of pGBDU vector (Clontech). The truncated GAI was cloned into the BamHI-PstI site of pGBU-C1 vector (Clontech). The truncated GAF1 was generated by PCR using this plasmid as template and cloned into the BamHI-PstI site of pGBDU vector (Clontech). The coding regions of IDD1 were cloned into the BamHI-PstI site of pGBDU vector (Clontech). The coding regions of GAI and RGA were cloned into the BamHI site of pGAD vector (Clontech). The coding regions of RGL1, RGL2, RGL3, and the N-terminal region of TPR4 were cloned into the SalI-BglII site of pGAD vector (Clontech).The primers used for all cloning are listed in Supplemental Table 1. Saccharomyces cerevisiae PJ69-4A was cotransformed with a bait and a prey plasmid. An Arabidopsis seedling cDNA library (Clontech) was screened on plates containing SD/-Leu/-Trp/-His with 3-aminotriazole (50 mM). About 2000 positive colonies were restreaked on plates containing SD/-Leu/-Trp/-His with 3-aminotriazole (50 mM). The −His plate assay and β-galactosidase assay were performed according to the manufacturer’s protocol for the Matchmaker Two Hybrid system (Clontech).
In Vitro Pull-Down Assay
The coding region of GAI was cloned into the BamHI site of pGEX4T-3 vector (GE Healthcare). The fusion proteins from Escherichia coli BL21 cells harboring pGEX4T-3-GAI and pGEX4T-3 plasmids were purified with Glutathione Sepharose 4B beads (GE Healthcare). The beads bound with GST-GAI and GST were washed with PBS buffer. The coding region of GAF1 was cloned into the NotI-XhoI site of pET30b vector. The recombinant protein from E. coli BL21 cells harboring pET30b-GAF1 plasmid was purified with chelating Sepharose Fast Flow (GE Healthcare). After being added to Glutathione Sepharose 4B beads carrying a GST fusion protein, purified GAF1 protein mixtures were incubated for 2 h at 4°C. After washing seven times with PBS, proteins were eluted with 10 mM reduced glutathione. Samples were separated by 10% SDS-PAGE and immunodetection with anti-GST antibody at 1:5000 dilution and anti-GAF1 antibody at 1:1000 dilution.
Application of GA4 to Col-0, ga1-3, and gaf1 idd1
Seven-day-old seedlings of Col-0, ga1-3, and gaf1 idd1 grown on half-strength Murashige and Skoog (MS) agar medium were transferred to soil and grown for an additional 2 months. To investigate GA sensitivity, plants were treated or not with 1 μM GA4. GA4 was applied by spraying once per week for 2 months.
Transactivation Assay
The 3-kb promoters of GA20ox2, RGL, GA3ox1, and GID1b from positions −1 to −3000 (where +1 is the initiation codon) were cloned into the SphI-SalI site of p-less GUS vector, which is a pUC18-based plasmid containing the GUS gene cassette of pBI101 (Takahashi et al., 1995). The 3-kb promoters of GID1a and MYB from positions −1 to −3000 (where +1 is the initiation codon) were cloned into the PstI-BamHI site of the p-less GUS vector using the In-Fusion cloning kit (Clontech). All primers used for transient assay analysis are shown in Supplemental Table 1. GAF1, truncated GAF1, IDD1, GAI, and TPR were cloned into the NotI-XhoI site of the pJ4 vector carrying the CaMV 35S promoter with a viral translation enhancer, the Ω sequence (Fukazawa et al., 2000), to be used as effectors. Protoplasts were prepared from T87 Arabidopsis cultured cells, and transfection of protoplasts was performed as described previously (Maliga et al., 1976; Satoh et al., 2004). Relative GUS activity was calculated by the normalization of LUC activity, and the data presented are averages of three independent biological replicates.
Gel Retardation Assay
The gel retardation assays were performed following the procedure described previously (Fukazawa et al., 2000, 2010). GAF1 was cloned into the NotI-XhoI site of pET30b vector (Novagen). Recombinant protein 6xHis-GAF1 was expressed and affinity-purified from E. coli BL21 (DE3) pLysE using Ni+ resin (Novagen). Nucleotide sequences of the double-stranded oligonucleotides used for the gel mobility shift assays are described in Supplemental Table 1. The oligonucleotides were annealed and then labeled using [α-32P]dCTP and the Klenow fragment of DNA polymerase I. Binding mixtures contained 50 fmol of labeled probe, 1 μg of purified recombinant GAF1 or 1 μg of control extract of E. coli, and 2 μg of poly(dI/dC). DNA competitor was used at 100-fold molar excess. The binding buffer consisted of 20 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 50 mM KCl, 1 mM EDTA, 10% (v/v) glycerol, and 2 μM ZnCl2. Reactions were incubated at 4°C for 30 min and loaded onto 4% polyacrylamide gels containing 6.7 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 3.3 mM sodium acetate.
Histochemical Staining
The promoter of GAF1 (−1500 to −1) was amplified by PCR from Arabidopsis genomic DNA and cloned into the HindIII-PstI site of binary vector pBI101 to generate a GUS fusion gene. The primers are listed in Supplemental Table 1. Kanamycin-resistant transgenic Arabidopsis plants were histochemically stained to detect GUS activity by immersing seedlings in a staining solution (100 mM sodium phosphate buffer, pH 7.0, with 50 mM NaCl, 1 mM potassium ferricyanide, 0.1% [v/v] Triton X-100 and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) overnight at 37°C. After staining, samples were immersed in a fixing solution (5% [v/v] formaldehyde, 5% [v/v] acetic acid, and 20% [v/v] ethanol), followed by dechlorophylation in 70% (v/v) ethanol.
Gene Expression Analysis
Total RNA from shoot, leaves, stems, flower, and root of Col-0 were extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. For the ga1-3, ga1-3/35S:GAF1, ga1-3/35S:ΔPAM, gai-1, gai-1/35S:GAF1, gai-1/35S:ΔPAM, and Col-0 lines, total RNA was isolated from 14-d-old plants. For the gaf1 and idd1 mutants and Col-0, total RNA was isolated from shoots of 10-d-old seedlings. A 1-μg aliquot of total RNA was used for cDNA synthesis using the QuantiTct Reverse Transcription Kit (Qiagen). Quantitative RT-PCR was performed on the ABI PRISM7000 sequence detection system using the THUNDERBIRD SYBER qPCR kit (Toyobo) according to the manufacturer’s instructions. UBIQUITIN11 expression was used as an internal standard.
BiFC
The YFPN and YFPC sequences were generated by PCR using EYFP DNA (Clontech) as a template and the primers listed in Supplemental Table 1. Each PCR product was cloned into pJ4 vector (Fukazawa et al., 2000). The full-length GAF1 cDNA was cloned into pJ4:YFPC, whereas GAI and TPR4 were cloned into pJ4:YFPN vector. Each plasmid was introduced into T87 Arabidopsis protoplast cells as described above. The transfected Arabidopsis protoplast cells were cultured for 24 h, and the YFP fluorescence was observed using a Zeiss LSM Pascal confocal microscope.
Preparation of Antibodies
The anti-GAF1 and anti-GAI antibodies were generated against the recombinant full-length GAF1 and GAI, respectively, in rabbits. The anti-TPR4 antibody was generated against the recombinant N-terminal 242 amino acids of TPR4 in rabbits. Recombinant 6xHis-GAF1, 6xHis-GAI, and 6xHis-N-terminal-TPR4 proteins as antigens were produced into E. coli BL21 (DE3) pLysE using pET30 (Novagen). The coding regions for GAI and the N-terminal region of TPR4 were cloned into the BamHI and SalI-NotI sites of pET30a vector, respectively (Novagen). The coding region of GAF1 was cloned into the NotI-XhoI site of pET30b vector (Novagen). The primer sets for cloning are listed in Supplemental Table 1.
Immunoblot Analysis
To investigate protein levels in 7- and 14-d-old seedlings of CaMV35S:GAF1 overexpressor transgenic plants, young leaves, mature leaves, flower buds, flowers, and roots of light-grown plants were harvested in liquid nitrogen. Total protein was extracted by SDS sample buffer and boiled for 10 min. For the gai-1, gai-1/35S:GAF1, gai-1/35S:ΔPAM, and Col-0 lines, total protein was isolated from 14-d-old plants. For the GA3- or paclobutrazol (PAC)-treated Col-0 plants, 7-d-old Col-0 seedlings were transferred to half-strength MS plates with or without 1 μM PAC or 1 μM PAC and 10 μM GA3, and these plant were incubated for 1 week. Proteins were separated by SDS-PAGE and immunoblotting with anti-GAF1 antibody at 1:1000 dilution.
Coimmunoprecipitation Assay
Coimmunoprecipitation assays of GAF1, GAI, and TPR4 were performed with 21-d-old 35S:4×myc-GAF1 seedlings. Plant materials were cross-linked with 1% formaldehyde, ground in liquid nitrogen, and then extracted with NEB buffer (20 mM HEPES-KOH, pH 7.5, 40 mM KCl, 1 mM EDTA, 0.5% Triton X-100, and 1× protease inhibitor cocktail [Roche]) at a ratio of 2 mL buffer/g tissue. After centrifugation, the supernatant was incubated with anti-myc antibody (MBL) and Protein G Sepharose beads (GE Healthcare). Proteins bound to the beads were resolved by SDS-PAGE and then detected by anti-myc HRP conjugate (MBL), anti-GAI (homemade), or anti-TPR4 (homemade) antibody. Anti-myc HRP conjugate was used at 1:5000. Anti-GAI and anti-TPR4 antibodies were used at 1:1000 dilution.
ChIP Assay
The ChIP experiment was performed following the procedure described previously (Fukazawa et al., 2010) with some modifications. In brief, 2-week-old 4×myc-GAF1 transgenic or Col-0 plants were cross-linked for 10 min in 1% (v/v) formaldehyde by vacuum filtration and incubated at 4°C for 1 h. Aliquots of each protein sample were immunoprecipitated with anti-GST (Santa Cruz Biotechnology), anti-myc (MBL International), anti-GAF1, anti-GAI, and anti-TPR (homemade) antibodies for 12 h at 4°C. The chromatin-antibody complexes were precipitated with salmon sperm DNA/protein G agarose beads for 2 h at 4°C. The amount of immunoprecipitated chromatin was determined by PCR. PCR cycles were 30 cycles for the GA20ox2 and GID1b promoters and 35 cycles for the GA20ox2 coding region. The PCR products were separated on 1% (w/v) agarose gels, blotted onto Hybond XL membranes (GE Healthcare), and hybridized with gene-specific DNA probes that were labeled using the AlkPhos Direct Labeling and Detection System (GE Healthcare). After hybridization, the signals were detected using the Image Reader LAS-3000 (Fujifilm). Primers used for ChIP analysis are listed in Supplemental Table 1. The coprecipitated level of each DNA fragment was quantified by real-time PCR using specific primer sets and normalized with input DNA level. The relative coprecipitated levels using preimmune serum or anti-GST antibody (immunoprecipitated DNA/input DNA) were set to 1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GAF1/IDD2 (At3g50700), IDD1/ENY (At5g66730), GAI (At1g14920), RGA (At2g01570), RGL1 (At1g66350), RGL2 (At3g03450), RGL3 (At5g63970), TPR1 (At1g80490), and TPR4 (At3g15880).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of GAF1 Overexpressor and the gaf1 Mutant.
Supplemental Figure 2. Relative Expression Levels of GAF1 and IDD1 in Several Plants.
Supplemental Figure 3. Complementation of gaf1 idd1 by GAF1pro:GAF1-GFP.
Supplemental Figure 4. gaf1 idd1-2 Mutant Exhibits Similar Phenotypes to the gaf1 idd1 Mutant.
Supplemental Figure 5. Binding Domain of GAF1 for GAI and Subcellular Localization of GAF1-GFP Protein.
Supplemental Figure 6. Domain of GAF1 Binding to TPR4 and Transactivation Assay Showing That GAI and TPR4 Do Not Exhibit Transcriptional Activity without GAF1.
Supplemental Table 1. Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank Kiminori Toyooka (RIKEN) for helpful discussions and support with confocal microscopy analysis. We thank Belay T. Ayele (University of Manitoba) for helpful comments on the article. We also thank Rie Yoshikawa for technical assistance. This study was supported by the Japan Society for the Promotion of Science (Grant 23770058 to J.F. and Grant 21370022 to Y.T.) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 24116525 to J.F. and Grant 24118004 to Y.T.).
AUTHOR CONTRIBUTIONS
J.F., Y.K., S.Y., and Y.T. designed the research. J.F., H.T., S.M., N.N., and K.N. performed research. J.F., M.Y., T.I., S.Y., and Y.T. analyzed data. J.F. and Y.T. wrote the article.
Glossary
- GA
gibberellin
- ChIP
chromatin immunoprecipitation
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- CaMV
cauliflower mosaic virus
- Col-0
Columbia-0
- SD
short-day
- GA3
gibberellic acid
- MS
Murashige and Skoog
- PAC
paclobutrazol
- LD
long-day
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Tremblay R., Wong A.Y., Coneva V., Kozaki A., Mable B.K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado J.A., Huang D., Wicki-Stordeur L., Hemstock L.E., Potentier M.S., Tsang E.W., Cutler A.J. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23: 1772–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J., Nakata M., Ito T., Yamaguchi S., Takahashi Y. (2010). The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J. 62: 1035–1045. [DOI] [PubMed] [Google Scholar]
- Fukazawa J., Sakai T., Ishida S., Yamaguchi I., Kamiya Y., Takahashi Y. (2000). Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale A.L., Ariizumi T., Steber C.M. (2012). Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol. 160: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Kouketu E., Katoh H., Aya K., Ueguchi-Tanaka M., Matsuoka M. (2012). The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 71: 443–453. [DOI] [PubMed] [Google Scholar]
- Hirst M., Ho C., Sabourin L., Rudnicki M., Penn L., Sadowski I. (2001). A two-hybrid system for transactivator bait proteins. Proc. Natl. Acad. Sci. USA 98: 8726–8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabet C., Sprague E.R., VanDemark A.P., Wolberger C. (2000). Characterization of the N-terminal domain of the yeast transcriptional repressor Tup1. Proposal for an association model of the repressor complex Tup1 x Ssn6. J. Biol. Chem. 275: 9011–9018. [DOI] [PubMed] [Google Scholar]
- Kozaki A., Hake S., Colasanti J. (2004). The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32: 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.T., Hogan K., Long J.A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., Kim J., Kang H., Kim D.H., Kawakami N., Choi G. (2013). ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13: 137–144. [DOI] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Maliga P., Lázár G., Sváb Z., Nagy F. (1976). Transient cycloheximide resistance in a tobacco cell line. Mol. Gen. Genet. 149: 267–271. [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E., Hedden P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60: 1979–1989. [DOI] [PubMed] [Google Scholar]
- Park J., Nguyen K.T., Park E., Jeon J.S., Choi G. (2013). DELLA proteins and their interacting RING finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D.E., Peng J., Harberd N.P. (2000). Plant GRAS and metazoan STATs: One family? BioEssays 22: 573–577. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M.G., Lunyak V.V., Glass C.K. (2006). Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20: 1405–1428. [DOI] [PubMed] [Google Scholar]
- Satoh R., Fujita Y., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. (2004). A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45: 309–317. [DOI] [PubMed] [Google Scholar]
- Singleton W.R. (1946). Inheritance of indeterminate growth in maize. J. Hered. 37: 61–64. [DOI] [PubMed] [Google Scholar]
- Sun T.P. (2010). Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 154: 567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223. [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Sakai T., Ishida S., Nagata T. (1995). Identification of auxin-responsive elements of parB and their expression in apices of shoot and root. Proc. Natl. Acad. Sci. USA 92: 6359–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135: 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2007a). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M. (2007b). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198. [DOI] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D.E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 110: 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.