Colocalization of cytochrome b6f and photosystem II complexes creates nanodomains that facilitate rapid exchange of the lipophilic electron carrier plastoquinone in the crowded thylakoid grana membrane.
Abstract
The cytochrome b6f (cytb6f) complex plays a central role in photosynthesis, coupling electron transport between photosystem II (PSII) and photosystem I to the generation of a transmembrane proton gradient used for the biosynthesis of ATP. Photosynthesis relies on rapid shuttling of electrons by plastoquinone (PQ) molecules between PSII and cytb6f complexes in the lipid phase of the thylakoid membrane. Thus, the relative membrane location of these complexes is crucial, yet remains unknown. Here, we exploit the selective binding of the electron transfer protein plastocyanin (Pc) to the lumenal membrane surface of the cytb6f complex using a Pc-functionalized atomic force microscope (AFM) probe to identify the position of cytb6f complexes in grana thylakoid membranes from spinach (Spinacia oleracea). This affinity-mapping AFM method directly correlates membrane surface topography with Pc-cytb6f interactions, allowing us to construct a map of the grana thylakoid membrane that reveals nanodomains of colocalized PSII and cytb6f complexes. We suggest that the close proximity between PSII and cytb6f complexes integrates solar energy conversion and electron transfer by fostering short-range diffusion of PQ in the protein-crowded thylakoid membrane, thereby optimizing photosynthetic efficiency.
INTRODUCTION
Photosynthesis in plants, algae, and cyanobacteria begins with the capture and trapping of solar energy by photosystem I and II (PSI and PSII). The cytochrome b6f (cytb6f) complex acts as the electrical connection between these two photosystems by oxidizing the PSII electron acceptor plastoquinone (PQ) and reducing the PSI electron donor plastocyanin (Pc; reviewed in Cramer et al., 2011). The electron transfer reactions performed by cytb6f are coupled to the generation of an electrochemical proton gradient across the chloroplast thylakoid membrane, which is harnessed by ATP synthase to form ATP. The membrane location and organization of cytb6f complexes is crucial to their function; it is known that the thylakoid membrane in vascular plants is divided into two domains, the grana stacks, which are enriched in PSII, and the interconnecting stromal lamellae, which are enriched in PSI and ATP synthase complexes (reviewed in Albertsson, 2001; Dekker and Boekema, 2005; Daum and Kühlbrandt, 2011; Nevo et al., 2012). The distribution of the cytb6f complexes between the grana and stromal lamellae is much less clear. Biochemical evidence obtained on thylakoids fractionated either mechanically or with the detergent digitonin suggested that cytb6f was distributed fairly evenly between the grana and stromal lamellae (Boardman and Anderson, 1967; Sane et al., 1970; Cox and Andersson, 1981; Anderson, 1982; Dunahay et al., 1984). This even distribution was supported by immunogold labeling of the cytb6f complex in intact thylakoids and by freeze-fracture electron microscopy studies comparing the wild type and a cytb6f-less mutant of the green alga Chlamydomonas reinhardtii (Allred and Staehelin, 1985; Olive et al., 1986; Vallon et al., 1991; Hinshaw and Miller, 1993). By contrast, fractionation with the detergents Triton X-100 or n-dodecyl-α-d-maltoside (α-DM) suggested that the grana were devoid of cytb6f and that this complex was confined to the stromal lamellae or grana margins (Berthold et al., 1981; Dunahay et al., 1984; Morrissey et al., 1986; van Roon et al., 2000). Exclusion of cytb6f from the grana would have significant consequences for photosynthetic electron transport since it would require long-range diffusion of PQ over hundreds of nanometers from PSII in the grana to cytb6f in the stromal lamellae. Yet, a range of evidence suggests that PQ diffusion is greatly restricted in the thylakoid membrane: first, PQ diffusion in thylakoid membranes is 1000-fold slower than in pure liposomes (Blackwell et al., 1994); second, spectroscopic data show that two pools of PQ with different rates of photoreduction by PSII exist in the thylakoid membrane and that equilibration between them is very slow (Joliot et al., 1992); finally, the rate of cytb6f reduction declines almost linearly in response to inhibition of PSII activity with the herbicide DCMU, suggesting that each cytb6f complex is connected to only a limited number of PQ molecules (Kirchhoff et al., 2000). Percolation theory suggests that protein crowding in the densely packed grana membrane, with 70 to 80% occupancy, is the likely cause of impaired PQ diffusion (Kirchhoff et al., 2002). These findings led to the hypothesis that microdomains of PSII, PQ, and cytb6f facilitate rapid diffusion of PQ over short (<20 nm) distances in the grana (Joliot et al., 1992; Kirchhoff et al., 2000). Crucially however, there is no direct structural evidence for such domains since no techniques are available to visualize the local arrangement of PSII and cytb6f complexes in the same membrane. Here, we describe the application of an affinity-mapping atomic force microscopy (AFM) technique (Vasilev et al., 2014) to uniquely identify and locate cytb6f complexes and to determine their proximity to PSII in grana membranes.
RESULTS
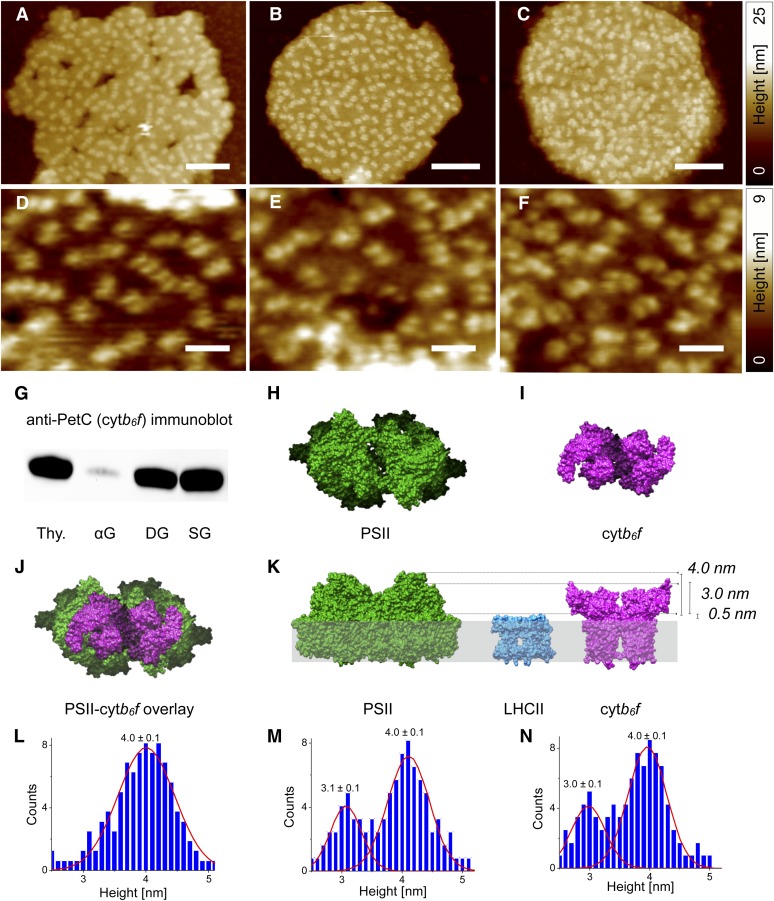
Two Major Types of Topographic Feature, Distinguished by Their Heights, Can Be Observed in AFM Images of Digitonin- and Sonication-Derived Grana Membranes
Thylakoid grana membranes were prepared by three different methods for interrogation by AFM: by limited solubilization with the detergents α-DM (van Roon et al., 2000) or digitonin (Anderson and Boardman, 1966) or alternatively mechanical fragmentation by sonication (Albertsson et al., 1994). Each of these methods is known to produce inside-out (i.e., lumen-side exposed) appressed pairs of membranes derived from the thylakoid grana stacks (Andersson et al., 1978; Dunahay et al., 1984; van Roon et al., 2000). SDS-PAGE analysis (Supplemental Figure 1) and chlorophyll a/b ratios (Table 1) confirmed that the three different methods produced membranes relatively enriched in PSII and light-harvesting complex II (LHCII) and depleted in PSI and ATP synthase compared with intact thylakoids. The chlorophyll a/b ratio and PSI contents of the α-DM grana (αG) were slightly lower than for grana prepared by digitonin (DG) or sonication (SG), consistent with previous reports (Anderson and Boardman, 1966; Albertsson et al., 1994; van Roon et al., 2000) (Table 1). AFM images of the three grana preparations recorded under liquid revealed appressed double membranes 400 to 500 nm in diameter and 18 to 20 nm in height. The grana membranes are covered with topographic features protruding 3 to 5 nm from the surrounding membrane surface that are 16 to 20 nm in width, consistent with the structure of the lumenal protrusions of the PSII core complex (Zouni et al., 2001; Ferreira et al., 2004; Loll et al., 2005; Guskov et al., 2009; Umena et al., 2011; Sznee et al., 2011) (Figures 1A to 1C). The holes and frayed edges often evident in αG membranes (Figure 1A) suggested a level of damage, as observed previously (van Roon et al., 2000; Sznee et al., 2011). Higher resolution images obtained on these membranes (Figures 1D to 1F) showed that the individual topographic features seen in Figures 1A to 1C are often resolved as two protrusions with maxima separated by ∼8 nm. Immunoblotting with an antibody specific to the PetC subunit of cytb6f indicated that this complex is not present in the αG membranes but is found in DG and SG membranes (Figure 1G). Spectroscopic assays confirmed this result; the DG and SG membranes contained 1.4 mmol cytf/mol chlorophyll, while none was detected in αG membranes (Table 1). The PSII/cytb6f ratio calculated from the spectroscopic data (Table 1) for the SG and DG membranes was 2.3, suggesting that some of the topographic features in these membranes likely belong to cytb6f that, like PSII, protrudes into the lumen space (Kurisu et al., 2003; Stroebel et al., 2003). The crystal structures of the dimeric cytb6f and PSII complexes reveal a similar lateral size and shape, with their long axes 16 and 20 nm, respectively, and with both dimers having two lumenal protrusions separated by ∼8 nm (Kurisu et al., 2003; Ferreira et al., 2004) (Figures 1H to 1J). These similarities hamper discrimination between these complexes in AFM images, given that probe sharpness limits the lateral resolution of the AFM to a typical value of ∼4 to 5 nm. However, the much higher (∼0.1 nm) z axis resolution of the AFM should resolve the 1-nm height difference between the lumenal protrusions of PSII and cytb6f complexes (Figure 1K). Accordingly, we measured the height of every protruding feature in these thylakoid membranes. The histograms of the height distributions clearly show two populations of topographic features (Figures 1L to 1N), a major peak centered on 4.0 ± 0.1 nm present in all the grana preparations and a minor peak corresponding to a 3.0/3.1 ± 0.1 nm height, which was only present in the DG and SG membranes. Assuming that we measure from the lumenal face of the surrounding LHCII complexes, which cover most of the grana membrane surface and protrude by ∼0.5 nm (Liu et al., 2004; Standfuss et al., 2005; Daum et al., 2010) (Figure 1K), the major and minor peaks are consistent with the predicted heights of the PSII and cytb6f complexes respectively. The area ratios of the 4.0- and 3.0/3.1-nm height distributions (Figures 1M and 1N) were 2.5:1, in reasonable agreement with the PSII/cytb6f ratio of 2.3 calculated by spectroscopy (Table 1). The absence of this secondary peak from analyses of AFM topographs for αG membranes correlates with the absence of cytb6f from the immunoblot data in Figure 1G and the spectroscopic data in Table 1, supporting the assignment of the 3-nm height features as cytb6f complexes.
Table 1. Chlorophyll and Cytochrome Content of Thylakoid Membrane Preparations.
| Membrane Preparation | Chlorophyll a/b Ratio | P700 (PSI)/Chlorophyll Ratio (mmol/mol) | Cytochrome f/Chlorophyll Ratio (mmol/mol) |
|---|---|---|---|
| α-DM grana | 2.23 ± 0.1 | 0.75 ± 0.1 | n.d. |
| Digitonin grana | 2.41 ± 0.1 | 1.49 ± 0.1 | 1.41 ± 0.1 |
| Sonication grana | 2.38 ± 0.1 | 1.42 ± 0.1 | 1.43 ± 0.1 |
| Intact thylakoids | 3.33 ± 0.1 | 2.63 ± 0.2 | 1.33 ± 0.1 |
P700 (PSI)/chlorophyll, cytochrome f/chlorophyll, and molar chlorophyll a/b ratio in grana membranes prepared by different techniques (±se). n.d., not detected.
Figure 1.
AFM Analysis of Grana Thylakoid Membranes Isolated from Spinach.
(A) to (F) AFM topographs of αG membranes ([A] and [D]), DG membranes ([B] and [E]), and SG membranes ([C] and [F]). Bars = 100 nm in (A) to (C) and 35 nm in (D) to (F).
(G) Immunoblot of the cytb6f (PetC) content of thylakoids and grana membrane preparations (thy. = intact thylakoids).
(H) to (J) Atomic models viewed from the thylakoid lumen membrane side of dimeric PSII (Protein Data Bank [PDB] code: 1S5L) (H), dimeric cytb6f (PDB code: 1UM3) (I), and overlay comparison (J).
(K) Atomic models of dimeric PSII, dimeric cytb6f, and trimeric LHCII (PDB code: 2BHW) viewed parallel to the thylakoid membrane. The dashed lines show the height difference between PSII and cytb6f of ∼1 nm and their heights above the LHCII complex, which protrudes by ∼0.5 nm from the luminal membrane surface.
(L) to (N) Histogram of maximum heights of protruding topological features above the surrounding area in αG (L), DG (M), and SG membranes (N).
Affinity-Mapping AFM Using Pc-Functionalized AFM Probes Allows the Position of cytb6f Complexes to Be Mapped in Grana Membranes
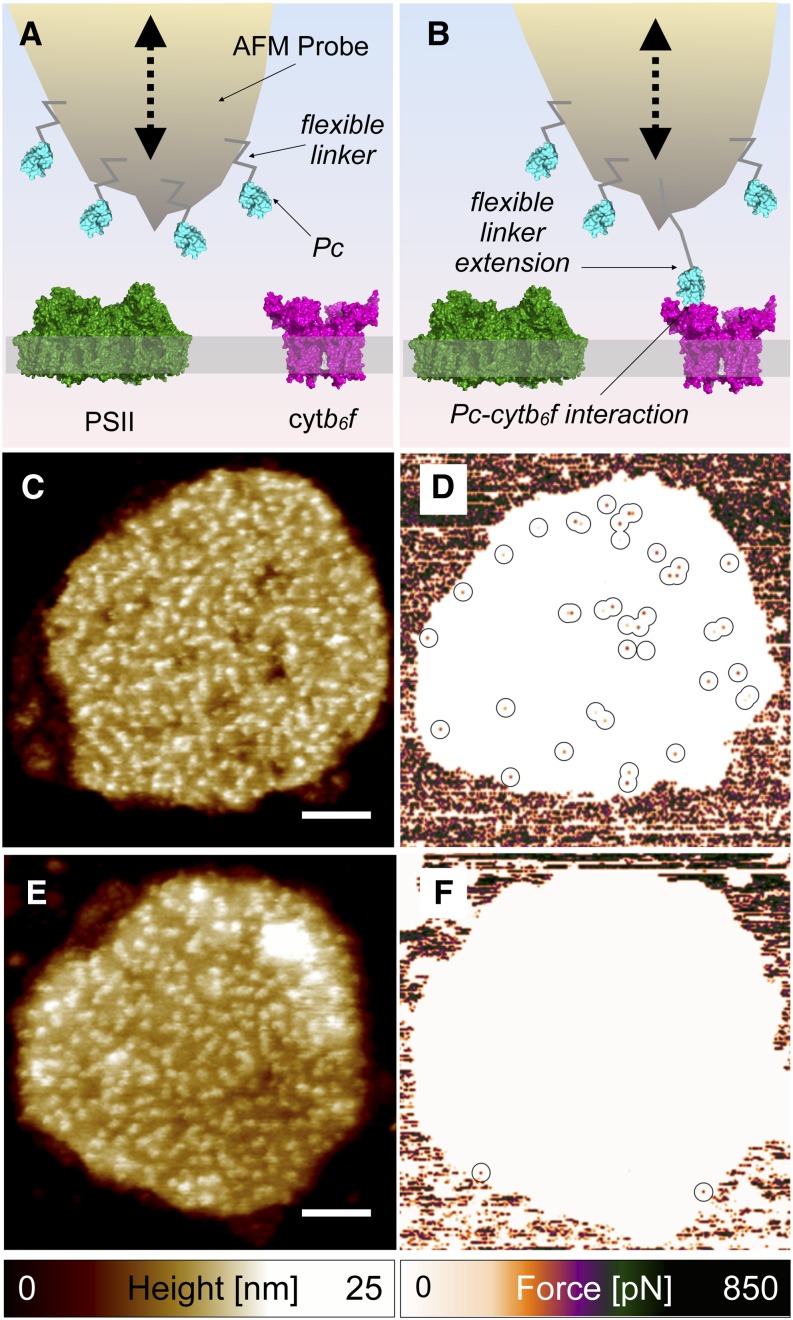
To positively confirm that these 3-nm protrusions correspond to the cytb6f complex, we exploited the recently described affinity-mapping AFM technique (Vasilev et al., 2014), this time utilizing the native interaction between the small soluble electron transfer protein Pc, which was attached to the AFM probe, and the exposed lumenal face of the cytb6f complexes in grana membranes adsorbed to a mica support. This method allows simultaneous recording and correlation of topographic and force (probe-sample adhesive interaction) data. The rationale of this experiment is shown in Figures 2A and 2B. Pc proteins purified from spinach (Spinacia oleracea) thylakoids (Supplemental Figures 2A to 2C) were attached by a flexible, 10-nm-long SM(PEG)24 linker molecule to an AFM probe (Figure 2A). No adhesive interactions should be observed when the Pc-functionalized probe interacts with a part of the membrane surface devoid of cytb6f complexes or with a PSII complex (Figure 2A). However, when the Pc-functionalized probe encounters a cytb6f complex at the membrane surface, a specific Pc-cytb6f interaction can occur; subsequently, the upward movement of the probe fully extends the linker molecule before rupturing this interaction (Figure 2B). To validate detection of specific Pc-cytb6f interactions using our affinity-mapping AFM method, we nanopatterned 300-nm-wide lines of purified cytb6f complexes on a glass substrate. The correspondence between the topographic and adhesion images in Supplemental Figures 3E and 3F, respectively, shows that a Pc-functionalized probe, preoxidized in 10 mM potassium ferricyanide, reliably detects multiple, specific Pc-cytb6f associations, as well as retaining the ability to record surface topography. Pc-functionalized probes were then used to investigate the organization of cytb6f complexes in purified grana membranes illuminated under white light with a power density of 11 W m−2 (measured at the sample surface) to ensure PSII-mediated formation of reduced PQ and, thus, reduction of cytb6f. When DG membranes were scanned with the preoxidized Pc-functionalized probes, the majority of the topographic features on the membrane surface were still well resolved (Figure 2C). Simultaneously with the topology image the forces required to rupture probe-sample interactions were recorded as an adhesion image (Figure 2D). We observed a number of locations (interaction hot spots) on the membrane surface where high (>50 pN) unbinding forces were recorded between the preoxidized Pc probe and the sample (Figure 2D). Crucially, when the DG membranes were scanned with Pc-functionalized probes prereduced in 10 mM sodium dithionite, the topographic features were still observed (Figure 2E), but now there were very few interaction hot spots observed on the membrane surface in the adhesion image (Figure 2F). The force-distance curves associated with such interaction hotspots on the membrane surface (see example in Figure 3A, cyan curve) show unbinding forces in the ∼200 to 500 pN range. The probe sample rupture length of ∼10 nm, obtained from inspection of the horizontal axis of Figure 3A (cyan curve), indicates that the linker fully extended before rupture of this interaction. Figure 3A also shows a typical force-distance curve for the majority of the membrane area (red curve), where we observed very low unbinding forces between the probe and the membrane surface that were comparable to the noise level of the measurement. Finally, Figure 3A shows an example of a force-distance curve recorded on the mica surface (orange curve) where we observed unbinding forces in the 300 to 500 pN range but importantly with a rupture length very close to 0 nm. Such events correspond to a nonspecific and direct interaction between the apex of the AFM probe and the mica, with no involvement of the linker or the attached Pc.
Figure 2.
Affinity-Mapping AFM of DG Membranes Isolated from Spinach.
(A) Cartoon depicting the principle of affinity-mapping AFM. The AFM probe laterally images the topography of the thylakoid lumen membrane surface via a tapping motion (dashed black arrow), while simultaneously recording probe-sample interaction forces for every pixel; the AFM probe is functionalized with Pc proteins attached via a flexible 10-nm-long SM(PEG)24 linker.
(B) Cartoon depicting the specific interaction of one of the Pc proteins attached to the AFM probe with a cytb6f complex on the membrane surface; when the AFM probe is withdrawn from the surface during the upward part of the tapping motion, the flexible linker is extended to its full 10 nm length before Pc-cytb6f unbinding occurs.
(C) and (D) The AFM probe was functionalized with preoxidized Pc: topography image (C) and adhesion image (D).
(E) and (F) AFM probe functionalized with prereduced Pc: topography image (E) and adhesion image (F).
Interaction hot spots where high (>50 pN) unbinding forces were recorded on the membrane surface are highlighted with circles. Bars = 100 nm in (C) and (E).
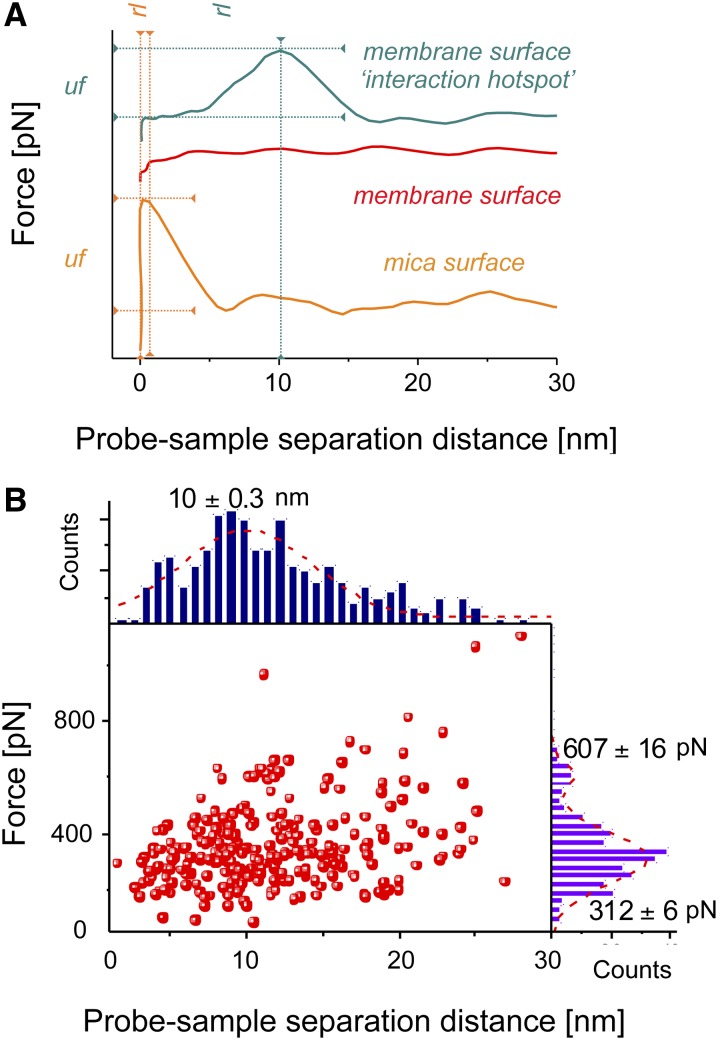
Figure 3.
Analysis and Quantification of Force-Distance Data Obtained on Grana Thylakoid Membranes Isolated from Spinach by Affinity-Mapping AFM.
(A) Typical retract regions of the force-distance curves recorded using preoxidized Pc-functionalized probes on three different regions of the DG membrane sample attached to the mica. The orange curve was recorded on the mica surface with a peak unbinding force (uf) of ∼450 pN at a probe-sample rupture length (rl) of ∼0 to 1 nm indicating a nonspecific interaction between the main body of the AFM probe and the mica. The red curve was recorded on the membrane surface and shows no distinct peak, indicating that there is little interaction with the AFM probe as it is retracted. The cyan curve was recorded at one of the interaction hot spots on the membrane surface where a high (>50 pN) unbinding force was recorded, in this case ∼300 pN, at a probe-sample rupture length of ∼10 nm, which is consistent with the length of the SM(PEG)24 linker with which the Pc is attached to the probe, indicating the detection of a specific Pc-cytb6f interaction.
(B) Peak unbinding force versus rupture length for 276 force-distance curves recorded at membrane locations with high (>50 pN) unbinding force (red squares) and associated histograms for force (purple) and rupture length (blue) with average values shown using preoxidized Pc-functionalized probes on DG membranes.
In order to obtain robust statistics for the interaction hotspots on the membrane surface, we analyzed 276 force-distance curves recorded on 14 different DG samples interrogated with preoxidized probes. The graph in Figure 3B shows a most probable unbinding force of 312 ± 5 pN (Figure 3B) and an average rupture length of 10.0 ± 0.3 nm, consistent with the length of the flexible SM(PEG)24 linker molecule used to attach the Pc to the AFM probe. A second smaller peak in the force distribution was centered on 607 ± 15 pN, which likely results from the rupture of the interaction between two Pc molecules on the functionalized probe and two cytb6f complexes on the surface.
The Frequency of Interaction between the Pc-Functionalized AFM Probe and cytb6f Complexes on the Membrane Surface Depends on the Redox Conditions
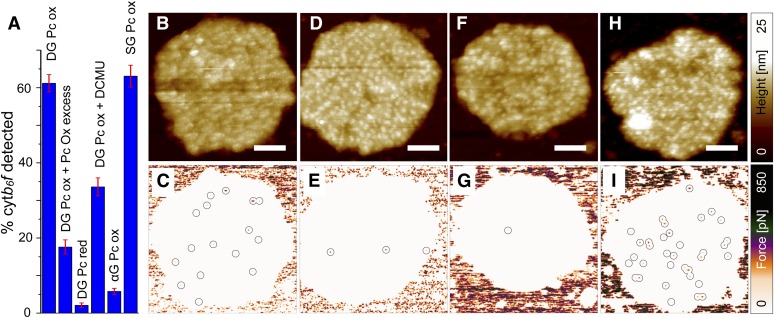
The estimated number of cytb6f complexes determined by spectroscopy (Table 1) suggests that the DG and SG membranes contain 51.5 dimeric complexes per 500-nm-diameter circular membrane disc, i.e., ∼262 cytb6f dimers/μm2. The average number of interaction hot spots detected with preoxidized Pc functionalized probes was 158 ± 12/μm2, suggesting around 60% of cytb6f dimers are detected by our method (Figure 4A). When the experiment was performed with prereduced Pc functionalized probes the number of interaction hotspots on the membrane surface decreased by 96%, indicating the preference for oxidized Pc to interact with cytb6f (Figure 4A). When PSII was inhibited by addition of 10 μΜ DCMU to lower the production of reduced PQ and the membrane imaged 10 min later, the number of interaction hotspots detected on the membrane surface decreased by 44%, indicating that the cytb6f redox state also affects the number of cytb6f complexes detected (Figures 4A to 4C). Addition of 50 μM excess oxidized Pc (twice the reported Kd of 23 μM; Meyer et al., 1993) to the sample prior to scanning, thus blocking potential cytb6f binding sites for the oxidized Pc-functionalized probe, lowered the number of interaction hotspots by 72% (Figures 4A, 4D, and 4E). Affinity-mapping AFM of αG membranes was instructive since the immunoblot and spectroscopic assays had indicated that these membranes contained little or no cytb6f. Scanning αG membranes with the oxidized Pc probes gave 91% fewer interaction hotspots than the DG membranes, consistent with the virtual absence of the cytb6f complex from the αG preparation (Figures 4A, 4F, and 4G). As expected, the SG membranes gave a very similar number of interaction hotspots as the DG membranes (Figures 4A, 4H, and 4I).
Figure 4.
Affinity-Mapping AFM of Grana Membranes Prepared under Different Conditions Using a Probe Functionalized with Pc.
Estimated percentage of cytb6f dimers detected by affinity-mapping AFM using various conditions as indicated on the graph (±se) (A). DG membranes were incubated with 10 μM DCMU to lower production of reduced PQ: topography image (B) and adhesion image (C). DG membranes were incubated with 50 μM preoxidized Pc to block cytb6f binding sites on the membrane surface: topography image (D) and adhesion image (E). αG membranes: topography image (F) and adhesion image (G). SG membranes: topography image (H) and adhesion image (I). Interaction hot spots where high (>50 pN) unbinding forces were recorded on the membrane surface are highlighted with circles. Bars = 100 nm.
Pc-cytb6f Interactions Detected by Affinity-Mapping AFM Specifically Label the 3-nm-High Topographic Features in AFM Images of Grana Membranes
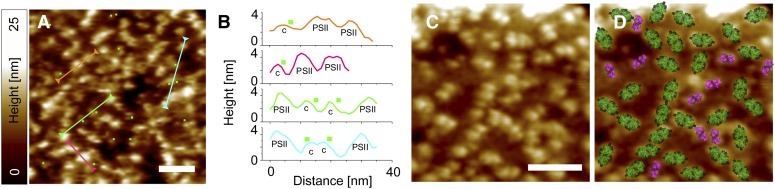
The interaction hotspots recorded on the surface of DG and SG membranes with preoxidized Pc-functionalized probes reliably identified topographic features that protrude ∼3 nm from the membrane surface at their highest point (Figures 5A and 5B), supporting the assignment of these features as cytb6f complexes. By contrast, the 4- to 5-nm protrusions from the membrane surface, consistent with the height of PSII, showed no high unbinding force signals, but were found in close proximity to the 3-nm topographic features (Figures 5A and 5B). These assignments led us to fit a representative area of our highest resolution AFM image (Figure 5C) with the crystal structures of the dimeric cytb6f and PSII complexes (Figure 5D). The resulting membrane model in Figure 5D suggests that cytb6f complexes (magenta) are often surrounded by PSII complexes (green) in small domains as originally suggested (Joliot et al., 1992; Kirchhoff et al., 2000).
Figure 5.
cytb6f/PSII Nanodomain Structure Revealed by Affinity-Mapping AFM of DG Membranes Isolated from Spinach.
(A) to (C) Topography image overlaid with the simultaneously recorded adhesion image; the interaction hot spots due to Pc-cytb6f binding are shown as green squares (A). The colored lines denote topographic cross sections, also shown in (B), and the green squares denote the interaction hotspots along these cross-sections; the topographic features are assigned to either cytb6f (C) or PSII.
(D) High-resolution AFM image of a cytb6f/PSII nanodomain fitted with the membrane-extrinsic parts of the crystal structures of the cytb6f (purple) and PSII (green) dimers (PDB codes: 1S5L and 1UM3, respectively).
Bars = 50 nm in (A) and 50 nm in (C).
DISCUSSION
Affinity-Mapping AFM Is a Useful Tool for Mapping Membrane Organization at the Nanoscale
Our affinity-mapping AFM approach shows how a biological membrane can be mapped on the nanoscale, by exploiting the specific binding of a membrane-extrinsic probe onto a membrane-bound target protein. Here, we made use of a transient interaction between two electron transfer proteins, the extrinsic electron acceptor Pc and the cytb6f donor, an integral membrane protein complex. When oxidized Pc, attached to an AFM probe by a 10-nm flexible linker, scans across the thylakoid membrane surface it specifically detects reduced cytb6f complexes. By simultaneously recording surface topology and Pc-cytb6f interactions, we were able to distinguish between cytb6f and PSII by their extents of protrusion from the membrane and by transient protein-protein interactions. The colocation and organization of the PSII and cytb6f complexes in grana thylakoid membranes in our combined topographic and affinity maps reveals a close association between these complexes and shows how the excitation energy and electron transfer events of photosynthesis are linked in the thylakoid membrane.
Affinity-Mapping AFM Using Pc-Functionalized AFM Probes Allows Pc-cytb6f Interactions to Be Quantified at the Single-Molecule Level under Near Native Conditions
The large number of interaction hotspots recorded in each force map also produced robust statistics for in situ quantification of the Pc-cytb6f interaction unbinding force, at the single-molecule level. The unbinding force of 312 ± 5 pN compares with a value of 480 pN measured by the same method, albeit in vitro, for the interaction between probe-attached cytochrome c2 (cytc2) and surface-bound reaction center (RC) complexes from the purple photosynthetic bacterium Rhodobacter sphaeroides (Vasilev et al., 2014). The latter value agreed with theory-based estimates of the cytc2-RC interaction, which calculated an unbinding force of 600 to 1000 pN (Pogorelov et al., 2007). By measuring an in situ membrane protein complex stabilized by the membrane bilayer and by interactions with neighbors, repeated measurements are possible that report functional interactions, with no extraction or unfolding of the cytb6f complex. Such stabilizing influences allow measurement of the ∼300 pN unbinding forces, which are comparable with those obtained using conventional force spectroscopy for a range of other interactions including 340 to 454 pN for streptavidin-biotin (Lee et al., 1994; Stevens et al., 2002), 507 pN for the hCG-anti-hCG pair (Stevens et al., 2002), 512 pN for the MPAD-anti-MPAD interaction (Kaur et al., 2004), and 420 pN for citrate synthase-GroEL (Vinckier et al., 1998). The forces we measure likely sample the binding interaction prior to electron transfer, given the requirement for both reduced cytb6f and oxidized Pc, as well as the similarity of the dwell time (∼160 μs at 1.0 kHz force curve repetition rate) of the AFM probe on the membrane surface to the ∼70 to 130 μs electron transfer between Pc and cytb6f (Haehnel et al., 1980; Delosme, 1991). Thus, multiple Pc-cytb6f encounters enable the mapping of most of the cytb6f complexes in the thylakoid membrane.
Earlier in vitro studies using site-directed mutants of Pc and the soluble domain of cytochrome f or the purified cytb6f complex concluded that interaction between these proteins depends largely on electrostatic interactions between the partners (Lee et al., 1995; Kannt et al., 1996; Illerhaus et al., 2000). However, mutagenesis of cytochrome f in C. reinhardtii found that electrostatic interactions exerted a much weaker influence on cytochrome f oxidation kinetics in vivo (Soriano et al., 1996, 1998), and other factors undoubtedly come into play in chloroplasts such as restricted diffusional space and the higher osmotic strength found in the thylakoid lumen. An NMR study of the complex between cytochrome f and plastocyanin from the cyanobacterium Nostoc showed that the binding interface involves hydrophobic residues with charged residues at the edge (Díaz-Moreno et al., 2005). A more recent NMR study of the Pc-cytb6f interaction using spinach components, including SG membranes, showed that there is an initial, loose electrostatic interaction and the subsequent formation of an electron transfer complex relies on hydrophobic interactions (Ueda et al., 2012).
The single-molecule affinity-mapping AFM approach taken in this work approximates the in vivo situation due to our use of cytb6f in situ in native membranes, rather than solubilized complexes. Thus, we expect that electrostatic interactions are involved in the initial encounter phase of the Pc-cytb6f interaction and that hydrophobic interactions then guide the formation of an electron transfer complex. The use of Pc tethered to an AFM probe with its attachment site distal to the Pc-cytf contact region steers the tethered Pc toward its binding site, likely bypassing the multiple binding/unbinding encounters and reorientations possible when two soluble components interact in solution. Rather, our AFM measurements reflect the aftermath of Pc-cytb6f complex formation, and the ∼300 pN unbinding forces we measure (Figure 3) reflect the factors that stabilize the Pc-cytb6f complex and quantify the forces required to disrupt it. Preparing the reduced cytb6f and oxidized Pc reactants prior to bringing them into contact might accelerate formation of a stable electron transfer complex and increase the likelihood that we have to apply an ∼300 pN unbinding force to pull it apart. As already noted, the duration of the 160-μs dwell time of Pc at the membrane surface might be sufficient to allow electron transfer, which takes ∼70 to 130 μs (Haehnel et al., 1980; Delosme, 1991); thus, our experiments could reflect the stability of the cytb6f (reduced)-Pc(oxidized) complex just prior to electron transfer or of the cytb6f (oxidized)-Pc(reduced) complex following electron transfer. In this respect, this work differs from the forces required to disrupt the in vitro interaction between probe-attached cytc2 and surface-bound RC complexes (Vasilev et al., 2014); in this case the cytc2-RC electron transfer is much faster, in the low microsecond range (Overfield et al., 1979; Moser and Dutton, 1988). Thus, the reduced cytc2 is rapidly oxidized and the unbinding events we measured likely reflected the interaction between reduced RC and oxidized cytc2, i.e., the aftermath of the electron transfer event (Table 1).
Our preparations contain PSI, so it is possible that some of the interaction hot spots we detect with our affinity-mapping AFM method could correspond to Pc-PSI interactions. However, we consider this to be unlikely for several reasons: First, the crystal structure of PSI (Amunts et al., 2007) shows that the lumenal protrusions of this complex are <1 nm, whereas the interaction hot spots we detect consistently label features 3 nm in height, a difference easily resolved by the 0.1 nm z-resolution of the AFM; second, in the event of binding to PSI, we would predict that the prereduced Pc-functionalized AFM probes would detect a similar number of interactions when in fact they were greatly decreased compared with the preoxidized probes. The fact we observed very few interactions of prereduced Pc-functionalized probes with the PSI complexes in DG and SG membranes suggests we could not maintain a photo-oxidized state, likely because we did not include a PSI electron acceptor.
cytb6f Complexes Are Distributed throughout the Grana Membranes among PSII Complexes
The colocation of the cytb6f and PSII complexes has important implications for photosynthesis and provides direct evidence for the microdomain hypothesis (Joliot et al., 1992; Kirchhoff et al., 2000). The proximity of these complexes, in regions more appropriately termed “nanodomains,” allows PQ diffusion over short (<20 nm) distances. Affinity-mapping AFM shows that cytb6f complexes are interspersed throughout the grana regions, in the vicinity of PSII complexes but not tightly bound with fixed stoichiometry in a supercomplex. Recent cryo-electron microscopy studies on DG membranes and intact thylakoids revealed the presence of numerous smaller densities alongside those assigned to PSII, which the authors suggested could belong to monomeric PSII or cytb6f complexes (Daum et al., 2010; Kouřil et al., 2011). Given the data reported in this article and the fact that PSII exists almost entirely as a dimer in the thylakoid membrane (Dekker and Boekema, 2005), these unassigned densities likely belong to cytb6f. As in our data, these densities were distributed throughout the PSII complexes in grana membrane (Daum et al., 2010; Kouřil et al., 2011). However, it is likely that cytb6f complexes are excluded from the 5 to 10% of grana membranes in wild-type plants that form ordered semicrystalline PSII arrays (Dekker and Boekema, 2005). It is reasonable to conclude that the granal cytb6f complexes we have colocated with PSII facilitate linear electron transport, while the remaining 40 to 50% located in the stromal lamellae facilitate cyclic electron transport as previously suggested (Albertsson, 2001). Indeed, there is evidence that the distribution of cytb6f complexes is dynamically responsive to the PQ redox state, with an increase in the proportion found in the stromal lamellae under conditions where the PQ pool is reduced (Vallon et al., 1991). The absence of cytb6f complexes from Triton- and α-DM-derived grana membranes may be due to their selective solubilization by Triton and α-DM detergents, which could explain the holes frequently observed in these membranes and their generally ragged appearance (Dunahay et al., 1984; van Roon et al., 2000; Sznee et al., 2011). Indeed, cytb6f was solubilized from the membrane at lower concentrations of Triton than used for either PSI or PSII complexes (Morrissey et al., 1986).
Nanodomains of cytb6f and PSII Complexes Ensure Rapid Exchange of PQ Molecules and, Thus, Efficient Electron Transport
There are some parallels between the colocation of cytb6f and PSII complexes shown in this work and recent study that used electron microscopy and AFM to demonstrate proximity between the dimeric cytochrome bc1 and dimeric RC complexes in the photosynthetic membrane of the bacterium R. sphaeroides (Cartron et al., 2014). The close proximity of cytochrome complexes and Type II (quinone acceptor) RCs, such as PSII, may therefore be advantageous for facilitating rapid exchange of quinones. Evolution has reconciled the high density of pigment binding proteins required to ensure the efficient transfer and trapping of solar energy with the need for sufficient diffusion space for quinone traffic between RC and cytochrome complexes; the solution revealed by this work is the distribution of cytb6f complexes among PSII supercomplexes in the grana, minimizing the average distance traversed by PQ molecules in the crowded membrane environment and promoting rapid exchange of PQ between cytb6f and PSII complexes.
METHODS
Membrane/Protein Isolation
Spinach (Spinacia oleracea) thylakoid membranes (Albertsson et al., 1994), αG membranes (van Roon et al., 2000), DG membranes (Anderson and Boardman, 1966), SG membranes (Albertsson et al., 1994), Pc (Morand and Krogmann, 1993), and cytb6f (Zhang et al., 2001) were prepared as previously described. SDS-PAGE and immunoblot analysis was performed as previously described (Ruban et al., 2006).
Spectroscopic Determination of Chlorophyll and Cytochrome Contents
Spectroscopic assays of P700 and cytb6f content were performed according the methods of Melis and Brown (1980) and Bendall et al. (1971), respectively. The obtained value of 1.4 mmol cytochrome f/mol chlorophyll is similar to that calculated using this method by Albertsson et al. (1994). To calculate number of cytb6f complexes per 500-nm grana disc, the following chlorophyll composition was assumed based on the antenna sizes and PSI/PSII ratios given by Danielsson et al. (2004) for grana derived by the same method from thylakoid membranes with similar chlorophyll a/b ratio: 120 C2S2 type PSII (120 × 204 = 24,480 Chls), 602 LHCII trimers (602 × 42 = 25,284 Chls), 122 PSI-LHCI (122 × 168 = 20,496 Chls), 240 CP24 (13 × 240 = 3120); total Chls = 73380; hence, 0.0014 × 73,380 = 102.7 cytochrome f molecules per single 500-nm grana disc (∼51.5 dimers).
AFM/Affinity-Mapping AFM
Grana membranes were adsorbed to the mica substrate as previously described (Sznee et al., 2011) and imaged in a buffer containing 10 mM HEPES-NaOH, pH 8.0, 5 mM NaCl, and 5 mM MgCl2. Imaging was performed by PeakForce quantitative nanomechanical mapping mode using a Bruker Multimode AFM. The spring constants of the AFM cantilevers used were individually determined to accurately quantify the force data and varied between 0.06 and 0.24 Nm−1. The Z-modulation amplitude was adjusted to values in the range 15 to 25 nm to allow enough probe-sample separation in order to fully stretch the SM(PEG)24 linker molecule on the AFM probe and to separate the Pc from the cytb6f molecules during each ramp cycle. Before starting the measurements, the Pc molecules on the AFM probes were either preoxidized by incubation in oxidizing buffer (imaging buffer supplemented with 10 mM potassium ferricyanide) or prereduced with reducing buffer (imaging buffer supplemented with 10 mM sodium dithionite) with a subsequent wash in imaging buffer. The contact probe-sample force was kept in the range 100 to 200 pN, and the imaging rate was adjusted (depending on the scan size and pixel density of the scan) in a way that ensured a 256 × 256 pixel PeakForce Capture image could be acquired at a modulation frequency of 1 kHz. During the affinity mapping, the sample was illuminated from a white light source through an optical fiber (Fiber-Lite MI-150; Dolan-Jener), and the power density of the illumination at the sample surface was measured with a Newport 842-PE meter. In addition, some partial illumination most probably occurred from the 670-nm laser used in the optical lever detection system for the AFM. For the control experiments, excess Pc or DCMU was added directly to the liquid cell by pipette at the concentrations described in the text. Subsequent image analysis was performed using Bruker Nanoscope Analysis v1.42 and OriginPro v8.5.1 software (OriginLab). The Nanoscope Analysis was used for the extraction and analysis of the PeakForce quantitative nanomechanical mapping spectroscopy data, and OriginPro 8.5 was used for the statistical analysis of all the force spectroscopy data and for all the calculations and curve fitting. Data reduction (positive identification of specific rupture events) was based on the analysis of the rupture events with probe-sample separations in the range 3 to 30 nm. The most probable values for the unbinding force and the rupture length were obtained from the maximum of the Gaussian fit to the force and rupture length distribution combined in a statistical histogram. The unbinding forces and rupture lengths of 276 rupture events were compiled in force or length distribution histograms.
Preparation of Functionalized Probes for Affinity-Mapping AFM
Hybrid AFM probes, Si tips mounted on Si3N4 rectangular or triangular cantilevers, model BL-AC40TS (Olympus Probes), or SNL (Bruker) were first cleaned by washing in acetone (HPLC grade from Fisher Scientific) and then cleaned in a home-built UV/Ozone cleaner (LSP035 Pen-Raylight source; LOT-Oriel) for 45 min. Immediately after the cleaning step, the AFM probes were placed into a glass desiccator purged with pure nitrogen for 10 min and then 20 μL (3-mercaptopropyl)trimethoxysilane was introduced into the desiccator. After another 5-min purge, the desiccator was evacuated down to a pressure of ∼0.3 kPa using a dry mechanical pump (Welch model 2027) and then sealed for 6 to 8 h to facilitate the deposition of the self-assembled monolayer (SAM). The next step in the functionalization of the AFM probes, immediately after the SAM formation, was to attach the Pc proteins. An amine-to-sulfhydryl heterobifunctional cross-linker with a 9.5-nm polyethylene glycol (PEG) spacer arm, terminated at one end with N-hydroxysuccinimide ester group and at the other with maleimide group [SM(PEG)24; Pierce Biotechnology] was used in order to attach an exposed lysine residue on the surface of the Pc molecule to the AFM probe. Both the amine-targeted and thiol-targeted reactions were accomplished simultaneously in sodium phosphate buffer, pH 7.4, at a final SM(PEG)24 concentration of 1 mM and a final Pc concentration of 50 nM for 40 min. Then, the AFM probes were gently washed (4 times) in 10 mM HEPES pH 8.0 buffer and stored in the same buffer for further use.
Preparation of Patterned Surfaces of cytb6f
The linear patterns of cytb6f were prepared by chemical patterning of glass substrates using a combination of reverse nanoimprint lithography and a wet lift-off and transfer of a thin polymer film replica together with a SAM of silanes to which the cytb6f molecules were selectively attached.
Accession Numbers
Structure data used in this work are available at the Protein Data Bank under the following accession numbers: Pc (1YLB), PSII (1S5L), cytb6f (1UM3), and LHCII (2BHW)
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Coomassie-Stained SDS-PAGE Gel of Protein Composition of Membrane Preparations.
Supplemental Figure 2. Preparation of Pc from Spinach Thylakoids.
Supplemental Figure 3. Preparation of cytb6f from Spinach Thylakoids and Patterning on Glass for Detection by Affinity-Mapping AFM.
Supplementary Material
Acknowledgments
M.P.J. acknowledges funding from the Leverhulme Trust, the Krebs Institute at the University of Sheffield, and Project Sunshine, University of Sheffield. C.N.H., J.D.O., and C.V. gratefully acknowledge funding from the Biotechnology and Biological Sciences Research Council (UK). This work was also supported as part of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences under Award Number DE-SC0001035. PARC’s role was to partially fund the Multimode VIII AFM system and to provide partial support for C.N.H.
AUTHOR CONTRIBUTIONS
M.P.J. performed the purification of membranes and isolated complexes and characterized them by spectroscopy. C.V. prepared the functionalized AFM probes. M.P.J. and C.V. performed the AFM experiments and data analysis. J.D.O. provided advice and support to AFM experiments. M.P.J., C.V., and C.N.H. designed the study and wrote the article. All authors discussed the results and commented on the article.
Glossary
- PSI
photosystem I
- PSII
photosystem II
- PQ
plastoquinone
- Pc
plastocyanin
- α-DM
n-dodecyl-α-d-maltoside
- AFM
atomic force microscopy
- DG
grana prepared by digitonin
- SG
grana prepared by sonication
- RC
reaction center
- SAM
self-assembled monolayer
- PDB
Protein Data Bank
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Albertsson P. (2001). A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci. 6: 349–358. [DOI] [PubMed] [Google Scholar]
- Albertsson P.Å., Andreasson E., Stefansson H., Wollenberger L. (1994). Fractionation of the thylakoid membrane. Methods Enzymol. 228: 469–482. [Google Scholar]
- Allred D.R., Staehelin L.A. (1985). Lateral distribution of the cytochrome b6/f and coupling factor ATP synthetase complexes of chloroplast thylakoid membranes. Plant Physiol. 78: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A., Drory O., Nelson N. (2007). The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447: 58–63. [DOI] [PubMed] [Google Scholar]
- Anderson J.M. (1982). Distribution of the cytochromes of spinach chloroplasts between the appressed membranes of grana stacks and stroma-exposed thylakoid regions. FEBS Lett. 138: 62–66. [Google Scholar]
- Andersson B., Simpson D.J., Høyer-Hansen G. (1978). Freeze-fracture evidence for the isolation of inside-out spinach thylakoid vesicles. Carlsberg Res. Commun. 43: 77–89. [Google Scholar]
- Blackwell M., Gibas C., Gygax S., Roman D., Wagner B. (1994). The plastoquinone diffusion coefficient in chloroplasts and its mechanistic implications. Biochim. Biophys. Acta 1183: 533–543. [Google Scholar]
- Bendall D.S., Davenport H.E., Hill R. (1971). Cytochrome components in chloroplasts of the higher plants. Methods Enzymol. 23: 327–344. [Google Scholar]
- Berthold D.A., Babcock G.T., Yocum C.F. (1981). A highly resolved, oxygen-evolving Photosystem II preparation from spinach thylakoid membranes. EPR and electron transport properties. FEBS Lett. 134: 231–234. [Google Scholar]
- Anderson J.M., Boardman N.K. (1966). Fractionation of the photochemical systems of photosynthesis. I. Chlorophyll contents and photochemical activities of particles isolated from spinach chloroplasts. Bibl. Laeger 112: 403–421. [DOI] [PubMed] [Google Scholar]
- Boardman N.K., Anderson J.M. (1967). Fractionation of the photochemical systems of photosynthesis. II. Cytochrome and carotenoid contents of particles isolated from spinach chloroplasts. Biochim. Biophys. Acta 143: 187–203. [DOI] [PubMed] [Google Scholar]
- Cartron M.L., Olsen J.D., Sener M., Jackson P.J., Brindley A.A., Qian P., Dickman M.J., Leggett G.J., Schulten K., Neil Hunter C. (2014). Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Biochim. Biophys. Acta http//dx..org/10.1016/j.bbabio.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.P., Andersson B. (1981). Lateral and transverse organisation of cytochromes in the chloroplast thylakoid membrane. Biochem. Biophys. Res. Commun. 103: 1336–1342. [DOI] [PubMed] [Google Scholar]
- Cramer W.A., Hasan S.S., Yamashita E. (2011). The Q cycle of cytochrome bc complexes: a structure perspective. Biochim. Biophys. Acta 1807: 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson R., Albertsson P.Å., Mamedov F., Styring S. (2004). Quantification of photosystem I and II in different parts of the thylakoid membrane from spinach. Biochim. Biophys. Acta 1608: 53–61. [DOI] [PubMed] [Google Scholar]
- Daum B., Kühlbrandt W. (2011). Electron tomography of plant thylakoid membranes. J. Exp. Bot. 62: 2393–2402. [DOI] [PubMed] [Google Scholar]
- Daum B., Nicastro D., Austin J., II, McIntosh J.R., Kühlbrandt W. (2010). Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22: 1299–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J.P., Boekema E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706: 12–39. [DOI] [PubMed] [Google Scholar]
- Delosme R. (1991). Electron transfer from cytochrome f to photosystem I in green algae. Photosynth. Res. 29: 45–54. [DOI] [PubMed] [Google Scholar]
- Díaz-Moreno I., Díaz-Quintana A., De la Rosa M.A., Ubbink M. (2005). Structure of the complex between plastocyanin and cytochrome f from the cyanobacterium Nostoc sp. PCC 7119 as determined by paramagnetic NMR. The balance between electrostatic and hydrophobic interactions within the transient complex determines the relative orientation of the two proteins. J. Biol. Chem. 280: 18908–18915. [DOI] [PubMed] [Google Scholar]
- Dunahay T.G., Staehelin L.A., Seibert M., Ogilvie P.D., Berg S.P. (1984). Structural, biochemical and biophysical characterization of four oxygen-evolving photosystem II preparations from spinach. Biochim. Biophys. Acta 764: 179–193. [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342. [DOI] [PubMed] [Google Scholar]
- Haehnel W., Pröpper A., Krause H. (1980). Evidence for complexed plastocyanin as the immediate electron donor of P-700. Biochim. Biophys. Acta 593: 384–399. [DOI] [PubMed] [Google Scholar]
- Hinshaw J.E., Miller K.R. (1993). Mapping the lateral distribution of photosystem II and the cytochrome b6/f complex by direct immune labeling of the thylakoid membrane. J. Struct. Biol. 111: 1–8. [DOI] [PubMed] [Google Scholar]
- Illerhaus J., Altschmied L., Reichert J., Zak E., Herrmann R.G., Haehnel W. (2000). Dynamic interaction of plastocyanin with the cytochrome bf complex. J. Biol. Chem. 275: 17590–17595. [DOI] [PubMed] [Google Scholar]
- Joliot P., Lavergne J., Bouchaud J.P. (1992). Plastoquinone compartmentation in chloroplasts I. Evidence for domains with different rates of photoreduction. Biochim. Biophys. Acta 1101: 1–12. [Google Scholar]
- Kannt A., Young S., Bendall D.S. (1996). The role of acidic residues of plastocyanin in its interaction with cytochrome f. Biochim. Biophys. Acta 1277: 115–126. [PubMed] [Google Scholar]
- Kaur J., Singh K.V., Schmid A.H., Varshney G.C., Suri C.R., Raje M. (2004). Atomic force spectroscopy-based study of antibody pesticide interactions for characterization of immunosensor surface. Biosens. Bioelectron. 20: 284–293. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Horstmann S., Weis E. (2000). Control of the photosynthetic electron transport by PQ diffusion microdomains in thylakoids of higher plants. Biochim. Biophys. Acta 1459: 148–168. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Mukherjee U., Galla H.J. (2002). Molecular architecture of the thylakoid membrane: lipid diffusion space for plastoquinone. Biochemistry 41: 4872–4882. [DOI] [PubMed] [Google Scholar]
- Kouřil R., Oostergetel G.T., Boekema E.J. (2011). Fine structure of granal thylakoid membrane organization using cryo electron tomography. Biochim. Biophys. Acta 1807: 368–374. [DOI] [PubMed] [Google Scholar]
- Kurisu G., Zhang H., Smith J.L., Cramer W.A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302: 1009–1014. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Hibino T., Takabe T., Weisbeek P.J., Takabe T. (1995). Site-directed mutagenetic study on the role of negative patches on silene plastocyanin in the interactions with cytochrome f and photosystem I. J. Biochem. 117: 1209–1217. [DOI] [PubMed] [Google Scholar]
- Lee G.U., Kidwell D.A., Colton R.J. (1994). Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir 10: 354–357. [Google Scholar]
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005). Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature 438: 1040–1044. [DOI] [PubMed] [Google Scholar]
- Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292. [DOI] [PubMed] [Google Scholar]
- Melis A., Brown J.S. (1980). Stoichiometry of system I and system II reaction centers and of plastoquinone in different photosynthetic membranes. Proc. Natl. Acad. Sci. USA 77: 4712–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T.E., Zhao Z.G., Cusanovich M.A., Tollin G. (1993). Transient kinetics of electron transfer from a variety of c-type cytochromes to plastocyanin. Biochemistry 32: 4552–4559. [DOI] [PubMed] [Google Scholar]
- Morand L.Z., Krogmann D.W. (1993). Large-scale preparation of pure plastocyanin from spinach. Biochim. Biophys. Acta 1141: 105–106. [Google Scholar]
- Morrissey P.J., McCauley S.W., Melis A. (1986). Differential detergent-solubilization of integral thylakoid membrane complexes in spinach chloroplasts. Localization of photosystem II, cytochrome b6-f complex and photosystem I. Eur. J. Biochem. 160: 389–393. [DOI] [PubMed] [Google Scholar]
- Moser C.C., Dutton P.L. (1988). Cytochrome c and c2 binding dynamics and electron transfer with photosynthetic reaction center protein and other integral membrane redox proteins. Biochemistry 27: 2450–2461. [DOI] [PubMed] [Google Scholar]
- Nevo R., Charuvi D., Tsabari O., Reich Z. (2012). Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 70: 157–176. [DOI] [PubMed] [Google Scholar]
- Olive J., Vallon O., Wollman F.A., Recouvreur M., Bennoun P. (1986). Studies on the cytochrome b6/f complex. II. Localization of the complexin the thylakoid membranes from spinach and Chlamydomonas reinhardtii by immunocytochemistry and freeze-fracture analysis of b6/f mutants. Biochim. Biophys. Acta 851: 239–248. [Google Scholar]
- Overfield R.E., Wraight C.A., Devault D. (1979). Microsecond photooxidation kinetics of cytochrome c2 from Rhodopseudomonas sphaeroides: in vivo and solution studies. FEBS Lett. 105: 137–142. [DOI] [PubMed] [Google Scholar]
- Pogorelov T.V., Autenrieth F., Roberts E., Luthey-Schulten Z.A. (2007). Cytochrome c2 exit strategy: dissociation studies and evolutionary implications. J. Phys. Chem. B 111: 618–634. [DOI] [PubMed] [Google Scholar]
- Ruban A.V., Solovieva S., Lee P.J., Ilioaia C., Wentworth M., Ganeteg U., Klimmek F., Chow W.S., Anderson J.M., Jansson S., Horton P. (2006). Plasticity in the composition of the light harvesting antenna of higher plants preserves structural integrity and biological function. J. Biol. Chem. 281: 14981–14990. [DOI] [PubMed] [Google Scholar]
- Sane P.V., Goodchild D.J., Park R.B. (1970). Characterization of chloroplast photosystems 1 and 2 separated by a non-detergent method. Biochim. Biophys. Acta 216: 162–178. [DOI] [PubMed] [Google Scholar]
- Soriano G.M., Ponamarev M.V., Tae G.-S., Cramer W.A. (1996). Effect of the interdomain basic region of cytochrome f on its redox reactions in vivo. Biochemistry 35: 14590–14598. [DOI] [PubMed] [Google Scholar]
- Soriano G.M., Ponamarev M.V., Piskorowski R.A., Cramer W.A. (1998). Identification of the basic residues of cytochrome f responsible for electrostatic docking interactions with plastocyanin in vitro: relevance to the electron transfer reaction in vivo. Biochemistry 37: 15120–15128. [DOI] [PubMed] [Google Scholar]
- Standfuss J., Terwisscha van Scheltinga A.C., Lamborghini M., Kühlbrandt W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 24: 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M.M., Allen S., Davies M.C., Roberts C.J., Schacht E., Tendler S.J.B., van Steenkiste S., Williams P.M. (2002). The development, characterization, and demonstration of a versatile immobilization strategy for biomolecular force measurements. Langmuir 18: 6659–6665. [Google Scholar]
- Stroebel D., Choquet Y., Popot J.L., Picot D. (2003). An atypical haem in the cytochrome b6f complex. Nature 426: 413–418. [DOI] [PubMed] [Google Scholar]
- Sznee K., Dekker J.P., Dame R.T., van Roon H., Wuite G.J.L., Frese R.N. (2011). Jumping mode atomic force microscopy on grana membranes from spinach. J. Biol. Chem. 286: 39164–39171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Nomoto N., Koga M., Ogasa H., Ogawa Y., Matsumoto M., Stampoulis P., Sode K., Terasawa H., Shimada I. (2012). Structural basis of efficient electron transport between photosynthetic membrane proteins and plastocyanin in spinach revealed using nuclear magnetic resonance. Plant Cell 24: 4173–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya K. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9Å. 473: 55–60. [DOI] [PubMed] [Google Scholar]
- Vallon O., Bulte L., Dainese P., Olive J., Bassi R., Wollman F.A. (1991). Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc. Natl. Acad. Sci. USA 88: 8262–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roon H., van Breemen J.F., de Weerd F.L., Dekker J.P., Boekema E.J. (2000). Solubilization of green plant thylakoid membranes with n-dodecyl-α,D-maltoside. Implications for the structural organization of the Photosystem II, Photosystem I, ATP synthase and cytochrome b6 f complexes. Photosynth. Res. 64: 155–166. [DOI] [PubMed] [Google Scholar]
- Vasilev C., Brindley A.A., Olsen J.D., Saer R.G., Beatty J.T., Hunter C.N. (2014). Nano-mechanical mapping of the interactions between surface-bound RC-LH1-PufX core complexes and cytochrome c2 attached to an AFM probe. Photosynth. Res. 120: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckier A., Gervasoni P., Zaugg F., Ziegler U., Lindner P., Groscurth P., Plückthun A., Semenza G. (1998). Atomic force microscopy detects changes in the interaction forces between GroEL and substrate proteins. Biophys. J. 74: 3256–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Whitelegge J.P., Cramer W.A. (2001). Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem. 276: 38159–38165. [DOI] [PubMed] [Google Scholar]
- Zouni A., Witt H.T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409: 739–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.