Plants use phenylalanine to produce abundant and diverse phenylpropanoid compounds, such as flavonoids, tannins, and lignin. Through phylogenetic, bioinformatic, and biochemical analyses of prephenate aminotransferase enzymes from deep taxonomic lineages, this study revealed unique evolutionary history and molecular changes of key enzymes responsible for phenylalanine biosynthesis in plants.
Abstract
The aromatic amino acid Phe is required for protein synthesis and serves as the precursor of abundant phenylpropanoid plant natural products. While Phe is synthesized from prephenate exclusively via a phenylpyruvate intermediate in model microbes, the alternative pathway via arogenate is predominant in plant Phe biosynthesis. However, the molecular and biochemical evolution of the plant arogenate pathway is currently unknown. Here, we conducted phylogenetically informed biochemical characterization of prephenate aminotransferases (PPA-ATs) that belong to class-Ib aspartate aminotransferases (AspAT Ibs) and catalyze the first committed step of the arogenate pathway in plants. Plant PPA-ATs and succeeding arogenate dehydratases (ADTs) were found to be most closely related to homologs from Chlorobi/Bacteroidetes bacteria. The Chlorobium tepidum PPA-AT and ADT homologs indeed efficiently converted prephenate and arogenate into arogenate and Phe, respectively. A subset of AspAT Ib enzymes exhibiting PPA-AT activity was further identified from both Plantae and prokaryotes and, together with site-directed mutagenesis, showed that Thr-84 and Lys-169 play key roles in specific recognition of dicarboxylic keto (prephenate) and amino (aspartate) acid substrates. The results suggest that, along with ADT, a gene encoding prephenate-specific PPA-AT was transferred from a Chlorobi/Bacteroidetes ancestor to a eukaryotic ancestor of Plantae, allowing efficient Phe and phenylpropanoid production via arogenate in plants today.
INTRODUCTION
Phe and Tyr are aromatic amino acids required for protein biosynthesis in all organisms and are produced by bacteria, fungi, and plants, but not in animals. These amino acids are essential nutrients in the animal diets, and their biosynthetic pathways are prime targets of antimicrobial drugs and plant herbicides (e.g., glyphosate) (Schönbrunn et al., 2001; Reichau et al., 2011). Phe and Tyr also serve as precursors of various aromatic natural products such as pigments, antibiotics, and alkaloids (Bentley, 1990; Maeda and Dudareva, 2012). In vascular plants, up to 30% of photosynthetically fixed carbon is directed toward Phe biosynthesis for the production of abundant phenylpropanoid compounds such as lignin (Weiss, 1986; Razal et al., 1996). However, it is currently unknown how Phe and Try biosynthetic pathways have evolved to support the production of diverse and abundant aromatic natural products in plants.
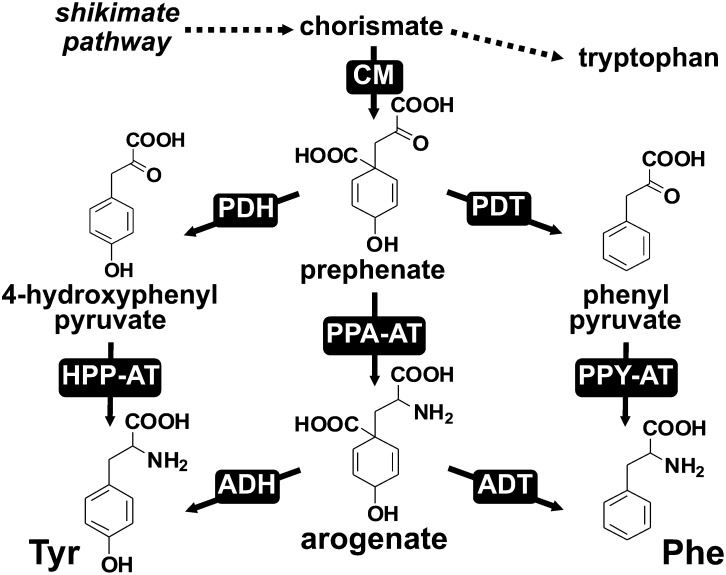
Phe and Tyr are synthesized from chorismate, the final product of the shikimate pathway, which is converted to prephenate by chorismate mutase (CM; Figure 1). In model microbes (e.g., Escherichia coli and Saccharomyces cerevisiae), prephenate is first converted by prephenate dehydratase (PDT) and prephenate dehydrogenase (PDH) to phenylpyruvate or 4-hydroxyphenylpyruvate, which are then transaminated to Phe and Tyr, respectively (the phenylpyruvate and 4-hydroxyphenylpyruvate pathways; Figure 1; Bentley, 1990). In E. coli, these final transamination steps in both Phe and Tyr biosynthesis are catalyzed by any of three distinct aminotransferases, tyrosine aminotransferase, aspartate aminotransferase, and branched-chain aminotransferase (Gelfand and Steinberg, 1977). By contrast, plants and some other microbes have evolved an alternative pathway to synthesize Phe and/or Tyr via the arogenate intermediate (Stenmark et al., 1974; Fazel et al., 1980; Jung et al., 1986). In this arogenate pathway, prephenate is first transaminated by prephenate aminotransferase (PPA-AT) to arogenate, which is then converted to Phe and Tyr by arogenate dehydratase (ADT) and arogenate dehydrogenase (ADH), respectively (Figure 1; Herrmann and Weaver, 1999; Tzin and Galili, 2010; Maeda and Dudareva, 2012).
Figure 1.
Different Pathways for Phe and Tyr Biosynthesis.
Phe and Tyr can each be synthesized from prephenate via two alternative routes, the arogenate pathway and/or the phenylpyruvate/4-hydroxyphenylpyruvate pathways. ADH (EC 1.3.1.79); ADT (EC 4.2.1.91); CM (EC 5.4.99.5); HPP-AT, 4-hydroxyphenylpyruvate aminotransferase; PDH (EC 1.3.1.12); PDT (EC 4.2.1.51); PPA-AT (EC 2.6.1.79); PPY-AT, phenylpyruvate aminotransferase.
ADH activity and ADH genes have been reported in plants as well as some bacteria (Stenmark et al., 1974; Fazel et al., 1980; Connelly and Conn, 1986; Rippert and Matringe, 2002; Legrand et al., 2006), suggesting that the arogenate pathway for Tyr biosynthesis is distributed among different kingdoms. Similarly, ADT activity responsible for the final step of the arogenate pathway for Phe biosynthesis has been detected in many plant tissues and some bacteria (Jung et al., 1986; Fischer and Jensen, 1987). The corresponding ADT genes from plants have been identified based on genetic screening (Yamada et al., 2008; Huang et al., 2010) or similarity to microbial PDTs (Cho et al., 2007; Maeda et al., 2010). Biochemical characterization of recombinant plant ADT enzymes showed that they have substrate preference toward arogenate over prephenate (Cho et al., 2007; Yamada et al., 2008; Maeda et al., 2010). Genetic analysis of plant ADTs further demonstrated that Phe is predominantly synthesized via the arogenate in plants (Maeda et al., 2010; Corea et al., 2012a, 2012b). However, phylogenetic analyses showed that the arogenate and prephenate specificity of ADH/PDH and ADT/PDT do not map tidily onto a single clade of the gene tree, suggesting that their substrate specificity has been gained and/or lost multiple times in the gene family’s evolution (Jensen, 1985; Song et al., 2005). Thus, the presence of sequences similar to known ADT or ADH does not necessarily infer the presence of the arogenate pathway.
PPA-AT catalyzes the first committed step of the arogenate pathway, the conversion of prephenate to arogenate (Figure 1). PPA-AT activities have been detected in plants and some microbes (Stenmark et al., 1974; Fazel and Jensen, 1979; Bonner and Jensen, 1985; Siehl et al., 1986; Abou-Zeid et al., 1995). Three types of aminotransferases, including class-Ib aspartate aminotransferases (AspAT Ibs), branched-chain aminotransferases (BCATs), and N-succinyl-diaminopimelate aminotransferases (S-DAPAT), were recently shown to have PPA-AT activity in different microbes (Graindorge et al., 2014). Several lines of evidence support that AspAT Ib-type PPA-AT is the major enzyme responsible for PPA-AT activity in plants. (1) PPA-AT activity detected in various plant tissues was associated with Asp-AT activity during purification (Bonner and Jensen, 1985; Siehl et al., 1986). (2) PPA-AT activity from Arabidopsis thaliana culture cells was purified to a single peak corresponding to AspAT Ib, encoded by At2g22250 (Graindorge et al., 2010). (3) Plant AspAT Ib genes are strongly coexpressed with shikimate, Phe, and phenylpropanoid pathway genes (Dal Cin et al., 2011; Maeda et al., 2011), which is not the case for plant homologs of microbial BCAT- and S-DAPAT-type PPA-ATs (Supplemental Data Set 1). (4) Genetic suppression of PPA-AT in petunia (Petunia hybrida) flowers eliminated the majority of PPA-AT activity (Maeda et al., 2011). (5) Finally, plant PPA-AT activity and the recombinant plant PPA-AT enzymes share unique substrate preference, specifically using acidic amino donors (i.e., aspartate and glutamate) and prephenate keto acceptor, but not phenylpyruvate or 4-hydroxyphenylpyruvate, the other aromatic keto acid substrates within the Phe and Tyr biosynthetic pathways (Bonner and Jensen, 1985; Siehl et al., 1986; Graindorge et al., 2010; Maeda et al., 2011).
Unlike the major effect of ADT suppression on the levels of Phe and Phe-derived compounds (Maeda et al., 2010, Corea et al., 2012a, 2012b), PPA-AT suppression led to a minor reduction in Phe levels (Maeda et al., 2011; de la Torre et al., 2014). However, this was due to compensatory operation of Phe biosynthesis via alternative phenylpyruvate pathway mediated by a separate phenylpyruvate aminotransferase (Yoo et al., 2013), which is homologous to tyrosine aminotransferase and has a broad substrate specificity (Gelfand and Steinberg, 1977; Gonda et al., 2010; Lee and Facchini, 2011; Riewe et al., 2012). Thus, besides having a role in plastidic aspartate metabolism owing to its Asp-AT activity (de la Torre et al., 2014), the identified plant PPA-ATs are indeed involved in Phe biosynthesis (Maeda et al., 2011; Yoo et al., 2013; de la Torre et al., 2014). The strict substrate specificity of plant PPA-ATs toward prephenate among the three aromatic keto acid substrates in Phe and Tyr biosynthesis (Graindorge et al., 2010; Maeda et al., 2011) also suggest that plant PPA-AT directs carbon flow specifically from prephenate toward arogenate and hence serves as an ideal marker to investigate the molecular and biochemical evolution of the plant arogenate pathway.
In this study, we conducted “phylobiochemical” characterization of homologs of plant PPA-ATs from a broad range of organisms. By combining intensive phylogenetic analysis with functional protein association analysis, recombinant enzyme kinetics, and site-directed mutagenesis, we found that plant PPA-ATs are phylogenetically and biochemically closely related to homologs from Chlorobi/Bacteroidetes. Our analysis further identified a subset of AspAT Ib enzymes having PPA-AT activity from a broad range of microbes, which enabled determination of two amino acid residues crucial for the substrate specificity of plant PPA-AT enzymes.
RESULTS
Plantae and Chlorobi/Bacteroidetes Share Common Ancestors of PPA-AT and ADT
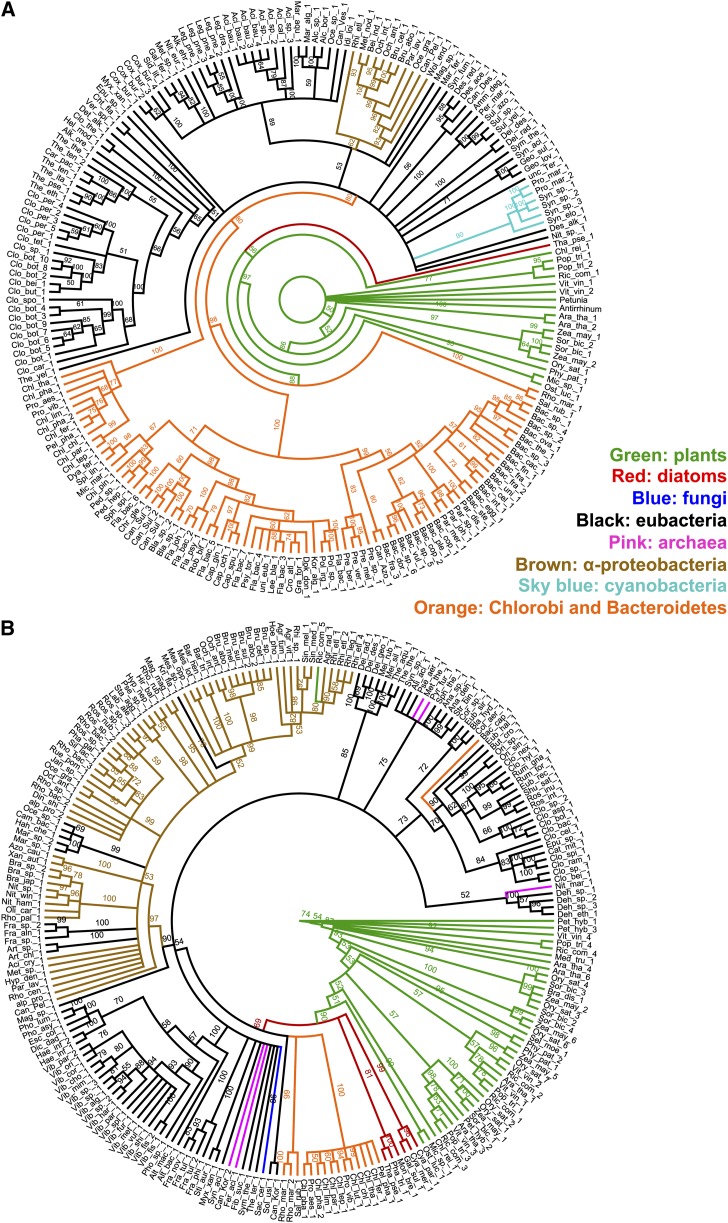
Maximum likelihood (ML) phylogenetic analyses were performed to identify which extant homologs in the Tree of Life are most closely associated with plant enzymes involved in Phe and Tyr biosynthesis. Homologs of PPA-AT and ADT (Figure 2) as well as ADH and CM (Supplemental Figure 1) were not identified in Metazoa by BLASTP (Supplemental Data Set 2), consistent with the absence of aromatic amino acid biosynthesis in animals. Besides photosynthetic protists (i.e., diatoms), the closest relatives of plant CMs were found in fungi (Supplemental Figure 1 and Supplemental Data Set 2), suggesting that plant CMs have eukaryotic origin. The inferred unrooted tree of ADH showed that the relationship of the plant/diatom clade to prokaryotic homologs was not well enough sampled and resolved to suggest a particular origin (Supplemental Figure 1).
Figure 2.
ML Phylogenetic Analyses of PPA-AT and ADT.
Node labels are nonparametric bootstrap percentage values. Colored bars and labels over phylograms indicate monophyletic groups. Trees for PPA-AT (A) and ADT (B) are arbitrarily rooted on the plant homolog used in the similarity searches.
The PPA-AT and ADT phylogenetic analyses showed unexpected phylogenetic distributions: the closest homologs to both plant PPA-ATs and ADTs were found in the bacterial phyla Bacteroidetes and Chlorobi (Figure 2; Supplemental Data Set 2). This is not likely to be an artifact caused by long-branch attraction since branch length heterogeneity was minimal (Supplemental Figure 2). Multiple ADT isoforms found in land plants (e.g., six Arabidopsis ADTs; Cho et al., 2007) were all within the monophyletic plant clade (Figure 2B) in agreement with their recent duplications of a single common ancestor. The alternative hypotheses that these plant PPA-AT and ADT genes were acquired via mitochondrial or plastid endosymbiosis were tested by constraining plant homologs to the cluster with α-proteobacteria or cyanobacteria homologs and evaluating them using an approximately unbiased (AU) test (α = 0.05). The data were significantly better explained by the unconstrained tree than by any of the trees constrained to α-proteobacteria or cyanobacteria homologs (Supplemental Table 1 and Supplemental Figure 3). Together, the phylogenetic affinity of PPA-AT and ADT homologs in Plantae and the Bacteroidetes/Chlorobi bacteria suggest that they shared close common ancestors.
A Chlorobium tepidum PPA-AT Homolog Can Efficiently Convert Prephenate to Arogenate
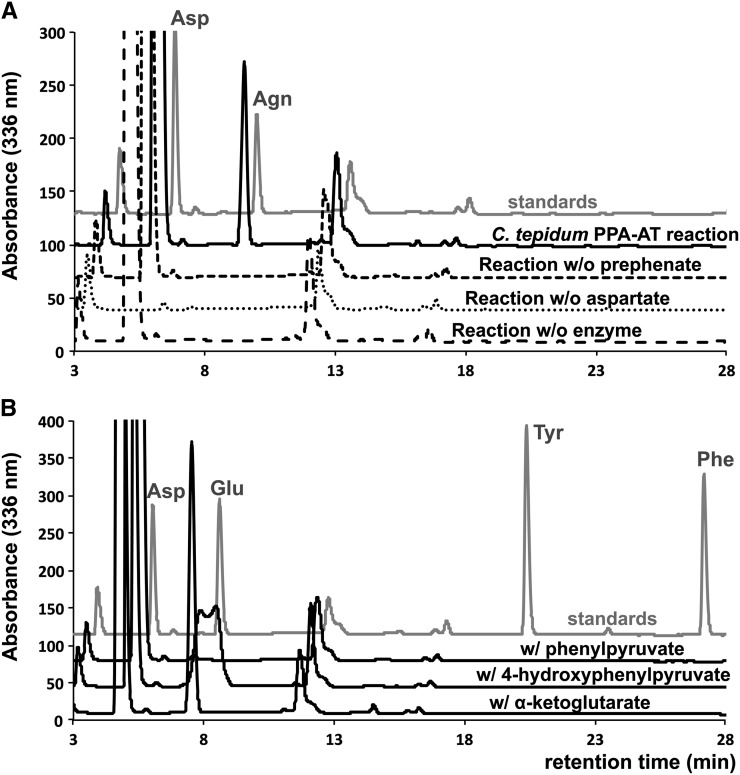
To examine if the phylogenetically close PPA-AT homolog of Chlorobi bacteria indeed has PPA-AT catalytic activity, the corresponding gene of green-sulfur bacterium Chlorobium tepidum was cloned and expressed in E. coli, and the recombinant protein was purified (Supplemental Figure 4) for biochemical characterization. The C. tepidum enzyme converted prephenate into arogenate when incubated with prephenate in the presence of pyridoxal 5′-phosphate (PLP) cofactor and aspartate amino donor (Figure 3A). No arogenate production was observed in the absence of prephenate, aspartate, or the enzyme (Figure 3A). Similar to plant PPA-ATs (Graindorge et al., 2010; Maeda et al., 2011), the C. tepidum enzyme displayed Asp-AT activity (conversion of α-ketoglutarate into glutamate using aspartate amino donor) and was unable to convert other aromatic keto acid substrates, phenylpyruvate and 4-hydroxyphenylpyruvate, into Phe and Tyr, respectively (PPY-AT and HPP-AT activity; Figure 3B). These results show that the C. tepidum homolog has PPA-AT activity and specifically recognizes prephenate among aromatic keto acid substrates involved in Phe and Tyr biosynthesis.
Figure 3.
PPA-AT, AspAT, PPY-AT, and HPP-AT Activity Assays Using C. tepidum PPA-AT Homolog.
(A) Arogenate (Agn) was produced after 15 min incubation of the recombinant C. tepidum enzyme (20 μg/mL) with 1 mM prephenate substrate, 5 mM aspartate (Asp) amino donor, and 200 μM PLP cofactor at 37°C (solid line). The reactions lacking prephenate, Asp, or the enzyme did not produce Agn (dotted lines).
(B) The C. tepidum enzyme converted α-ketoglutarate into glutamate (Asp-AT activity) but did not use phenylpyruvate (PPY-AT activity) and 4-hydroxyphenylpyruvate (HPP-AT activity).
To compare the biochemical properties of the C. tepidum PPA-AT homolog with those of plant PPA-ATs, kinetic analysis was conducted for PPA-AT and Asp-AT activities of the C. tepidum enzymes. Apparent Km toward prephenate and α-ketoglutarate were 464 and 949 μM, respectively, in PPA-AT and AspAT assays using aspartate amino donor (Table 1), which are comparable to those of Arabidopsis PPA-AT (355 and 691 μM, respectively; Maeda et al., 2011). A similar Km (346 μM) was also observed for prephenate when glutamate was used as an amino donor, consistent with prior reports (Graindorge et al., 2010; Maeda et al., 2011). The turnover rates (kcat) of PPA-AT and Asp-AT activity were also comparable between C. tepidum (12 to 17 s−1) and Arabidopsis (5 to 17 s−1) enzymes (Table 1; Maeda et al., 2011).
Table 1. Kinetic Analysis of C. tepidum and Synechocystis sp PCC6803 PPA-AT Homologs.
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| C. tepidum PPA-AT homolog | |||
| Prephenate | 12.3 ± 0.2 | 464 ± 39 | 26.8 ± 2.5 |
| α-Ketoglutarate | 17.2 ± 1.9 | 949 ± 102 | 18.9 ± 3.9 |
| Synechocystis sp PCC6803 PPA-AT homolog | |||
| Prephenate | 1.1 ± 0.1 | 2539 ± 254 | 0.44 ± 0.02 |
| α-Ketoglutarate | 54.2 ± 3.7 | 486 ± 33 | 111.7 ± 5.7 |
Kinetic parameters were obtained for prephenate and α-ketoglutarate in 5-min reactions using 0.5 and 2 μg/mL recombinant enzymes, respectively, for C. tepidum PPA-AT homolog and 8 and 0.5 μg/mL for Synechocystis homolog. Data are means ± se (n = 3 independent experiments). 20 mM aspartate and 200 μM PLP were used as an amino donor and a cofactor, respectively.
The purified recombinant PPA-AT homolog of cyanobacterium Synechocystis sp PCC6803, which was more distantly related to Arabidopsis PPA-AT enzyme than C. tepidum PPA-AT homologs (Figure 2A), was also generated (Supplemental Figure 4), biochemically characterized, and compared with the C. tepidum and Arabidopsis enzymes. The Synechocystis enzyme also displayed strong Asp-AT activity (Km of 486 μM, kcat of 54.2 s−1; Table 1) similar to the C. tepidum and Arabidopsis enzymes. Although the same Synechocystis enzyme was reported to be devoid of PPA-AT activity in a previous study based on a coupled spectrophotometric assay (Graindorge et al., 2014), we detected some PPA-AT activity through direct detection of the arogenate product by HPLC; however, its Km (2.5 mM) and kcat (1 s−1) with prephenate were much higher and lower, respectively, than those of C. tepidum and Arabidopsis enzymes (Table 1). Taken together, these results show that, while all three enzymes exhibit comparable Asp-AT activity, the PPA-AT activity of the C. tepidum enzyme is more similar to that of Arabidopsis PPA-AT than that of the Synechocystis enzyme.
C. tepidum ADT Homolog Converts Arogenate into Phe
The C. tepidum homolog of plant ADTs was previously reported as PDT, although only weak PDT activity was detected (Tan et al., 2008). To test if the C. tepidum ADT homolog is capable of converting arogenate into Phe, the corresponding gene was cloned and its purified recombinant enzyme (Supplemental Figure 4) was subjected to ADT and PDT assays. Since E. coli crude extract contains endogenous PDT activity derived from a bifunctional CM-PDT enzyme (Davidson et al., 1972; Zhang et al., 1998), CM activity was analyzed in the purified fraction to ensure the absence of E. coli CM-PDT contamination. Consistent with the prior report (Tan et al., 2008), PDT activity was detectable only at high enzyme concentrations and prolonged reaction times (Supplemental Figure 5). When incubated with arogenate substrate, however, the C. tepidum ADT homolog was able to produce Phe efficiently (Supplemental Figure 5).
Kinetic analysis of the C. tepidum enzyme revealed that the apparent Km toward arogenate and prephenate were 1544 and 827 μM, respectively (Table 2). Moreover, kcat with arogenate was 20-fold higher than that with prephenate substrate (5.4 and 0.27 s−1, respectively; Table 2). These parameters were within the range of previously reported Arabidopsis ADTs that use both arogenate and, to a lesser extent, prephenate substrates (ADT1, ADT2, and ADT6, Km and kcat with arogenate ranging from 0.8 to 3 mM and 3.2 to 6.1 s−1, respectively; Cho et al., 2007). The 10-fold higher catalytic efficiency of the C. tepidum enzyme for arogenate than prephenate (Table 2) suggests that, similar to the Arabidopsis ADTs, the C. tepidum ADT homolog exhibits stronger ADT activity than PDT activity.
Table 2. Kinetic Analysis of PDT and ADT Activity of the C. tepidum ADT Homolog.
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Arogenate | 5.4 ± 0.2 | 1544 ± 122 | 3.5 ± 0.2 |
| Prephenate | 0.27 ± 0.04 | 827 ± 87 | 0.32 ± 0.02 |
Identification of Enzymes Homologous to Plant PPA-ATs and Having PPA-AT Activity
Above results showed that a distantly related sequence (i.e., from Synechocystis) to plant PPA-ATs still exhibits PPA-AT activity, though much weaker than the C. tepidum PPA-AT homolog (Table 1). Thus, we further examined the phylogenetic distribution of homologs of plant PPA-ATs that actually exhibit PPA-AT activity. A deeper ML phylogenetic analysis oriented around the transitive nature of molecular evolution (Pearson and Lipman, 1988) was performed (Supplemental Figure 6) and used as a framework to identify the transition between prephenate utilizing and non-utilizing aminotransferases. To construct a manageable-sized tree that included homologs from deep taxonomic lineages, one out of every 13 sequences was arbitrarily selected (see Methods). Thus, it is important to note that the absence of a sequence in a given organism does not necessarily infer the absence of a homolog of plant PPA-ATs.
For an initial prediction of a set of aminotransferases that are likely involved in Phe and Tyr biosynthesis, a functional protein association analysis was conducted for representative PPA-AT homologs (Supplemental Figure 6) using STRING (Supplemental Table 2; Franceschini et al., 2013; http://string-db.org), which integrates various database information (e.g., co-occurrence in genome, coexpression data, and physical protein-protein interaction). The PPA-AT homologs of Rhodothermus marinus (ZP_04423142.1), C. tepidum, Thermus thermophilus, Synechocystis sp PCC6803, and Dickeya dadantii, located within a monophyletic clade containing Plantae, bacteria, and archaea (Supplemental Figure 6), showed some functional associations with enzymes catalyzing the proceeding and/or subsequent steps of the PPA-AT reaction, PDT/ADT, PDH/ADH, and CM (Supplemental Table 2). By contrast, more distantly related homologs, such as the Alicyclobacillus acidocaldarius sequence and the second most similar R. marinus sequence (YP_003290582.1) to Arabidopsis PPA-AT, did not display such functional association (Supplemental Table 2). These results suggest that AspAT Ib enzymes having PPA-AT activity may be present within a clade containing the D. dadantii sequence and other homologs more closely related to plant PPA-ATs (Supplemental Figure 6).
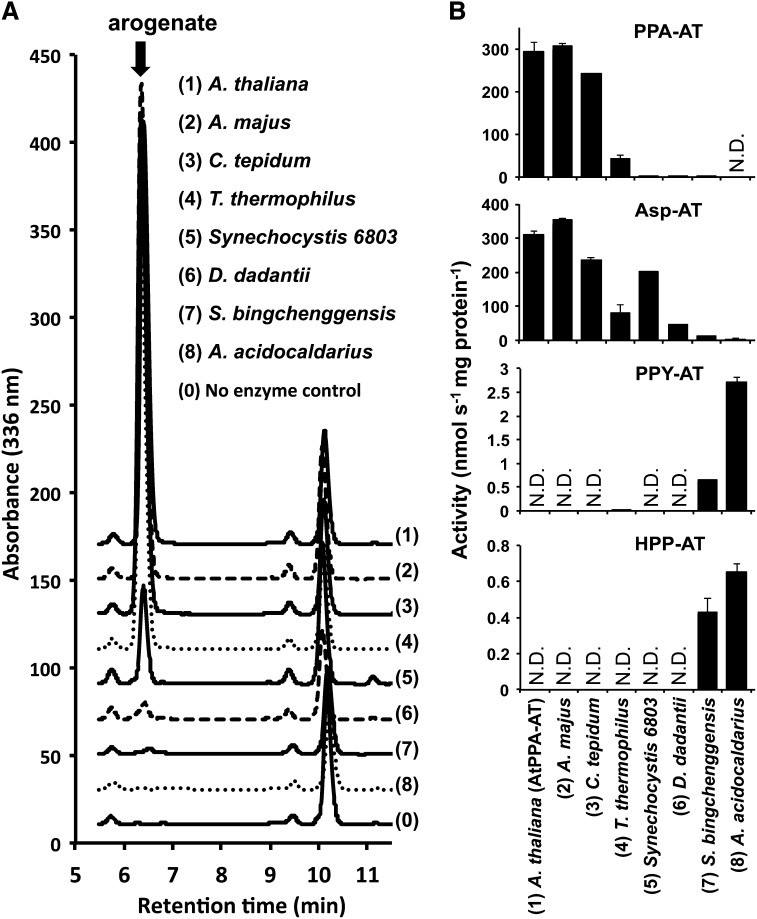
To experimentally test this prediction, the representative PPA-AT homologs around the predicted boundary (Supplemental Figure 6) were cloned and the corresponding recombinant enzymes were purified (Supplemental Figure 4) and biochemically characterized. Besides PPA-AT homologs from Arabidopsis, C. tepidum, and Synechocystis (Figure 3, Table 1; Maeda et al., 2011), enzymes from Antirrhinum majus (vascular plant), T. thermophilus (Deinococcus-Thermus bacterium), D. dadantii Ech703 (γ-proteobacterium), Streptomyces bingchenggensis (actinobacterium), and A. acidocaldarius (Firmicutes bacterium) were analyzed. PPA-AT activity was detected in the enzymes of A. majus, T. thermophilus, Synechocystis sp PCC6803, and D. dadantii (Figure 4A). The specific activities of these enzymes declined as their similarity to plant PPA-ATs decreased (Figure 4; Supplemental Table 3). The S. bingchenggensis enzyme showed minor PPA-AT activity only when high concentrations of enzyme were used, while no PPA-AT activity was detected in the A. acidocaldarius enzyme analyzed (Figure 4; Supplemental Table 3). Although the result does not necessarily infer that S. bingchenggensis and A. acidocaldarius do not possess AspAT Ib enzymes having PPA-AT activity (as noted above), it indicates that the particular S. bingchenggensis and A. acidocaldarius enzymes, which were located near the predicted boundary and thus characterized in this study (Supplemental Figure 6), lack significant PPA-AT activity. These biochemical analyses suggest that the enzymes exhibiting PPA-AT activity are present within a clade containing the D. dadantii sequence and more closely related homologs to plant PPA-ATs, whereas ones in outside groups (e.g., the A. acidocaldarius sequence) do not exhibit detectable PPA-AT activity.
Figure 4.
Aminotransferase Activity of PPA-AT Homologs from Different Organisms.
(A) Significant arogenate production was observed in PPA-AT reactions containing (1) Arabidopsis, (2) A. majus, (3) C. tepidum, (4) T. thermophilus, (5) Synechocystis sp PCC6803, or (6) D. dadantii enzymes, but not with (7) S. bingchenggensis, (8) A. acidocaldarius, or (0) no enzyme control, after incubation with 1 mM prephenate, 5 mM l-aspartate, and 200 μM PLP for 35 min at 37°C.
(B) The activity of PPA-AT, Asp-AT, PPY-AT, and HPP-AT for individual enzymes was measured after incubation with 1 mM keto acid substrate, 5 mM l-aspartate, and 200 μM PLP for 15 min at 37°C. N.D., below detection limit. Data are means ± se (n ≥ 3).
To test the substrate specificity of these PPA-AT homologs, other aromatic keto acid substrates, such as phenylpyruvate and 4-hydroxyphenylpyruvate, were used in aminotransferase assays. Similar to the C. tepidum enzyme (Figure 3, Table 1) and previously characterized plant enzymes (Graindorge et al., 2010; Maeda et al., 2011), both plant enzymes from Arabidopsis and A. majus as well as T. thermophilus, Synechocystis, and D. dadantii enzymes were unable to use phenylpyruvate and 4-hydroxyphenylpyruvate, with an exception of minor phenylpyruvate aminotransferase activity detected in the T. thermophilus enzyme (Figure 4B; Supplemental Table 3). By contrast, the S. bingchenggensis and A. acidocaldarius enzymes, with very low to no PPA-AT activity, were able to convert phenylpyruvate and 4-hydroxyphenylpyruvate into Phe and Tyr, respectively (Figure 4B; Supplemental Table 3). Thus, the characterized aminotransferases with PPA-AT activity use exclusively prephenate out of three aromatic keto acids substrates within the Phe and Tyr pathways.
Identification of Highly Conserved Amino Acid Residues Involved in the Substrate Specificity of Arabidopsis PPA-AT Enzyme
Using the defined set of aminotransferase sequences having PPA-AT activity, we then searched for amino acid residues potentially responsible for PPA-AT activity by comparing the peptide sequences of enzymes with PPA-AT activity and closely related, but prephenate non-utilizing aminotransferases (e.g., the A. acidocaldarius enzyme). The sequence alignment showed that two amino acid residues, Thr-84 and Lys-169, were conserved among all sequences within the boundary exhibiting PPA-AT activity but were absent in outgroup sequences (Figure 5; Supplemental Data Set 3), including the A. acidocaldarius enzyme, which was devoid of PPA-AT activity (Figure 4; Supplemental Table 3).
Figure 5.
Identification of Thr-84 and Lys-169 as Highly Conserved among AspAT Ib Enzymes Having PPA-AT Activity.
Primary peptide sequence alignment of prephenate utilizing and not utilizing PPA-AT homologs (above and below the dotted line) revealed Thr-84 and Lys-169 are highly conserved only among AspAT Ib enzymes exhibiting PPA-AT activity. Phylogenetic relationships of individual enzymes are shown in Supplemental Figure 6, except for PPA-AT homologs of Petunia hybrida (HM635905.1), Salinibacter ruber (YP_445071.1), and Ostreococcus lucimarinus (XP_001421566.1; Figure 2). Alignments were performed by ClustalW with default settings and shaded using the BoxShade Version 3.21 software program.
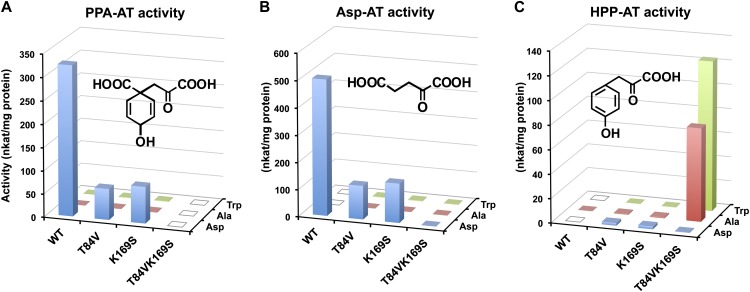
Amino acid residues corresponding to Thr-84 and Lys-169 were previously shown to be located in the substrate binding pocket playing key roles in dicarboxylic acid amino donor (i.e., aspartate and glutamate) specificity of the T. thermophilus PPA-AT homolog (Nobe et al., 1998; Nakai et al., 1999; Ura et al., 2001), which was previously reported as an AspAT Ib enzyme (Okamoto et al., 1996). However, prephenate, also a dicarboxylic acid substrate for PPA-AT activity (Figure 6), had not been tested as a keto acceptor. To examine the role of the Thr-84 and Lys-169 residues in plant PPA-ATs, mutant versions of Arabidopsis PPA-AT with substitution of Thr-84 or/and Lys-169 by valine and serine, respectively, were generated by site-directed mutagenesis (Supplemental Figure 4). The purified recombinant enzymes of both T84V and K169S single mutants still exhibited significant PPA-AT and Asp-AT activity (with aspartate amino donor) but their specific activities were greatly reduced compared with the wild type (blue bars in Figures 6A and 6B; Supplemental Table 4). The double T84VK169S mutant completely lost PPA-AT activity (Figure 6A; Supplemental Table 4), although it still maintained protein stability, shown by similar thermo-denaturation responses to the wild-type enzyme (Supplemental Figure 7). Kinetic analysis of PPA-AT activity of these mutant and wild-type enzymes revealed that both T84V and K169S single mutants had significantly reduced apparent Km value toward prephenate (Table 3). As the turnover rates (kcat) were also reduced by each mutation, the catalytic efficiencies (kcat/Km) of both single mutants were 10-fold lower than that of the wild type (Table 3).
Figure 6.
Amino Donor and Keto Acceptor Preference of Arabidopsis PPA-AT Wild Type and Mutants.
PPA-AT (A), Asp-AT (B), and HPP-AT (C) activities were analyzed in the wild type, T84V and K169S single, and T84VK169S double mutants using l-aspartate (Asp), l-alanine (Ala), or l-tryptophan (Trp) as an amino donor. The structures of respective keto acid substrates, prephenate (A), α-ketoglutarate (B), and 4-hydroxyphenylpyruvate (C), are shown in each panel. The reaction mixtures containing 3 mM keto acid substrate, 20 mM amino donor, 200 μM PLP, and 0.5 to 20 μg/mL purified recombinant enzyme were incubated at 37°C for 10 min. Data are means of three independent experiments, and individual standard errors are shown in Supplemental Table 4. Open squares: activity was not detectable (Supplemental Table 4).
Table 3. Kinetic Analysis of Arabidopsis PPA-AT Wild Type and Mutants.
| Enzymes | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Prephenate (Asp amino donor) | |||
| Wild type | 21.3 ± 1.0 | 312 ± 30 | 69.0 ± 4.2 |
| T84V | 4.5 ± 1.2** | 676 ± 96* | 6.5 ± 0.9** |
| K169S | 5.5 ± 1.5** | 791 ± 97* | 6.7 ± 1.1** |
| T84VK169S | N.D. | N.D. | N.D. |
| 4-Hydroxyphenylpyruvate (Trp amino donor) | |||
| T84VK169S | 5.0 ± 0.3 | 288 ± 15 | 17.2 ± 0.7 |
Kinetic parameters were obtained for prephenate using aspartate amino donor (20 mM) in 10-min reactions using 0.5 and 2 μg/mL recombinant enzymes for wild type and mutants of Arabidopsis PPA-AT enzymes, respectively. For the T84VK169S double mutant, PPA-AT activity was not detectable (N.D.); instead, kinetic parameters were obtained for 4-hydroxyphenylpyruvate using Trp amino donor (20 mM) in 3-min reactions using 4 μg/mL recombinant enzyme. PLP (200 μM) was used as a cofactor. Data are means ± se (n ≥ 3). Significant difference from the corresponding wild type value is indicated (*P < 0.05, **P < 0.01, Student’s t test).
The other aromatic keto acid substrate, 4-hydroxyphenylpyruvate, has a similar structure as prephenate but is not a dicarboxylic acid (Figure 6). Interestingly, when 4-hydroxyphenylpyruvate was used as a keto acceptor (still with aspartate amino donor), all three mutants including the double mutant, but not the wild type, started to show HPP-AT activity, albeit with much lower activity than their PPA-AT and Asp-AT activities (blue bars in Figure 6C). In addition to aspartate, alanine and tryptophan (nondicarboxylic and, thus, neutral amino acids) were also used to analyze PPA-AT, Asp-AT, and HPP-AT activities (red and green bars in Figure 6). PPA-AT and Asp-AT activity were very low to below detection even with alanine or tryptophan in both the wild type and all three mutants (Figures 6A and 6B; Supplemental Table 4). By contrast, strong HPP-AT activity was observed in the T84VK169S double mutant when alanine or tryptophan was used as an amino donor together with 4-hydroxyphenylpyruvate keto acceptor (Figure 6C). Thus, dicarboxylic acid substrates of both amino donor (aspartate) and keto acceptor (prephenate or α-ketoglutarate) need to be replaced with neutral ones (alanine or tryptophan, and 4-hydroxyphenylpyruvate) for the T84VK169S mutant of Arabidopsis PPA-AT to function efficiently.
Further kinetic analysis showed that the catalytic efficiency of HPP-AT activity of the T84VK169S mutant was in a similar range as that of the PPA-AT activity of the wild-type enzyme (17.2 versus 69 mM−1 s−1; Table 3). Moreover, the apparent Km of the T84VK169S mutant toward 4-hydroxyphenylpyruvate (with tryptophan) was almost identical to that of the wild type toward prephenate (with aspartate, 288 versus 312 μM; Table 3), indicating that the two mutations at Thr-84 and Lys-169 switched the substrate specificity of Arabidopsis PPA-AT from dicarboxylic acids to nondicarboxylic acids.
DISCUSSION
A broad range of organisms, with the exception of animals, possess the aromatic amino acid pathways and thus synthesize Phe, Tyr, and Trp. A prior molecular evolution study revealed that plant enzymes involved in the shikimate pathway were acquired from multiple prokaryotes including cyanobacteria and other eubacteria (Richards et al., 2006). In this study, we investigated the molecular evolution of enzymes involved in plant Phe and Tyr biosynthesis, whose pathway structures are highly diverse among different organisms having the alternative arogenate and/or phenylpyruvate/4-hydroxyphenylpyruvate pathways (Bentley, 1990; Herrmann and Weaver, 1999; Tzin and Galili, 2010; Maeda and Dudareva, 2012).
Evolutionary History of Plant Phe Biosynthesis
The phylogenetic distribution of CMs, which catalyze the initial step of all Phe and Tyr biosynthetic pathways (Figure 1), suggests that plants retained a CM enzyme derived from their nuclear, eukaryotic history (Supplemental Figure 1). By contrast, plant PPA-ATs and ADTs, but not ADHs, showed close phylogenetic affinity with homologs in the Bacteroidetes (nonphotoautotrophic) and the Chlorobi (photolithotrophic; Figure 2), which are well-documented closely aligned bacterial phyla (Gupta, 2004). A prior study suggested that plant PPA-ATs (reported as prokaryotic-type aspartate aminotransferases that belong to AspAT Ib type) are closely related to cyanobacterial enzymes and may have an endosymbiotic origin (de la Torre et al., 2006). However, in this study, cyanobacterial homologs were more distantly related to plant PPA-ATs and ADTs than were Chlorobi/Bacteroidetes homologs (Figure 2). Hypotheses of PPA-AT and ADT lateral gene transfer (LGT) associated with the endosymbiotic origins of the mitochondria or chloroplast were not supported by the AU test (Supplemental Table 1). In addition, detailed biochemical characterizations of C. tepidum (Chlorobi) and Synechocystis (cyanobacterium) PPA-AT homologs showed that the C. tepidum homolog exhibits biochemical properties more similar to plant PPA-ATs (e.g., strong PPA-AT activity; Graindorge et al., 2010; Maeda et al., 2011) than does the Synechocystis counterpart (Table 1). The weak to no PPA-AT activity detected in the Synechocystis AspAT Ib enzyme (Table 1; Graindorge et al., 2014) is consistent with the presence of BCAT-type enzymes having strong PPA-AT activity with broad substrate specificity in cyanobacteria (Stenmark et al., 1974; Jensen and Stenmark, 1975; Graindorge et al., 2014). Also, the ADT homolog of C. tepidum, previously reported as PDT (Tan et al., 2008), showed 10-fold higher catalytic efficiency for ADT than PDT activity (Table 2), whereas cyanobacteria such as Synechocystis process a bifunctional CM-PDT enzyme (Kaneko et al., 1996) and lack ADT activity (Stenmark et al., 1974; Hall et al., 1982). These phylogenetic and biochemical results together suggest that both PPA-AT and ADT, and thus the arogenate Phe pathway, were acquired by a eukaryotic ancestor of plants from a bacterial lineage that gave rise to sister phyla Chlorobi and Bacteroidetes.
A remaining question is whether the gene transfer of PPA-AT and ADT occurred directly or indirectly from an ancestor of Chlorobi/Bacteroidetes bacteria to a Plantae ancestor. The simplest hypothesis is that LGT occurred directly from Chlorobi/Bacteroidetes to Plantae ancestors. Although less likely, an alternative hypothesis is that LGT occurred from Chlorobi/Bacteroidetes to a specific lineage of cyanobacterial ancestors that is most closely related to the plastid ancestor but became extinct or has not been discovered. This possibility was previously proposed by Gross et al. (2008) when the gene cluster of menaquinone/phylloquinone biosynthetic enzymes showed close phylogenetic relationship between Plantae and Chlorobi/γ-Proteobacteria. However, the discovery of extant cyanobacteria that contain Chlorobi/γ-Proteobacteria-like menaquinone/phylloquinone enzymes and/or Chlorobi/Bacteroidetes-like PPA-AT and ADT is required to support the latter hypothesis.
Sequences homologous to both plant PPA-ATs and ADTs were found in all analyzed photosynthetic eukaryotes, including autotrophic diatoms (e.g., Thalassiosira pseudonana) whose plastids are derived from red algae (Oudot-Le Secq et al., 2007). These results suggest that the PPA-AT and ADT genes derived from Chlorobi/Bacteroidetes were acquired by a common ancestor of Plantae (including land plants and red and green algae) and have been maintained in all Plantae lineages. In the case of ADTs, the acquired ancestral gene was further duplicated multiple times within land plants (as all algae have a single ADT gene; Tohge et al., 2013), and some isoforms became very specific to arogenate over prephenate substrate (e.g., Arabidopsis ADT3, 4, and 5 [Cho et al., 2007] and petunia ADT1 [Maeda et al., 2010]). One exception is the lack of a PPA-AT gene in red algae (e.g., Cyanidioschyzon merolae; Matsuzaki et al., 2004) and glaucophytes (i.e., Cyanophora paradoxa; Price et al., 2012), which may be due to the recent loss of PPA-AT (at least AspAT Ib type) in the sequenced red alga lineages that might have co-opted another aminotransferase(s) for Phe and Tyr biosynthesis.
Phylogenetic Distribution of PPA-AT Enzymes
E. coli and yeast, two extensively studied model microbes, use exclusively the phenylpyruvate/4-hydroxyphenylpyruvate pathways for Phe/Tyr biosynthesis and do not have PPA-AT genes and activity (Gelfand and Steinberg, 1977; Urrestarazu et al., 1998). However, our analysis and the most recent study (Graindorge et al., 2014) revealed that aminotransferases having PPA-AT activity are present not only in plants but also in a broader range of microbes. Plant PPA-ATs belong to the AspAT Ib class (Graindorge et al., 2010; Dal Cin et al., 2011; Maeda et al., 2011), which falls within subfamily Iγ (Mehta et al., 1993; Jensen and Gu, 1996) and is distantly related to class-Ia AspATs (subfamily Iα) commonly present in all organisms (Alfano and Kahn, 1993; Okamoto et al., 1996; Nobe et al., 1998). In addition to plants, AspAT Ib-type enzymes having strong PPA-AT activity were found in Chlorobi bacteria (C. tepidum; Figure 3, Table 1) and α-proteobacteria (Rhizobium meliloti and Rhodobacter sphaeroides; Graindorge et al., 2014). The most extensively studied AspAT Ib enzyme from T. thermophilus (Nobe et al., 1998; Nakai et al., 1999; Ura et al., 2001) also showed a comparable level of PPA-AT activity to plant and C. tepidum enzymes (Figure 4; Supplemental Table 3). Similar to PPA-AT activity detected in plant tissues (Bonner and Jensen, 1985; Siehl et al., 1986), the purified recombinant enzymes of these AspAT Ibs specifically used aspartate and glutamate amino donors and, besides α-ketoglutarate and oxaloacetate (for Asp-AT activity), efficiently used prephenate keto acceptor, but not phenylpyruvate or 4-hydroxyphenylpyruvate (Figures 3B and 4B; Graindorge et al., 2010; Maeda et al., 2011; Graindorge et al., 2014). AspAT Ib enzymes from other bacteria, such as Synechocystis (Table 1), D. dadantii (Figure 4), and Nitrosomonas europaea (β-proteobacteria; Graindorge et al., 2014), also showed PPA-AT activity but at much lower rates, which appears to be due to the presence of additional enzyme(s) efficiently catalyzing the PPA-AT reaction (e.g., BCAT and S-DAPAT types in Synechocystis and N. europaea, respectively; Graindorge et al., 2014) and/or the operation of an alternative pathway(s) for Phe and/or Tyr biosynthesis. Although further genetic analyses are required to determine relative contributions of alternative pathways and/or different types of PPA-AT enzymes to overall Phe and Tyr biosynthesis, these results suggest that the arogenate pathway is distributed in Plantae as well as in a wide range of prokaryotes.
Role of Thr-84 and Lys-169 in the Substrate Specificity of Plant PPA-AT Enzyme
Aminotransferases catalyze a PLP-dependent transamination reaction between an α-amino acid donor and an α-keto acid amino acceptor via the “ping-pong bi-bi” mechanism (Kirsch et al., 1984; Toney, 2014). The AspAT Ib enzymes were previously shown to preferentially use dicarboxylic acid amino donors (i.e., aspartate and glutamate), in contrast to class Ia AspATs that use both acidic and neutral amino donors (Kuramitsu et al., 1990; Nakai et al., 1999). Structural analyses of AspAT Ib enzymes from T. thermophilus and plants identified several amino acid residues responsible for their Asp-AT activity (Nobe et al., 1998; Nakai et al., 1999; Ura et al., 2001; de la Torre et al., 2009). Lys-109 of the T. thermophilus enzyme, which corresponds to Lys-169 of Arabidopsis PPA-AT (Figure 5), was shown to be critical for specific recognition of dicarboxylic acids by forming salt bridges with their distal carboxyl group (Nobe et al., 1998; Nakai et al., 1999). However, prephenate was not used as a keto acid substrate, as the involvement of AspAT Ib enzymes in catalysis of PPA-AT reaction was shown only recently (Graindorge et al., 2010; Maeda et al., 2011; Graindorge et al., 2014).
In this study, biochemical characterizations of various homologs of plant PPA-ATs guided by phylogenetic analysis defined a subset of AspAT Ib enzymes having PPA-AT activity, which further allowed us to identify Thr-84 and Lys-169 being potentially important residues for their PPA-AT activity. In the T. thermophilus AspAT Ib enzyme, the single K109S mutation almost completely abolished Asp-AT activity (<104-fold lower activity than the wild type), indicating that Lys-109 is a major determinant of the dicarboxylic acid substrate specificity of the T. thermophilus AspAT Ib enzyme (Nobe et al., 1998). Although the Thr-17 residues in the T. thermophilus enzyme, which corresponds to Thr-84 of Arabidopsis PPA-AT, also interacts with the distal carboxyl group of a dicarboxylic substrate (Nakai et al., 1999), addition of T17V together with other mutations in the T. thermophilus K109S mutant did not further reduce the acidic substrate specificity (Ura et al., 2001). On the contrary, the K169S mutant of Arabidopsis PPA-AT still maintained significant Asp-AT and PPA-AT activity and lost these activities completely only when it was combined with the T84V mutation (Figures 6A and 6B, Table 3). These results suggest that, in Arabidopsis PPA-AT, Lys-169 and Thr-84 have additive roles in recognition of dicarboxylic substrates (e.g., prephenate, α-ketoglutarate, and aspartate) for both PPA-AT and Asp-AT activity.
Along with the loss of acidic amino donor specificity, the introduction of K109S mutation in the T. thermophilus AspAT Ib enzyme alone or together with T17V and other mutations increased Asp-AT activity with neutral and hydrophobic amino donors (Nobe et al., 1998; Ura et al., 2001). However, in Arabidopsis PPA-AT, the T84V and K169S mutations, even in combination, did not substantially increase Asp-AT and PPA-AT activity with alanine or tryptophan (Figures 6A and 6B). Only when neutral substrates were used for both keto acceptor (4-hydroxyphenylpyruvate) together with amino donor (alanine or tryptophan), the T84VK169S double mutants of Arabidopsis PPA-AT showed strong aminotransferase activity (HPP-AT activity; Figure 6C, Table 3), demonstrating that the simultaneous mutation of Thr-84 and Lys-169 can shift the enzyme specificity from dicarboxylic acid to neutral nondicarboxylic acid substrates. Thus, our results indicate that both Thr-84 and Lys-169 residues of plant PPA-AT play crucial roles not only in acidic amino donor specificity but also in specific recognition of dicarboxylic keto acid substrates, including prephenate. This study also demonstrates how the phylobiochemical characterization of enzymes from deep taxonomic lineages can be used to determine key molecular changes that alter enzyme substrate specificity.
Conclusions
In summary, based on the current results combined with a previous hypothesis for the evolutionary development of Phe and Tyr biosynthesis (Jensen and Pierson, 1975), we propose a model of the evolution of the arogenate pathway for plant Phe biosynthesis: A PPA-AT enzyme evolved from an AspAT Ib-type aminotransferase, possibly having activities with neutral substrates (e.g., HPP-AT activity observed in the A. acidocaldarius enzyme [Figure 4B] and Arabidopsis PPA-AT double mutant [Figure 6]), through acquisition of Thr-84 and Lys-169 and most likely additional permissive mutations. This led to the cellular accumulation of arogenate and the alteration of PDT/ADT substrate preference from prephenate to arogenate. The eukaryotic host of the plant ancestor, or the free-living cyanobacterium that gave rise to the primary plastid, acquired both PPA-AT and ADT from an ancestor of Chlorobi/Bacteroidetes bacteria. The introduction of the PPA-AT and ADT enzymes allowed plants to synthesize Phe predominantly via the arogenate pathway (Jung et al., 1986; Maeda et al., 2010; Yoo et al., 2013), which currently supports the production of diverse and abundant phenylpropanoid natural products, including lignin (Corea et al., 2012b).
METHODS
Analysis of Molecular Evolution
For phylogenetic analyses shown in Figure 2 (Supplemental Data Set 4), homologous PPA-AT- and ADT-like sequences were identified by a protein BLASTP search of Arabidopsis thaliana PPA-AT (NP_565529.1) and ADT1 (NP_172644.1), respectively, on the NCBI refseq database with the BLOSUM45 matrix. Approximately 200 to 250 most similar sequences (e-value cutoff of 1e−40) were selected and aligned using MAFFT E-INS-i (Katoh et al., 2002). A majority rule consensus 100 bootstrap neighbor-joining phylogeny was initially generated using PAUP* 4.0b10 (http://paup.csit.fsu.edu), and redundant sequences forming a same-species polytomy at branch tips were pruned. The new data set was then aligned using MAFFT FFT-NS-i (Katoh et al., 2002), and gaps with <2% of total sequences were trimmed. A ProtTest 3.2.2 analysis (Darriba et al., 2011) identified LG+I+G to be the optimal model. ML 1024 replicate nonparametric bootstrap and 1024 replicate ML search GARLI 2.0 analyses were performed (http://garli.googlecode.com). A majority rule consensus tree was generated using the SumTrees 3.3.1 program of the DendroPy 3.12.0 package (Sukumaran and Holder, 2010). The best likelihood tree was selected and bootstrap support values were mapped on nodes using SumTrees.
Phylogenetic analysis of CM and ADH (Supplemental Figure 1 and Supplemental Data Set 4) was conducted as described above for PPA-AT and ADT (Figure 2) using Arabidopsis CM1 (NP_566846.1) and ADH1 (NP_198343.1) as queries in the BLASTP similarity searches (e-value cut off of 1e−20).
A deep phylogenetic analysis of PPA-AT (Supplemental Figure 9 and Supplemental Data Set 4) was performed on more distantly related PPA-AT-like sequences as determined as follows. Shared hits in reciprocal similarity searches can indicate common ancestry between two query sequences (Pearson, 1996). Therefore, four amino acid sequences representing various PPA-AT-like activities (Arabidopsis PPA-AT [NP_565529.1/AT2G22250], Arabidopsis AGD2/DAT [AT4G33680], Arabidopsis CORI3/CL [AT4G23600], and Homo sapiens kyurenine-oxoglutarate transaminase [CAA57702]) were selected for Smith-Waterman similarity searches (Smith and Waterman, 1981) using SSEARCH v35 (Pearson and Lipman, 1988). Searches were performed on the NCBI nr database with an e-value cutoff of 1e−6. A total of 3913 sequences were in common between the four searches, thereby establishing deep common ancestry. To reduce the data set to a manageable size for ML analyses, these were pruned down to 301 sequences by sampling every 13th sequence. The resulting data set was then edited so that the four query sequences and characterized sequences (e.g., Chlorobium tepidum and Thermus thermophilus) were included. This data set was aligned using MAFFT FFT-NS-i, and gaps were trimmed. ProtTest 3.2.2 analysis identified LG+I+G as the best-fitting model.
AU Test of PPA-AT and ADT
For PPA-AT and ADT hypothesis testing (Supplemental Table 1 and Supplemental Figure 3), two additional constrained 1024 replicate ML search GARLI analyses were also performed constraining plants with either α-proteobacteria or cyanobacteria. The program CONSEL (Shimodaira and Hasegawa, 2001) was used to perform the AU test (Shimodaira, 2002) on comparisons between the best likelihood ML tree for each hypothesis. Given the lower expectation value cutoff criterion used in the PPA-AT SSEARCHs, there were several different orthologous aminotransferase groups, sharing common ancient ancestry, present in the PPA-AT analyses. Therefore, when assigning plant PPA-ATs constrained with either cyanobacterial or α-proteobacterial orthologs, only homologs from the previously stated eubacterial phyla present in PPA-AT proximal basal clades (i.e., presumably matched-orthologous eubacterial enzymes) were selected for the constraint ML analyses.
Functional Protein Association Analysis
Functionality of PPA-AT homologs were predicted using STRING (http://string-db.org) (Franceschini et al., 2013). Amino acid sequences of individual PPA-AT homologs (Supplemental Data Set 2) were used as queries to identify the top three proteins that are predicted to be functionally associated with them based on, e.g., genome colocalization, coexpression, and physical protein-protein interactions (Supplemental Table 2; Franceschini et al., 2013). Only PPA-AT homologs that were deposited in the STRING database were used.
Recombinant Enzyme Expression and Purification
PPA-AT and ADT homologs were first PCR amplified using primers described in Supplemental Table 5 and the following templates: cDNA for Antirrhinum majus homolog (KF726139); genomic DNAs for C. tepidum PPA-AT (NP_661859.1) and ADT (NP_662549.1) homologs, Dickeya dadantii Ech703 (YP_002987080.1), and Alicyclobacillus acidocaldarius (ZP_03494134.1) homologs; cells for Synechocystis sp PCC6803 homolog (NP_442191.1, by colony PCR); plasmids containing the target genes for T. thermophilus (YP_143312.1) and Streptomyces bingchenggensis (ADI11720.1) homologs were obtained from RIKEN BRC Japan Collection of Microorganisms and GENEWIZ, respectively. For A. majus, C. tepidum, and T. thermophilus, Platinum Pfx DNA polymerase (Invitrogen) was used for PCR amplification. For A. majus, the DNA fragment encoding the mature protein of PPA-AT (lacking the first 66 amino acids predicted to be a plastid transit peptide by ChloroP) was subcloned into the pET28a vector at NdeI and BamHI sites. The PCR fragments of the C. tepidum and T. thermophilus genes were first cloned into the pENTR/DTOPO vector and then transferred to pDEST17 using LR clonase (Invitrogen). For the remaining PPA-AT homologs (i.e., Synechocystis sp PCC6803, D. dadantii Ech703, S. bingchenggensis, and Alicyclobacillus acidocaldarius), Phusion DNA Polymerase (Fermentas) was used to amplify DNA fragments, which were inserted into pET28a (Novagen) linearized by NdeI and EcoRI enzymes using the InFusion Cloning protocol (Clontech). Restriction digestion and sequencing confirmed that no errors were introduced during PCR amplification and cloning.
For recombinant protein expression, the cloned plasmids were first transformed into Rosetta-2 Escherichia coli. The E. coli cultures grown overnight at 18°C after isopropyl β-d-1-thiogalactopyranoside (400 mM) induction were harvested, resuspended in the lysis buffer (50 mM sodium phosphate, pH 8.0, 0.1 mM PLP, 1 mM DTT, 1 mM EDTA, 1 mM PMSF, and 10% glycerol), and sonicated. The supernatant of the cell lysate was then desalted by G-50-80 gel filtration resin (Sigma-Aldrich) and applied to a column containing Ni-NTN resin (5 PRIME) equilibrated with the binding/wash buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 10 mM imidazole). After washing the column with 15 mL binding/wash buffer, the recombinant proteins were eluted with the elution buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 500 mM imidazole), desalted, and eluted with the reaction buffer (50 mM sodium phosphate, pH 8.0, 0.1 mM PLP, and 10% glycerol). For the purification of C. tepidum ADT homolog, an additional wash was performed with 5 mL 40 mM imidazole buffer (in 50 mM sodium phosphate, pH 8.0, and 300 mM NaCl), and the purified enzyme was desalted into 50 mM sodium phosphate (pH 8.0), 1 mM EDTA, and 10% glycerol. The expression and the purity of recombinant enzymes were confirmed by SDS-PAGE stained with Coomassie Brilliant Blue R 250 (Supplemental Figure 4).
Enzyme Assays
The aminotransferase reactions were conducted in mixtures containing 50 mM sodium phosphate (pH 8.0) and 200 μM PLP together with an amino donor and a keto acid acceptor (Maeda et al., 2011). The ADT reactions were performed in 20 mM Tris-HCl (pH 8.0) and 1 mM EDTA together with arogenate (Maeda et al., 2010). The reactions were initiated by adding the enzymes. After incubation at 37°C for the time periods indicated in each figure or table legend, the reactions were stopped by adding methanol (v/v 66%), with the exception for PPA-AT assays using glutamate donor, which was stopped by 0.5 n HCl, further incubated at 37°C to convert arogenate into Phe, and neutralized with 0.5 n NaOH. The reaction products, arogenate, aspartate, glutamate, Tyr, and Phe, were quantified by HPLC (Agilent 1260 equipped with an Eclipse Plus XDB-C18 column) as o-phthaldialdehyde derivatives (Maeda et al., 2010). Arogenate, aspartate, and glutamate were separated at a flow rate of 0.5 mL/min with a 10-min linear gradient of 10 to 30% methanol in 0.1% (v/v) ammonium acetate (pH 6.8), whereas 0.8 mL/min with a 30-min linear gradient of 20 to 45% methanol was used for Tyr. Phe was separated at 0.5 mL/min with a 30-min linear gradient of 10 to 70% methanol in 20 mM sodium phosphate buffer (pH 6.8). The PDT and CM activities were measured as described previously (Maeda et al., 2010) by spectrophotometrically analyzing phenylpyruvate production in alkaline conditions (Cotton and Gibson, 1965). The quantifications were based on standard curves generated by respective authentic standards. Kinetic analyses were conducted by hyperbolic regression analysis using HYPER.EXE (version 1.01). All enzyme assays, with the exceptions of qualitative analyses presented in Figures 3 and 4 that aimed to detect any detectable enzyme activity, were performed at an appropriate enzyme concentration and reaction time, so that reaction velocity was proportional to enzyme concentration and linear during the incubation time period.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis PPA-AT (At2g22250/NP_565529.1), PPA-AT homologs of A. majus (KF726139), C. tepidum (NP_661859.1), Synechocystis sp PCC6803 (NP_442191.1), T. thermophilus (YP_143312.1), D. dadantii Ech703 (YP_002987080.1), S. bingchenggensis (ADI11720.1), A. acidocaldarius (ZP_03494134.1), and C. tepidum ADT homolog (NP_662549.1). Accession numbers of sequences that were used only in phylogenetic analyses are listed in Supplemental Data Set 2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ML Phylogenetic Nonparametric Bootstrap Consensus Analyses of CM and ADH.
Supplemental Figure 2. PPA-AT Optimal ML Search Trees Showing Estimated Branch Lengths.
Supplemental Figure 3. PPA-AT and ADT Optimal ML Search Trees Used for the Approximately Unbiased Test Shown in Supplemental Table 1.
Supplemental Figure 4. SDS-PAGE of Purified Recombinant Enzymes.
Supplemental Figure 5. Time-Dependent ADT and PDT Activity of C. tepidum ADT Homolog.
Supplemental Figure 6. Prediction of the Transition between Functional and Nonfunctional PPA-ATs Using a Deep ML Phylogenetic Tree.
Supplemental Figure 7. Representative Thermo-Denaturation Responses of Wild Type and Mutants of Arabidopsis PPA-AT (AtPPA-AT) Enzymes.
Supplemental Table 1. Approximately Unbiased Test of ADT and PPA-AT Phylogenies.
Supplemental Table 2. Functional Protein Association Prediction of PPA-AT Homologs.
Supplemental Table 3. Aminotransferase Activities of PPA-AT Homologs with Different Keto Acid Acceptors.
Supplemental Table 4. PPA-AT, Asp-AT, and HPP-AT Activities of Arabidopsis PPA-AT Wild Type and Mutants with Different Amino Donors.
Supplemental Table 5. Primer Sequences Used for Cloning of PPA-AT Homologs.
Supplemental Data Set 1. Coexpression Analysis of Plant Homologs of AspAT Ib, BCAT, and S-DAPAT Type PPA-ATs.
Supplemental Data Set 2. Taxon Mapping of PPA-AT, ADT, CM, and ADH Phylogenetic Trees.
Supplemental Data Set 3. Multiple Sequence Alignment of Functional and Nonfunctional PPA-ATs.
Supplemental Data Set 4. Sequence Alignments Corresponding to All Phylogenetic Analyses Presented in the Study.
Supplementary Material
Acknowledgments
We thank Kajetan Vogl and Donald Bryant (Pennsylvania State University) for providing C. tepidum DNA, Radin Sadre and Dean DellaPenna (Michigan State University) for Synechocystis sp PCC6803 cells, RIKEN BRC JCM for T. thermophilus HB8 DNA, Venkatesh Balakrishnan and Nicole Perna (University of Wisconsin-Madison) for D. dadantii strain Ech703 DNA, and Colleen Drinkwater and David Mead (Lucigen) for A. acidocaldarius LAA1 DNA. The ML analyses were performed on the Virginia Tech Advanced Research Computing clusters. We also thank Jiameng Zheng for generating the A. majus PPA-AT construct, Conor Meehan for helpful phylogenetic discussions, and David Baum, Eran Pichersky, Ayumi Minoda, and members of the Maeda laboratory for critical reading of the article. This work was supported by start-up funds from the Graduate School, the College of Letters and Science, and the Department of Botany, University of Wisconsin-Madison to H.A.M. and by the Agricultural and Food Research Initiative Competitive Grant 2010-65115-20385 from the USDA National Institute of Food and Agriculture and Grant MCB-0919987 from the National Science Foundation to N.D.
AUTHOR CONTRIBUTIONS
H.A.M. and J.G.J. designed the research. A.J.W., R.K.C., and J.G.J. performed phylogenetic analyses. C.D. and H.A.M. performed the rest of the experiments. C.D., A.J.W., J.G.J., N.D., and H.A.M. analyzed data. H.A.M. wrote the article with contributions from all other authors.
Glossary
- PDT
prephenate dehydratase
- CM
chorismate mutase
- PDH
prephenate dehydrogenase
- PPA-AT
prephenate aminotransferase
- ADT
arogenate dehydratase
- ADH
arogenate dehydrogenase
- AspAT Ib
class Ib aspartate aminotransferase
- BCAT
branched-chain aminotransferase
- S-DAPAT
N-succinyl-diaminopimelate aminotransferases
- ML
maximum likelihood
- AU
approximately unbiased
- PLP
pyridoxal 5′-phosphate
- LGT
lateral gene transfer
Footnotes
Online version contains Web-only data.
References
- Abou-Zeid A., Euverink G., Hessels G.I., Jensen R.A., Dijkhuizen L. (1995). Biosynthesis of l-phenylalanine and l-tyrosine in the actinomycete Amycolatopsis methanolica. Appl. Environ. Microbiol. 61: 1298–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano J.R., Kahn M.L. (1993). Isolation and characterization of a gene coding for a novel aspartate aminotransferase from Rhizobium meliloti. J. Bacteriol. 175: 4186–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R. (1990). The shikimate pathway—a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25: 307–384. [DOI] [PubMed] [Google Scholar]
- Bonner C.A., Jensen R.A. (1985). Novel features of prephenate aminotransferase from cell cultures of Nicotiana silvestris. Arch. Biochem. Biophys. 238: 237–246. [DOI] [PubMed] [Google Scholar]
- Cho M.H., et al. (2007). Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. J. Biol. Chem. 282: 30827–30835. [DOI] [PubMed] [Google Scholar]
- Connelly J.A., Conn E.E. (1986). Tyrosine biosynthesis in Sorghum bicolor: isolation and regulatory properties of arogenate dehydrogenase. Z. Naturforsch., C, J. Biosci. 41: 69–78. [DOI] [PubMed] [Google Scholar]
- Corea O.R., Bedgar D.L., Davin L.B., Lewis N.G. (2012a). The arogenate dehydratase gene family: towards understanding differential regulation of carbon flux through phenylalanine into primary versus secondary metabolic pathways. Phytochemistry 82: 22–37. [DOI] [PubMed] [Google Scholar]
- Corea O.R., Ki C., Cardenas C.L., Kim S.J., Brewer S.E., Patten A.M., Davin L.B., Lewis N.G. (2012b). Arogenate dehydratase isoenzymes profoundly and differentially modulate carbon flux into lignins. J. Biol. Chem. 287: 11446–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R.G.H., Gibson F. (1965). Biosynthesis of phenylalanine and tyrosine - Enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochim. Biophys. Acta 100: 76–88. [DOI] [PubMed] [Google Scholar]
- Dal Cin V., et al. (2011). Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23: 2738–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B.E., Blackburn E.H., Dopheide T.A. (1972). Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. I. Purification, molecular weight, and amino acid composition. J. Biol. Chem. 247: 4441–4446. [PubMed] [Google Scholar]
- de la Torre F., El-Azaz J., Avila C., Cánovas F.M. (2014). Deciphering the role of aspartate and prephenate aminotransferase activities in plastid nitrogen metabolism. Plant Physiol. 164: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F., Moya-García A.A., Suárez M.-F., Rodríguez-Caso C., Cañas R.A., Sánchez-Jiménez F., Cánovas F.M. (2009). Molecular modeling and site-directed mutagenesis reveal essential residues for catalysis in a prokaryote-type aspartate aminotransferase. Plant Physiol. 149: 1648–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F., De Santis L., Suárez M.F., Crespillo R., Cánovas F.M. (2006). Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: implications for plant amino acid metabolism. Plant J. 46: 414–425. [DOI] [PubMed] [Google Scholar]
- Fazel A.M., Bowen J.R., Jensen R.A. (1980). Arogenate (pretyrosine) is an obligatory intermediate of L-tyrosine biosynthesis: confirmation in a microbial mutant. Proc. Natl. Acad. Sci. USA 77: 1270–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazel A.M., Jensen R.A. (1979). Aromatic aminotransferases in coryneform bacteria. J. Bacteriol. 140: 580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R., Jensen R. (1987). Arogenate dehydratase. Methods Enzymol. 142: 495–502. [DOI] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L.J. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41: D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand D.H., Steinberg R.A. (1977). Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J. Bacteriol. 130: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda I., Bar E., Portnoy V., Lev S., Burger J., Schaffer A.A., Tadmor Y., Gepstein S., Giovannoni J.J., Katzir N., Lewinsohn E. (2010). Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. J. Exp. Bot. 61: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge M., Giustini C., Jacomin A.C., Kraut A., Curien G., Matringe M. (2010). Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: the last homeless enzyme of aromatic amino acids biosynthesis. FEBS Lett. 584: 4357–4360. [DOI] [PubMed] [Google Scholar]
- Graindorge M., Giustini C., Kraut A., Moyet L., Curien G., Matringe M. (2014). Three different classes of aminotransferases evolved prephenate aminotransferase functionality in arogenate-competent microorganisms. J. Biol. Chem. 289: 3198–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J., Meurer J., Bhattacharya D. (2008). Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol. Biol. 8: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.S. (2004). The phylogeny and signature sequences characteristics of Fibrobacteres, Chlorobi, and Bacteroidetes. Crit. Rev. Microbiol. 30: 123–143. [DOI] [PubMed] [Google Scholar]
- Hall G.C., Flick M.B., Gherna R.L., Jensen R.A. (1982). Biochemical diversity for biosynthesis of aromatic amino acids among the cyanobacteria. J. Bacteriol. 149: 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K.M., Weaver L.M. (1999). The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 473–503. [DOI] [PubMed] [Google Scholar]
- Huang T., Tohge T., Lytovchenko A., Fernie A.R., Jander G. (2010). Pleiotropic physiological consequences of feedback-insensitive phenylalanine biosynthesis in Arabidopsis thaliana. Plant J. 63: 823–835. [DOI] [PubMed] [Google Scholar]
- Jensen R.A. (1985). Biochemical pathways in prokaryotes can be traced backward through evolutionary time. Mol. Biol. Evol. 2: 92–108. [DOI] [PubMed] [Google Scholar]
- Jensen R.A., Gu W. (1996). Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J. Bacteriol. 178: 2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.A., Pierson D.L. (1975). Evolutionary implications of different types of microbial enzymology for L-tyrosine biosynthesis. Nature 254: 667–671. [DOI] [PubMed] [Google Scholar]
- Jensen R.A., Stenmark S.L. (1975). The ancient origin of a second microbial pathway for L-tyrosine biosynthesis in prokaryotes. J. Mol. Evol. 4: 249–259. [Google Scholar]
- Jung E., Zamir L.O., Jensen R.A. (1986). Chloroplasts of higher plants synthesize L-phenylalanine via L-arogenate. Proc. Natl. Acad. Sci. USA 83: 7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3: 109–136. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch J.F., Eichele G., Ford G.C., Vincent M.G., Jansonius J.N., Gehring H., Christen P. (1984). Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J. Mol. Biol. 174: 497–525. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Hiromi K., Hayashi H., Morino Y., Kagamiyama H. (1990). Pre-steady-state kinetics of Escherichia coli aspartate aminotransferase catalyzed reactions and thermodynamic aspects of its substrate specificity. Biochemistry 29: 5469–5476. [DOI] [PubMed] [Google Scholar]
- Lee E.J., Facchini P.J. (2011). Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol. 157: 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand P., Dumas R., Seux M., Rippert P., Ravelli R., Ferrer J.L., Matringe M. (2006). Biochemical characterization and crystal structure of Synechocystis arogenate dehydrogenase provide insights into catalytic reaction. Structure 14: 767–776. [DOI] [PubMed] [Google Scholar]
- Maeda H., Dudareva N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63: 73–105. [DOI] [PubMed] [Google Scholar]
- Maeda H., Shasany A.K., Schnepp J., Orlova I., Taguchi G., Cooper B.R., Rhodes D., Pichersky E., Dudareva N. (2010). RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 22: 832–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Yoo H., Dudareva N. (2011). Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat. Chem. Biol. 7: 19–21. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657. [DOI] [PubMed] [Google Scholar]
- Mehta P.K., Hale T.I., Christen P. (1993). Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214: 549–561. [DOI] [PubMed] [Google Scholar]
- Nakai T., Okada K., Akutsu S., Miyahara I., Kawaguchi S., Kato R., Kuramitsu S., Hirotsu K. (1999). Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate. Biochemistry 38: 2413–2424. [DOI] [PubMed] [Google Scholar]
- Nobe Y., Kawaguchi S., Ura H., Nakai T., Hirotsu K., Kato R., Kuramitsu S. (1998). The novel substrate recognition mechanism utilized by aspartate aminotransferase of the extreme thermophile Thermus thermophilus HB8. J. Biol. Chem. 273: 29554–29564. [DOI] [PubMed] [Google Scholar]
- Okamoto A., Kato R., Masui R., Yamagishi A., Oshima T., Kuramitsu S. (1996). An aspartate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. 119: 135–144. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M.P., Grimwood J., Shapiro H., Armbrust E.V., Bowler C., Green B.R. (2007). Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277: 427–439. [DOI] [PubMed] [Google Scholar]
- Pearson W.R. (1996). Effective protein sequence comparison. Methods Enzymol. 266: 227–258. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. (1988). Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85: 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.C., et al. (2012). Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335: 843–847. [DOI] [PubMed] [Google Scholar]
- Razal R.A., Ellis S., Singh S., Lewis N.G., Towers G.H.N. (1996). Nitrogen recycling in phenylpropanoid metabolism. Phytochemistry 41: 31–35. [Google Scholar]
- Reichau S., Jiao W., Walker S.R., Hutton R.D., Baker E.N., Parker E.J. (2011). Potent inhibitors of a shikimate pathway enzyme from Mycobacterium tuberculosis: combining mechanism- and modeling-based design. J. Biol. Chem. 286: 16197–16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T.A., Dacks J.B., Campbell S.A., Blanchard J.L., Foster P.G., McLeod R., Roberts C.W. (2006). Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5: 1517–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D., Koohi M., Lisec J., Pfeiffer M., Lippmann R., Schmeichel J., Willmitzer L., Altmann T. (2012). A tyrosine aminotransferase involved in tocopherol synthesis in Arabidopsis. Plant J. 71: 850–859. [DOI] [PubMed] [Google Scholar]
- Rippert P., Matringe M. (2002). Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly similar and active protein domains. Plant Mol. Biol. 48: 361–368. [DOI] [PubMed] [Google Scholar]
- Schönbrunn E., Eschenburg S., Shuttleworth W.A., Schloss J.V., Amrhein N., Evans J.N., Kabsch W. (2001). Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 98: 1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51: 492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247. [DOI] [PubMed] [Google Scholar]
- Siehl D.L., Connelly J.A., Conn E.E. (1986). Tyrosine biosynthesis in Sorghum bicolor: characteristics of prephenate aminotransferase. Z. Naturforsch., C, J. Biosci. 41: 79–86. [DOI] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S. (1981). Identification of common molecular subsequences. J. Mol. Biol. 147: 195–197. [DOI] [PubMed] [Google Scholar]
- Song J., Bonner C.A., Wolinsky M., Jensen R.A. (2005). The TyrA family of aromatic-pathway dehydrogenases in phylogenetic context. BMC Biol. 3: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark S.L., Pierson D.L., Jensen R.A., Glover G.I. (1974). Blue-green bacteria synthesise L-tyrosine by the pretyrosine pathway. Nature 247: 290–292. [DOI] [PubMed] [Google Scholar]
- Sukumaran J., Holder M.T. (2010). DendroPy: a Python library for phylogenetic computing. Bioinformatics 26: 1569–1571. [DOI] [PubMed] [Google Scholar]
- Tan K., Li H., Zhang R., Gu M., Clancy S.T., Joachimiak A. (2008). Structures of open (R) and close (T) states of prephenate dehydratase (PDT)—implication of allosteric regulation by L-phenylalanine. J. Struct. Biol. 162: 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Watanabe M., Hoefgen R., Fernie A.R. (2013). Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toney M.D. (2014). Aspartate aminotransferase: an old dog teaches new tricks. Arch. Biochem. Biophys. 544: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V., Galili G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3: 956–972. [DOI] [PubMed] [Google Scholar]
- Ura H., Nakai T., Kawaguchi S.I., Miyahara I., Hirotsu K., Kuramitsu S. (2001). Substrate recognition mechanism of thermophilic dual-substrate enzyme. J. Biochem. 130: 89–98. [DOI] [PubMed] [Google Scholar]
- Urrestarazu A., Vissers S., Iraqui I., Grenson M. (1998). Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol. Gen. Genet. 257: 230–237. [DOI] [PubMed] [Google Scholar]
- Weiss, U. (1986). Early research on the shikimate pathway: Some personal remarks and reminiscences. In The Shikimic Acid Pathway: Recent Advances in Phytochemistry, Vol. 20, E.E. Conn, ed (New York: Plenum Press), pp. 1–12. [Google Scholar]
- Yamada T., Matsuda F., Kasai K., Fukuoka S., Kitamura K., Tozawa Y., Miyagawa H., Wakasa K. (2008). Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell 20: 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H., Widhalm J.R., Qian Y., Maeda H., Cooper B.R., Jannasch A.S., Gonda I., Lewinsohn E., Rhodes D., Dudareva N. (2013). An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat. Commun. 4: 2833. [DOI] [PubMed] [Google Scholar]
- Zhang S., Pohnert G., Kongsaeree P., Wilson D.B., Clardy J., Ganem B. (1998). Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273: 6248–6253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.