This work reports the distinct mechanisms of two homologous NAC proteins of tomato, JA2 and JA2L, in regulating Pseudomonas syringae pv tomato DC3000–induced stomatal movement. Whereas JA2 acts in abscisic acid (ABA)–mediated stomatal closure by promoting ABA biosynthesis, JA2L functions in jasmonate/coronatine–mediated stomatal reopening by suppressing salicylic acid accumulation.
Abstract
To restrict pathogen entry, plants close stomata as an integral part of innate immunity. To counteract this defense, Pseudomonas syringae pv tomato produces coronatine (COR), which mimics jasmonic acid (JA), to reopen stomata for bacterial entry. It is believed that abscisic acid (ABA) plays a central role in regulating bacteria-triggered stomatal closure and that stomatal reopening requires the JA/COR pathway, but the downstream signaling events remain unclear. We studied the stomatal immunity of tomato (Solanum lycopersicum) and report here the distinct roles of two homologous NAC (for NAM, ATAF1,2, and CUC2) transcription factors, JA2 (for jasmonic acid2) and JA2L (for JA2-like), in regulating pathogen-triggered stomatal movement. ABA activates JA2 expression, and genetic manipulation of JA2 revealed its positive role in ABA-mediated stomatal closure. We show that JA2 exerts this effect by regulating the expression of an ABA biosynthetic gene. By contrast, JA and COR activate JA2L expression, and genetic manipulation of JA2L revealed its positive role in JA/COR-mediated stomatal reopening. We show that JA2L executes this effect by regulating the expression of genes involved in the metabolism of salicylic acid. Thus, these closely related NAC proteins differentially regulate pathogen-induced stomatal closure and reopening through distinct mechanisms.
INTRODUCTION
Plants have coexisted with microbes, including bacterial pathogens, for millions of years, and the interactions between these organisms have resulted in their coevolution. Pathogens have evolved multiple strategies to facilitate their virulence in their hosts, and plants have developed diverse and sophisticated systems of innate immunity to resist pathogen attack, including pathogen-associated molecular pattern (PAMP)–triggered immunity and effector-triggered immunity (Chisholm et al., 2006; Bent and Mackey, 2007; Faulkner and Robatzek, 2012; Xin and He, 2013).
Pathogenic microbes need to reach the interior of the plant to cause disease, and natural openings or accidental wounds provide portals for pathogen invasion (Melotto et al., 2008; Faulkner and Robatzek, 2012). Stomata are microscopic pores formed by pairs of guard cells in the epidermis of plants. Plants actively regulate stomatal aperture in response to abiotic environmental conditions (e.g., light, humidity, and CO2 concentration) to optimize gas exchange and water loss (Kim et al., 2010). Research on the molecular mechanisms underlying stomatal regulation in response to abiotic signals has uncovered a complex and dynamic regulatory network in which the plant hormone abscisic acid (ABA) plays a central role (Schroeder et al., 2001a, 2001b; Kim et al., 2010).
Historically, stomata were merely assumed to be passive ports for pathogen entry. However, recent studies revealed that stomatal openings are a major route of pathogen entry into the plant. Accordingly, plants have evolved mechanisms to actively regulate stomatal aperture as an integral part of innate immunity to prevent pathogen invasion (Melotto et al., 2006, 2008; Zhang et al., 2008; Liu et al., 2009; Zeng and He, 2010; Zeng et al., 2011; Desclos-Theveniau et al., 2012; Kumar et al., 2012; Singh et al., 2012; Montillet and Hirt, 2013; Montillet et al., 2013). The role of stomata in plant immunity was discovered during the susceptible interaction between the host Arabidopsis thaliana and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000, a model interaction system in which to investigate molecular mechanisms underlying pathogen virulence, host immunity, and host–pathogen coevolution (Chisholm et al., 2006; Bent and Mackey, 2007; Spoel and Dong, 2008; Dou and Zhou, 2012; Faulkner and Robatzek, 2012; Fu and Dong, 2013; Xin and He, 2013). In their pioneering investigation, Melotto et al. (2006) described two successive steps of stomatal movement at an early stage of P. s. tomato DC3000 infection of Arabidopsis leaves. First, upon the perception of pathogen infection and PAMP treatment, plants close stomata within 1 h to inhibit the entry of pathogen and host tissue colonization. Pathogen-induced stomatal closure requires the plant immune receptors (e.g., FLAGELLIN-SENSING2, which recognizes flg22, a biologically active peptide derived from bacterial flagellin) and the plant immune hormone salicylic acid (SA); thus, this response is referred to as the stomatal defense, an integral part of plant innate immunity (Melotto et al., 2006; Zeng and He, 2010; Zeng et al., 2011). Second, to counteract this stomatal defense, P. s. tomato DC3000 is able to reopen stomata after 3 to 4 h, to facilitate its entry into the plant leaf and therefore cause other aspects of virulence in the apoplast (Melotto et al., 2006).
The discovery of stomatal defense promoted further studies to uncover the downstream signal transduction pathways involved in bacteria-triggered stomatal closure and reopening. In addition to SA, the plant hormone ABA, which was known to be important for stomatal regulation in response to abiotic stresses, plays a central role in regulating P. s. tomato– and PAMP-triggered stomatal closure, because Arabidopsis mutants defective in ABA biosynthesis or signaling failed to close stomata in response to P. s. tomato infection or PAMP treatment (Melotto et al., 2006; Zhang et al., 2008; Zeng and He, 2010). These findings suggest that guard cell signal transduction in response to biotic and abiotic stresses share common steps. A recent investigation provided evidence that ABA signaling acts genetically downstream of SA signaling in the regulation of pathogen-triggered stomatal closure (Zeng and He, 2010). Moreover, it was recently shown that an oxylipin pathway converges with the ABA pathway at the level of an anion channel to regulate stomatal closure and plant immunity (Montillet et al., 2013). However, the signaling events further downstream of ABA remain largely unknown. Prominently, it is not clear whether pathogen infection leads to increased ABA biosynthesis in the stomata or, simply, whether pathogen/PAMP-triggered stomatal closure requires a basal level of ABA in the guard cells.
Melotto et al. (2006) also discovered that, to overcome stomatal defense, P. s. tomato DC3000 produces the toxin coronatine (COR) to reopen the stomata, which were closed upon pathogen infection. In addition to being essential for stomatal reopening, COR is also essential for the virulent pathogen to overcome the apoplastic defense, including SA-mediated defense (Kloek et al., 2001; Zhao et al., 2003; Brooks et al., 2005; Zeng et al., 2011; Zheng et al., 2012). Several lines of evidence led to the hypothesis that, by producing COR, P. s. tomato DC3000 hijacks the jasmonic acid (JA) signaling pathway to suppress plant defenses (including stomatal and apoplastic defenses) through hormone crosstalk (Spoel and Dong, 2008; Robert-Seilaniantz et al., 2011). First, COR is a structural and functional mimic of JA (Bender et al., 1999; Brooks et al., 2005; Uppalapati et al., 2005). In particular, COR is most similar to JA-Ile, the active form of the plant hormone JA (Katsir et al., 2008; Fonseca et al., 2009b). Second, the virulence function of COR requires the CORONATINE INSENSITIVE1 (COI1) F-box protein of the Skp/Cullin/F-box complex (SCFCOI1), which is involved in ubiquitin-mediated degradation of substrates, the JAZ (for jasmonate ZIM domain) repressor proteins (Xie et al., 1998; Chini et al., 2007; Thines et al., 2007). The SCFCOI1-JAZ complex was believed to be involved in the perception of JA-Ile and COR (Katsir et al., 2008; Fonseca et al., 2009b; Yan et al., 2009; Sheard et al., 2010). Third, like JA-Ile, COR directly targets the COI1-JAZ receptor complex to promote the degradation of the JAZ repressors (Katsir et al., 2008; Fonseca et al., 2009b; Yan et al., 2009; Sheard et al., 2010). Since JAZ proteins normally interact with and repress the function of JA signaling transcription factors (TFs), including the master regulator MYC2, degradation of JAZ repressors leads to the derepression of these TFs and, therefore, the transcriptional activation of JA/COR-responsive genes (Lorenzo et al., 2004; Chini et al., 2007; Dombrecht et al., 2007; Fonseca et al., 2009a; Fernández-Calvo et al., 2011; Qi et al., 2011; Song et al., 2011). Supporting this hypothesis, a signaling cascade by which COR and JA suppress the plant immune response through the SCFCOI1-JAZ-MYC2 signaling pathway was recently elucidated (Zheng et al., 2012). In this signaling cascade, COR and JA activate the expression of three homologous NAC (for NAM, ATAF1,2, and CUC2) family TFs through MYC2, which eventually inhibits SA accumulation and, therefore, suppresses SA-mediated plant immunity (Zheng et al., 2012).
We investigate the immune response of tomato (Solanum lycopersicum) and report here the molecular mechanisms by which JA2 (for jasmonic acid2) and JA2L (for JA2-like), two homologous NAC TFs in tomato, regulate P. s. tomato DC3000–triggered stomatal movement. We reveal that JA2 acts in ABA-mediated stomatal closure, whereas JA2L functions in COR/JA-mediated stomatal reopening. We further demonstrate that JA2 regulates ABA biosynthesis by selectively activating the transcription of an ABA biosynthetic gene and that JA2L suppresses SA accumulation by regulating genes involved in SA metabolism. Thus, signaling cascades by which plants promote stomatal closure through activating ABA biosynthesis and bacteria suppress host defense through inhibiting SA accumulation have been elucidated. Our results promise to shed new light on the molecular mechanisms underlying stomatal responses to pathogen infection.
RESULTS
P. s. tomato DC3000–Triggered Stomatal Movement of Tomato Leaves
Previous elegant studies revealed that, during early stages of the P. s. tomato DC3000 and Arabidopsis interaction, Arabidopsis plants actively close stomata to prevent bacterial invasion; as a counterdefense, P. s. tomato DC3000 uses its virulence factor COR to reopen stomata (Melotto et al., 2006; Zhang et al., 2008; Liu et al., 2009; Zeng and He, 2010; Zeng et al., 2011; Desclos-Theveniau et al., 2012; Kumar et al., 2012; Singh et al., 2012; Zheng et al., 2012; Montillet and Hirt, 2013). As a first step to investigate the signaling events involved in pathogen-triggered stomatal immunity of tomato plants, we dip-inoculated wild-type tomato leaves with P. s. tomato DC3000 and investigated the pathogen-induced stomatal response. For these experiments, tomato seedlings were light-adapted for at least 3 h to ensure that most of the stomata were open before bacterium infection. Within 1 h of incubation, we observed a marked reduction of stomatal aperture; at 4 h after incubation, however, the stomatal aperture had reverted to the prebacterial treatment state (Figures 1A to 1D). These results indicate that, in response to P. s. tomato DC3000 infection, the stomatal movement of tomato plants is largely similar to that of the intensively studied Arabidopsis plants.
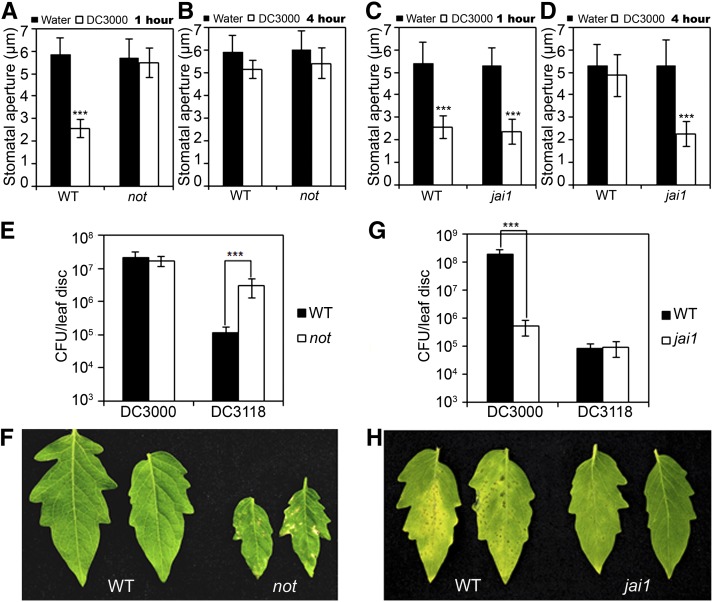
Figure 1.
Distinct Roles of ABA and JA in Regulating P. s. tomato DC3000–Triggered Stomatal Movement of Tomato Plants.
(A) and (B) Stomatal aperture response in leaf peels of wild-type (cv Ailsa Craig) and not plants after 1 h of incubation (A) or 4 h of incubation (B) with either water or P. s. tomato DC3000.
(C) and (D) Stomatal aperture response in leaf peels of wild-type (cv Castlemart) and jai1 plants after 1 h of incubation (C) or 4 h of incubation (D) with either water or P. s. tomato DC3000.
For (A) to (D), error bars represent se. Each experiment was repeated at least three times with similar results. Student’s t test was used to compare means between water and pathogen treatment of the same genotype (***P < 0.001).
(E) Wild-type and not plants were dip inoculated with suspensions of P. s. tomato DC3000 or P. s. tomato DC3118, and bacterial growth was measured at 3 DAI.
(F) Wild-type and not plants were dip inoculated with P. s. tomato DC3118, and photographs of the disease symptoms were taken at 3 DAI.
(G) Wild-type and jai1 plants were dip inoculated with P. s. tomato DC3000 or P. s. tomato DC3118, and bacterial growth was measured at 3 DAI.
(H) Wild-type and jai1 plants were dip inoculated with P. s. tomato DC3000, and photographs of the disease symptoms were taken at 3 DAI.
For (E) and (G), Student’s t test was used to compare the log-transformed data of different genotypes (***P < 0.001). Error bars represent sd of three replicates. CFU, colony-forming units.
NCED1-Dependent ABA Biosynthesis Is Required for P. s. tomato DC3000–Triggered Stomatal Closure but Not COR-Mediated Stomatal Reopening
Studies in the Arabidopsis–P. s. tomato DC3000 interaction system revealed that the phytohormones ABA and JA play distinct roles in regulating pathogen-triggered stomatal movement. These observations found that, whereas the ABA pathway is required for pathogen-induced stomatal closure (Melotto et al., 2006; Zhang et al., 2008; Zeng et al., 2011), the JA pathway is required for COR-mediated stomatal reopening (Melotto et al., 2006; Zheng et al., 2012). To test whether a similar scenario underlies the interactions between P. s. tomato DC3000 and tomato plants, we investigated the stomatal movement of ABA- or JA-related tomato mutants in response to the surface inoculation of P. s. tomato DC3000. notabilis (not) is a well-characterized ABA-deficient tomato mutant because this line contains a mutation in the NCED1 gene, which encodes 9-cis-epoxycarotenoid dioxygenase (NCED), a rate-limiting enzyme of ABA biosynthesis (Burbidge et al., 1999; Thompson et al., 2000a, 2000b, 2004). We found that, in the not mutants, P. s. tomato DC3000–induced stomatal closure at 1 h was largely compromised, while P. s. tomato DC3000–induced stomatal reopening at 4 h was not affected (Figures 1A and 1B). These results support that NCED1-dependent ABA biosynthesis is required for P. s. tomato DC3000–induced stomatal closure but not for COR-mediated stomatal reopening.
In P. s. tomato dip inoculation, plant mutants defective in ABA-mediated stomatal closure are more susceptible to COR-deficient (COR−) mutant bacteria (Melotto et al., 2006; Zeng and He, 2010). Since not plants are defective in P. s. tomato DC3000–triggered stomatal closure (Figures 1A and 1B), we hypothesized that this plant mutation might be able to rescue the virulence defect of the COR− mutant bacteria, which are unable to overcome stomatal defense. To test this hypothesis, we dip-inoculated not plants with P. s. tomato DC3118, a widely used COR− bacteria strain (Zhao et al., 2003). In control experiments, the COR-producing P. s. tomato DC3000 multiplied similarly in wild-type and not plants at 3 d after inoculation (DAI) (Figure 1E). By contrast, when the COR− P. s. tomato DC3118 were applied to the leaf surface, the multiplication of bacteria in wild-type leaves was greatly reduced compared with that of P. s. tomato DC3000, and no disease symptoms were observed (Figure 1F), confirming that the COR-mediated suppression of stomatal defense is critical for P. s. tomato DC3000 infection. Remarkably, in surface-inoculated not plants, the multiplication of the COR− P. s. tomato DC3118 at 3 DAI was much higher than that in wild-type leaves and caused prominent disease symptoms (Figures 1E and 1F), indicating that the not mutation indeed rescued the virulence defect of the COR− pathogen. Together, these data strengthen the notion that NCED1-dependent ABA biosynthesis is required for P. s. tomato DC3000–induced stomatal closure but not for COR-regulated stomatal reopening in tomato plants.
Jai1/Sl-COI1–Dependent JA Signaling of Tomato Is Required for COR-Regulated Stomatal Reopening but Not ABA-Mediated Stomatal Closure
To evaluate the role of JA in the pathogen-triggered stomatal movement of tomato, we examined the stomatal response of the JA-insensitive mutant jai1 (Li et al., 2002) in response to surface inoculation by P. s. tomato DC3000. The tomato jai1 mutant harbors a mutation in Sl-COI1, the tomato homolog of the Arabidopsis COI1 gene (Li et al., 2004). In response to P. s. tomato DC3000 infection, jai1 plants closed the stomata at 1 h (Figure 1C), demonstrating that Jai1/COI1-dependent JA signaling is not required for P. s. tomato DC3000–induced stomatal closure. However, P. s. tomato DC3000–induced stomatal reopening at 4 h was largely compromised in jai1 plants (Figure 1D), indicating that Jai1/Sl-COI1–dependent JA signaling is required for COR-regulated stomatal reopening.
Since jai1 plants are defective in P. s. tomato DC3000–triggered stomatal reopening (Figures 1C and 1D), we expect that these plants are more resistant to the COR-producing bacteria, which use the phytotoxin COR to reopen stomata and facilitate pathogen entry into the plant. Indeed, in our surface inoculation assays, the multiplication of the COR-producing P. s. tomato DC3000 in jai1 leaves at 3 DAI was greatly reduced as compared with that in wild-type leaves (Figure 1G), and the pathogen failed to cause disease symptoms (Figure 1H). These results are consistent with the long-standing observation that the Arabidopsis coi1 mutant and the tomato jai1 mutant show enhanced resistance to P. s. tomato DC3000 (Kloek et al., 2001; Zhao et al., 2003). Together, these results support the idea that the Jai1/COI1-dependent JA signaling pathway is required for pathogen-triggered reopening of the stomata.
Tomato JA2 and JA2L, Which Encode Closely Related NAC TFs, Were Differentially Regulated by ABA and JA in Guard Cells
The above-described investigations revealed distinct roles of ABA and JA in regulating P. s. tomato DC3000–triggered stomatal movement of tomato plants: whereas ABA is required for pathogen-induced stomatal closure, JA is required for JA/COR-regulated stomatal reopening. These results are largely consistent with the extensive observations performed in the model system of P. s. tomato DC3000–Arabidopsis interactions (Melotto et al., 2006, 2008; Zhang et al., 2008; Acharya and Assmann, 2009; Kim et al., 2010; Zeng and He, 2010; Zeng et al., 2011; Zheng et al., 2012). However, the downstream signal transduction pathways by which ABA promotes stomatal closure and JA/COR promotes stomatal reopening remain largely unknown.
To identify the signaling components involved in ABA-mediated stomatal closure as well as JA-mediated stomatal reopening of tomato plants, we focused on guard cell–specific TF genes whose expression was differentially regulated by ABA and JA. Several lines of evidence led us to the hypothesis that two closely related NAC family TF genes, JA2 (Solyc12g013620), and JA2L (Solyc07g063410), are plausible candidates (Figures 2A and 2B; Supplemental Figure 1). First, in response to P. s. tomato DC3000 infection, transcript levels of JA2 and JA2L showed a slight, yet significant, increases (Supplemental Figure 2), indicating that the two NAC genes were induced during the early phase of pathogen–tomato interaction. Second, JA2 and JA2L have three homologs in Arabidopsis, ANAC019, ANAC055, and ANAC072 (Figure 2B; Supplemental Figure 1B). Our previous work demonstrated that the expression of ANAC019 and ANAC055 was induced by both JA and ABA and that these TF genes are involved in the JA-mediated plant defense response to pathogen infection (Bu et al., 2008). Third, ANAC019, ANAC055, and ANAC072 are specifically involved in COR-regulated stomatal reopening but not ABA-mediated stomatal closure (Zheng et al., 2012).
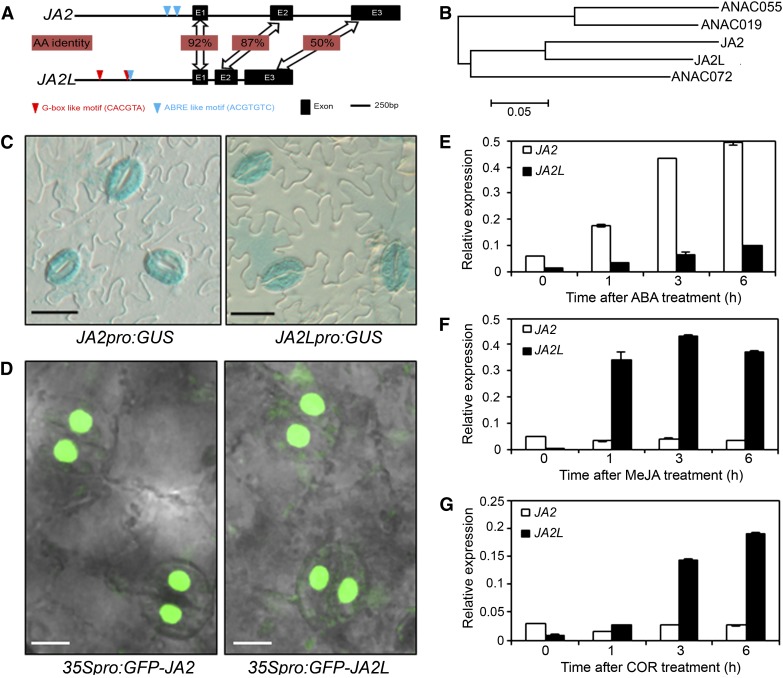
Figure 2.
JA2 and JA2L Are Differentially Regulated by ABA and JA/COR in Guard Cells.
(A) Genomic organization of JA2 and JA2L. The G-box–like motif (red triangles) and ABRE-like motif (blue triangles) are shown within the 1500-bp promoters; exons are indicated as black boxes.
(B) Phylogenetic analysis of JA2, JA2L, ANAC019, ANAC055, and ANAC072.
(C) Expression of JA2pro:GUS and JA2Lpro:GUS in the epidermis of transgenic tomato leaves. Bars = 25 μm.
(D) Expression of GFP-JA2 and GFP-JA2L fusion proteins in tomato leaves. Leaves of transgenic tomato plants expressing GFP-JA2 or GFP-JA2L were observed and imaged using a laser-scanning confocal microscope (Leica Microsystems). Bars = 10 μm.
(E) to (G) JA2 and JA2L transcripts were measured in wild-type (cv M82) seedlings treated with ABA (E), MeJA (F), or COR (G). Leaves at the indicated time points after treatment were harvested for RNA extraction and qRT-PCR analysis. Transcript levels of JA2 and JA2L were normalized to Actin2 expression. Error bars represent the sd of three technical replicates. Each experiment was repeated at least three times with similar results.
It is worth noting that JA2 and JA2L are the most closely related NAC TF members in the tomato genome; they show similar exon–intron gene structure and share 66% identity at the amino acid level (Figure 2A; Supplemental Figure 1A). To explore the possible functions of JA2 and JA2L in ABA-mediated stomatal closure and/or JA/COR-regulated stomatal reopening, we examined the tissue-specific expression patterns of the two genes. Promoter-GUS (for β-glucuronidase) fusion assays (Figure 2C; Supplemental Figure 3) and GFP (for green fluorescent protein)-protein fusion assays (Figure 2D) confirmed that both JA2 and JA2L were richly expressed in guard cells of tomato leaves. We then analyzed their induction by ABA, JA, and COR. Interestingly, our quantitative real-time PCR (qRT-PCR) assays with gene-specific primers revealed that the expression of JA2 was not induced by JA and COR (Figures 2F and 2G) but was quickly and strongly induced by ABA (Figure 2E). Parallel experiments indicated that ABA-induced expression of JA2 was not affected by the not mutation (Supplemental Figure 4A). By contrast, the expression of JA2L was only mildly induced by ABA (Figure 2E) but was strongly and quickly induced by JA and COR (Figures 2F and 2G). Closer examination revealed that the JA-induced expression of JA2L was largely abolished in the jail1 plants (Supplemental Figure 4B), indicating that JA induces the expression of JA2L in a Jai1/COI1-dependent manner. We also examined the possible induction of the two TF genes by abiotic cues and found that whereas JA2 expression is induced by dehydration (Supplemental Figure 5), JA2L is induced by mechanical wounding (Supplemental Figure 5). Considering that ABA plays a major role in regulating the plant dehydration response whereas JA plays a major role in regulating the plant wound response, these results are consistent with the above-described hormone induction patterns of the two TF genes. The hormone-specific induction patterns of JA2 and JA2L prompted us to examine the hormone-responsive cis-elements in their −2000 to −1 proximal promoter region. Our sequence analyses revealed the existence of two ABA-responsive element (ABRE)–like motifs (ACGTGTC) in the 2000-bp region of the JA2 promoter, but no G-box–like motif (CACGTA) was identified in this region (Figure 2A). Sequence analysis identified two G-box–like motifs and one ABRE motif in the parallel promoter region of JA2L (Figure 2A). The ABREs are required for ABA induction (Umezawa et al., 2010), and the G-box–like elements are required for JA/COR induction (Dombrecht et al., 2007; Figueroa and Browse, 2012). In this context, our results raised the possibility that JA2, which is specifically induced by ABA (Figure 2E), is involved in ABA-regulated stomatal closure and that JA2L, which is specifically induced by JA and COR (Figures 2F and 2G), is involved in JA/COR-mediated stomatal reopening.
Genetic Manipulation of JA2 or JA2L Differentially Alters P. s. tomato DC3000–Induced Stomatal Movement
To examine the role of JA2 in P. s. tomato DC3000–triggered stomatal movement, we applied the chimeric repressor silencing technology, in which JA2 was converted into a dominant negative form by fusing it with the SRDX repression domain (for SUPERMAN repression domain X) (Hiratsu et al., 2002, 2003). The chimeric repressor silencing technology can the overcome functional redundancy of TFs and effectively suppress their target genes with resultant loss-of-function phenotypes (Hiratsu et al., 2003; Ikeda et al., 2009). Recently, this technology was successfully applied to investigate the function of several TF genes in Arabidopsis (Heyman et al., 2013; Nakata et al., 2013). Transgenic tomato lines JA2-SRDX-1 and JA2-SRDX-2, in which the JA2-SRDX fusion gene was overexpressed (Supplemental Figure 6A), were selected for pathogen inoculation assays to study the role of JA2 in pathogen-triggered stomatal movement.
In our standard surface inoculation assays, the P. s. tomato DC3000–induced stomatal closure at 1 h was impaired in JA2-SRDX plants as compared with wild-type plants (Figure 3A), but the pathogen-triggered stomatal reopening at 4 h remained normal in these transgenic plants (Figure 3B), indicating that JA2 is required for P. s. tomato DC3000–induced stomatal closure but not stomatal reopening. Consistent with this observation, JA2-SRDX plants were also impaired in flg22-induced stomatal closure (Supplemental Figure 7A). Furthermore, in surface-inoculated plants, the COR− pathogen P. s. tomato DC3118 multiplied much better in JA2-SRDX plants than in wild-type plants (Figure 3E) and caused typical disease symptoms (Figure 3F), indicating that the JA2-SRDX plants effectively rescued the virulence deficiency of the COR− pathogen. Therefore, our pathogen-response assays revealed that, in response to the COR-producing or COR− pathogens, the stomatal response and disease symptom development of JA2-SRDX plants resembled those of the ABA-deficient not plants (Figures 1A to 1F). These results led us to the hypothesis that JA2 is specifically involved in ABA-mediated stomatal closure in response to P. s. tomato DC3000 infection.
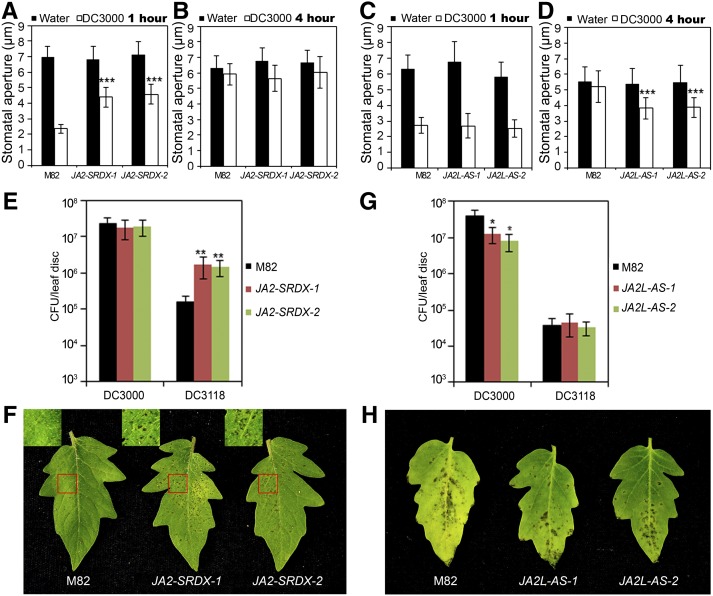
Figure 3.
Involvement of JA2 and JA2L in P. s. tomato–Induced Stomatal Movement.
(A) and (B) Stomatal aperture response in leaf peels of wild-type (cv M82) and JA2-SRDX plants after 1 h of incubation (A) or 4 h of incubation (B) with either water or P. s. tomato DC3000.
(C) and (D) Stomatal aperture response in leaf peels of wild-type (cv M82) and JA2L-AS plants after 1 h of incubation (C) or 4 h of incubation (D) with either water or P. s. tomato DC3000.
For (A) to (D), error bars represent se. Each experiment was repeated at least three times with similar results. Student’s t test was used to compare means between the wild type and each transgenic line after incubation with P. s. tomato DC3000 (***P < 0.001).
(E) Wild-type and JA2-SRDX plants were dip inoculated with suspensions of P. s. tomato DC3000 or P. s. tomato DC3118, and bacterial growth was measured at 3 DAI.
(F) Wild-type and JA2-SRDX plants were dip inoculated with P. s. tomato DC3118, and photographs of the disease symptoms were taken at 3 DAI.
(G) Wild-type and JA2L-AS plants were dip inoculated with P. s. tomato DC3000 or P. s. tomato DC3118, and bacterial growth was measured at 3 DAI.
(H) Wild-type and JA2L-AS plants were dip inoculated with P. s. tomato DC3000, and photographs of the disease symptoms were taken at 3 DAI.
For (E) and (G), Student’s t test was used to compare the log-transformed data of each transgenic line with those of the wild type (*P < 0.05 and **P < 0.01). Error bars represent sd of three replicates. CFU, colony-forming units.
To examine the function of JA2L in pathogen-induced stomatal movement, we generated transgenic tomato plants expressing an antisense version of the JA2L cDNA. JA2L-AS-1 and JA2L-AS-2, in which the expression levels of endogenous JA2L, but not those of JA2, were substantially reduced (Supplemental Figure 6B), were selected for further studies. In surface inoculation assays, P. s. tomato DC3000–induced stomatal closure at 1 h was largely normal in JA2L-AS plants (Figure 3C), but the pathogen-triggered stomatal reopening at 4 h was substantially impaired in these transgenic plants as compared with that in wild-type plants (Figure 3D), indicating that, in contrast with JA2, JA2L is required for COR-regulated stomatal reopening but not ABA-mediated stomatal closure. Consistent with this, at 3 DAI in surface-inoculated JA2L-AS plants, multiplication of P. s. tomato DC3000 and disease symptoms caused by this COR+ pathogen were significantly inhibited as compared with wild-type plants (Figures 3G and 3H). As expected, at 3 DAI, the multiplication of the COR− pathogen DC3118 was inhibited to a level similar to that in wild-type plants (Figure 3G). In summary, the pathogen-response phenotype of JA2L-AS plants in response to the COR+ or COR− pathogens showed similarity to that of the JA signaling mutant jai1 (Figures 1C, 1D, 1G, and 1H). Together, these results support the idea that JA2L is involved in JA/COR-mediated stomatal reopening in response to P. s. tomato DC3000 infection.
JA2 Is Required for NCED1 Expression and ABA Biosynthesis
Our results showing that JA2 is involved in ABA-mediated stomatal closure in response to P. s. tomato DC3000 infection prompted us to carry out an in-depth investigation of the underlying mechanisms. Several lines of evidence support the hypothesis that, similar to the not mutants, the JA2-SRDX plants are defective in ABA biosynthesis. First, as described above, JA2-SRDX plants are defective in P. s. tomato DC3000– and flg22-induced stomatal closure and these plants are able to rescue the virulence deficiency of the COR-deficient bacteria (Figures 3A, 3E, and 3F; Supplemental Figure 7A). Second, similar to the well-characterized not mutants, which were defective in NCED1-dependent ABA biosynthesis (Supplemental Figure 8; Burbidge et al., 1999; Thompson et al., 2004), JA2-SRDX plants are defective in dehydration-induced stomatal closure (Supplemental Figure 9). Third, the stomatal closure deficiency of not and JA2-SRDX plants is readily rescued by exogenous application of ABA (Supplemental Figure 7B). Fourth, soil-grown JA2-SRDX plants exhibit a dwarf phenotype with obviously shorter internodes as compared with their wild-type counterparts (Figures 4A to 4C). This dwarf phenotype was also observed in several classical tomato mutants defective in ABA biosynthesis (Nagel et al., 1994; Burbidge et al., 1999; Sharp et al., 2000; Thompson et al., 2004), including not (Supplemental Figure 10). Finally, our hormone measurement assays revealed that the endogenous ABA levels in JA2-SRDX-1 plants were substantially lower than those of the wild-type plants (Figure 4D; Supplemental Figure 8).
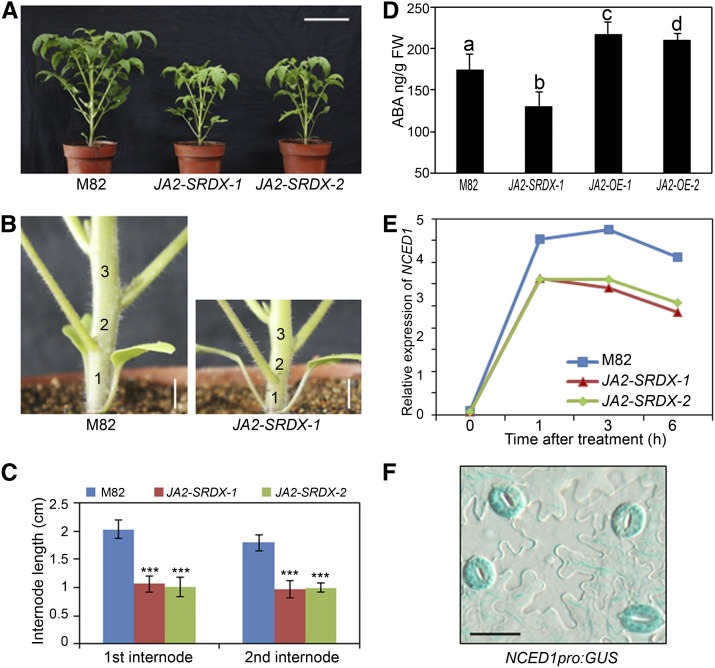
Figure 4.
JA2 Is Important for ABA Biosynthesis and NCED1 Expression.
(A) Phenotypic appearance of 6-week-old wild-type (cv M82) and JA2-SRDX transgenic plants. Bar = 10 cm.
(B) Representative images showing that JA2-SRDX transgenic plants have shorter internodes. Bars = 1 cm.
(C) Internode lengths of wild-type and JA2-SRDX transgenic plants. Error bars indicates se (n = 8). Student’s t test was used to compare means between the wild type and each transgenic line (***P < 0.001).
(D) ABA contents in wild-type, JA2-SRDX, and JA2-OE plants. Untreated healthy leaves from 4-week-old seedlings of the indicated genotypes were harvested for ABA measurement. Error bars represent sd of three replicates. ANOVA was performed for statistical analysis. FW, fresh weight.
(E) Dehydration-induced expression of NCED1 in wild-type and JA2-SRDX plants. Leaves from 4-week-old wild-type and JA2-SRDX plants were excised and dehydrated for the indicated times before total RNA was extracted for qRT-PCR analysis. Transcript levels of NCED1 were normalized to Actin2 expression. Error bars represent the sd of three technical replicates. This experiment was repeated three times with similar results.
(F) Expression of NCED1pro:GUS in the epidermis of transgenic tomato leaves. Bar = 25 μm.
The observation that JA2-SRDX-1 plants contain reduced ABA levels raised the possibility that, as a potential TF, JA2 might regulate ABA biosynthesis by targeting ABA biosynthetic genes. To identify the putative targets of JA2, the dehydration-induced expression patterns of several known ABA biosynthetic genes in tomato (Seo and Koshiba, 2002; Schwartz et al., 2003; Xiong and Zhu, 2003; Nambara and Marion-Poll, 2005) were compared between JA2-SRDX-1 and wild-type plants. Among them, ZEP1 encodes a zeaxanthin epoxidase that converts zeaxanthin to violaxanthin via the intermediate antheraxanthin (Thompson et al., 2000a). As described above, NCED1 encodes a rate-limiting enzyme of ABA biosynthesis. Mutation of NCED1 underlies the ABA-deficient phenotype of the classical not mutant in tomato (Burbidge et al., 1999; Thompson et al., 2000b, 2004). ABA2 encodes an alcohol dehydrogenase that converts xanthoxin into abscisic aldehyde (Schwartz et al., 2003). FLACCA (FLA) encodes a molybdenum cofactor sulfurase that catalyzes the oxidation of abscisic aldehyde to ABA (Sagi et al., 2002). SITIENS (SIT) encodes an aldehyde oxidase (AO) that catalyzes the oxidation of aldehyde to ABA (Harrison et al., 2011).
Our qRT-PCR assays revealed that genetic manipulation of JA2 had a minor, if any, effect on the dehydration-regulated expression of ZEP1, ABA2, FLA, and SIT (Supplemental Figure 11). By contrast, the dehydration-induced expression levels of NCED1 were substantially reduced in JA2-SRDX plants as compared with those in wild-type plants (Figure 4E). Closer observation revealed that the basal expression levels of NCED1 were already low in JA2-SRDX plants (Supplemental Figure 12). These results demonstrate that the NAC TF LeJA2 is important for the basal and dehydration-induced expression of NCED1.
The above-described results hint that NCED1 is a transcriptional target of JA2. We reasoned that, if this is the case, JA2 and NCED1 would exhibit similar expression patterns in response to intrinsic or extrinsic cues. Indeed, several lines of evidence support this hypothesis. First, promoter-GUS fusion assays revealed that, like JA2 (Figure 2C), NCED1 was richly expressed in guard cells (Figure 4F; Supplemental Figure 3). Second, in the classical ABA-deficient sit mutant of tomato (Stubbe, 1957, 1958, 1959; Taylor et al., 2000; Harrison et al., 2011), which harbors a mutation in an AO enzyme that catalyzes the final step of ABA biosynthesis, the steady state expression levels of NCED1 were substantially elevated over those in its wild-type counterpart (Thompson et al., 2000a; Supplemental Figure 13). Interestingly, the steady state expression levels of JA2 were also elevated in sit mutants as compared with wild-type plants (Supplemental Figure 13). Considering that NCED1 encodes a rate-limiting enzyme in ABA biosynthesis, our results revealed a possible compensation mechanism for ABA biosynthesis in ABA-deficient mutants, and it is most likely that JA2 and NCED1 are involved in this compensation mechanism. Third, the expression levels of both NCED1 and JA2 were strongly upregulated by dehydration in wild-type plants, and in sit mutants, dehydration-induced upregulation of NCED1 and JA2 expression was markedly reduced (Supplemental Figure 13), suggesting that the expression of both NCED1 and JA2 is activated by dehydration in an ABA-dependent manner.
Collectively, our data showing that genetic manipulation of JA2 affects the expression of NCED1 and that the two genes exhibit similar expression patterns in response to intrinsic or extrinsic cues strongly support the idea that JA2 regulates NCED1 expression for ABA biosynthesis in tomato.
JA2 Regulates ABA Biosynthesis by Activating the Expression of NCED1
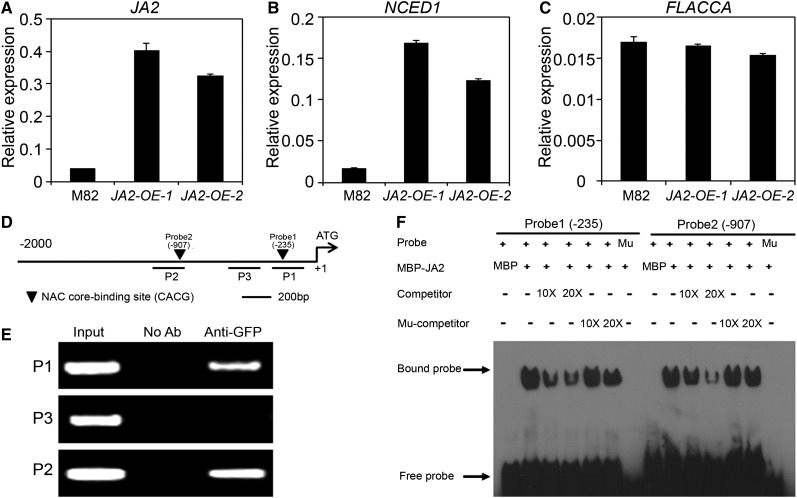
We then generated transgenic tomato lines that express the GFP-JA2 fusion protein under the control of the 35S promoter (JA2-OE plants). Two of these lines, JA2-OE-1 and JA2-OE-2, in which the high expression of the GFP-JA2 transgene was verified (Figure 5A), were selected for further analyses. qRT-PCR assays revealed that the expression levels of NCED1 (Figure 5B), but not those of FLA (Figure 5C), were markedly increased in JA2-OE lines as compared with wild-type plants, indicating that overexpression of JA2 selectively activates the expression of NCED1. In line with this observation, our hormone measurement assays revealed that the endogenous ABA levels were significantly higher in JA2-OE plants as compared with wild-type plants (Figure 4D). These results substantiated the scenario that JA2 regulates ABA biosynthesis by selectively activating the expression of NCED1.
Figure 5.
JA2 Regulates NCED1 Expression through Direct Interaction with Its Promoter.
(A) to (C) Basal transcript levels of JA2, NCED1, and FLA were measured in wild-type (cv M82) and JA2-OE plants. Leaves from 4-week-old plants grown under normal conditions were harvested at the same time for RNA extraction and qRT-PCR analyses. Transcript levels of each gene were normalized to Actin2 expression. Error bars represent the sd of three technical replicates. Shown are representative data from one biological replicate; three biological replicates were conducted, yielding similar results.
(D) Schematic diagram of the NCED1 promoter showing the presence of the NAC core binding site (black triangles). Short horizontal lines indicate DNA fragments (P1, P2, and P3) used for ChIP-PCR experiments. Shown are 2-kb upstream sequences of the NCED1 gene. The translational start site (ATG) is shown at position +1.
(E) Enrichment of the indicated DNA fragments (P1 and P2) following ChIP using anti-GFP antibodies. Chromatin of transgenic plants expressing GFP-JA2 was immunoprecipitated with anti-GFP antibodies, and the presence of the indicated DNA in the immune complex was determined by PCR. The DNA fragment P3 was used as a negative control. The experiment was repeated three times with similar results.
(F) EMSA showing that the MBP-JA2 fusion protein binds to DNA probes from the NCED1 promoter in vitro. Biotin-labeled probes were incubated with MBP-JA2 protein, and the free and bound DNAs (arrows) were separated on an acrylamide gel. As indicated, unlabeled probes were used as competitors. Mu, mutated probe in which the NAC core binding site (CACG) was deleted.
Next, we set up experiments to demonstrate that JA2 regulates NCED1 expression through direct interaction. It is well known that NAC TFs preferably bind to the so-called NAC core binding site (CACG) of their target promoters (Tran et al., 2004; Zheng et al., 2012). Indeed, our sequence analysis identified this type of NAC core binding sites in the P1 and P2 regions of the NCED1 promoter (Figure 5D). To test the in vivo interaction between JA2 and these NAC core binding sites, we performed chromatin immunoprecipitation (ChIP) assays using the aforementioned JA2-OE-1 plants, which express the GFP-JA2 fusion protein (Figure 2D). For these experiments, primers were designed to cover the NAC core binding sites (i.e., P1 and P2; Figure 5D). Primers covering the P3 region, where there is no NAC core binding site within a 295-bp stretch of sequence, were also included as a negative control (Figure 5D). As expected, the DNA samples precipitated by GFP-JA2 were enriched in the P1 and P2 regions, but not the P3 region, of the NCED1 promoter (Figure 5E). In the same ChIP experiment, GFP-JA2 was not enriched in the promoter region of JA2L targets (see below and Supplemental Figure 14). These results indicate a specific interaction between JA2 and the NAC core binding sites of the NCED1 promoter.
A DNA electrophoretic mobility-shift assay (EMSA) was conducted to confirm that JA2 binds these NAC core binding motifs in vitro. For these experiments, we designed DNA probes (i.e., Probe1 and Probe2) covering the NAC core binding sites in the P1 and P2 regions and purified JA2 as a JA2–maltose binding protein (MBP) fusion protein. As shown in Figure 5F, the MBP-JA2 fusion protein was able to bind Probe1, and this binding could be effectively competed by the addition of unlabeled DNA probe. By contrast, MBP-JA2 failed to bind the mutant Probe1, and the addition of unlabeled mutant DNA probe barely affected the interaction between MBP-JA2 and Probe1, indicating that JA2 binds Probe1 in a NAC core binding site–dependent manner. Parallel experiments indicated that JA2 also specifically bound to Probe2 in a NAC core binding site–dependent manner (Figure 5F). Taken together, our data support the notion that JA2 regulates ABA biosynthesis through direct binding to the promoter of NCED1, which encodes a rate-limiting enzyme of ABA biosynthesis.
We then investigated the induction of the JA2-NCED1 transcriptional module during the early phase of pathogen infection. In wild-type plants, both JA2 (Supplemental Figure 2A) and NCED1 (Supplemental Figure 2C) showed a slight yet significant induction by pathogen infection. The transcript levels of NCED1 in JA2-SRDX plants were much lower than those in the wild-type plants under control or pathogen infection conditions (Supplemental Figure 2C), suggesting that JA2 is important for maintaining the basal and pathogen-induced expression of NCED1. Consistently, the ABA levels from whole leaves of JA2-SRDX lines were much lower than those in wild-type plants under control or pathogen infection conditions (Supplemental Figure 2D). However, in our hormone measurement assays, which are based on mixed cell types (whole leaves), we failed to detect any significant difference of ABA levels between the two treatments (water control or DC3000 infection) in both wild-type and JA2-SRDX lines (Supplemental Figure 2D).
JA2L Is Important for COR-Mediated Virulence in the Apoplast
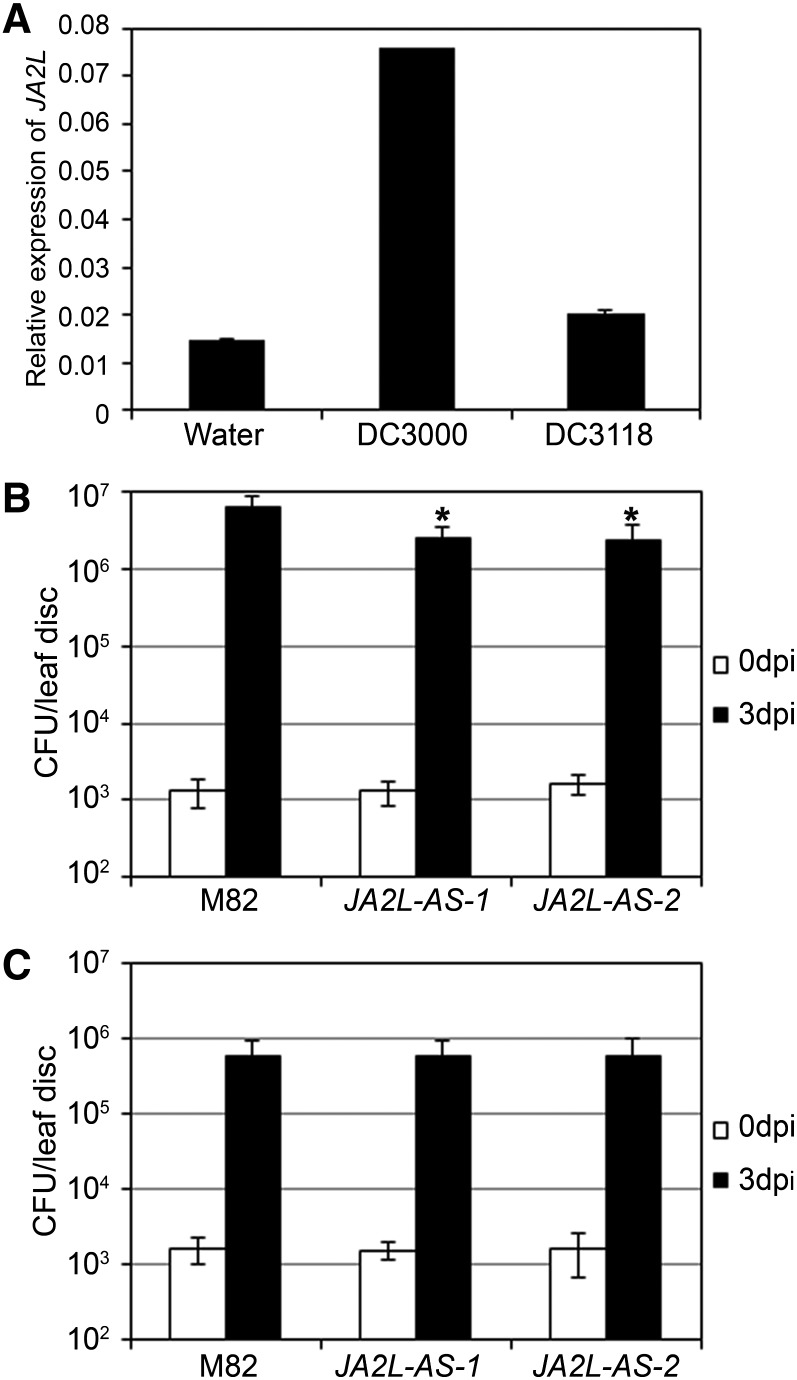
JA2L-AS plants are defective in P. s. tomato DC3000–triggered stomatal reopening (Figures 3D, 3G, and 3H), clearly demonstrating that JA2L is required for COR-mediated suppression of stomatal defense. It is known that, in addition to stomatal regulation, COR also regulates other aspects of P. s. tomato DC3000–induced virulence in the apoplast, including suppression of the SA-mediated defense, promotion of bacterial growth, and promotion of chlorotic disease symptom development (Mittal and Davis, 1995; Kloek et al., 2001; Brooks et al., 2005; Uppalapati et al., 2007; Zheng et al., 2012). We were interested in whether JA2L is also involved in COR-mediated virulence in the apoplast. To test this, we vacuum-infiltrated P. s. tomato DC3000 into tomato leaves to bypass the stomatal regulation and quantified the pathogen-induced expression of JA2L with qRT-PCR. In these assays, the COR-producing P. s. tomato DC3000, but not the COR− P. s. tomato DC3118, induces the expression of JA2L, indicating that pathogen infection activates the expression of JA2L in the apoplast in a COR-dependent manner (Figure 6A). This observation is consistent with the fact that the expression of JA2L is activated by exogenous application of both COR and JA (Figures 2F and 2G).
Figure 6.
JA2L Plays a Role in COR-Mediated Virulence in the Apoplast.
(A) Pathogen-induced expression of JA2L. Wild-type (cv M82) plants were vacuum-infiltrated with water, P. s. tomato DC3000, or P. s. tomato DC3118, and leaf tissues were collected for RNA extraction and RT-PCR assays at 24 HAI. Transcript levels of JA2L were normalized to Actin2 expression. Error bars represent the sd of three technical replicates. This experiment was repeated three times with similar results.
(B) and (C) Wild-type (cv M82) and JA2L-AS plants were vacuum-infiltrated with P. s. tomato DC3000 (B) or P. s. tomato DC3118 (C), and bacterial growth was measured at the indicated time points. Student’s t test was used to compare the log-transformed data of each transgenic line with those of the wild type (*P < 0.05). Error bars represent sd of three replicates. CFU, colony-forming units.
To validate that JA2L may play a role in the COR-mediated virulence in the apoplast, we vacuum-infiltrated JA2L-AS and wild-type plants with P. s. tomato DC3000 and measured bacterial growth afterward. At 3 DAI, the bacterial numbers present in the JA2L-AS plants were significantly lower than those in wild-type plants (Figure 6B), indicating that the JA2L-AS plants were more resistant than wild-type plants to P. s. tomato DC3000. On the contrary, in the same assays, JA2-SRDX plants and wild-type plants exhibited similar levels of bacterial growth (Supplemental Figure 15). These results demonstrate that JA2L, but not JA2, is required for P. s. tomato DC3000 virulence in the apoplast. When the COR-deficient P. s. tomato DC3118 was vacuum-infiltrated, JA2L-AS plants and wild-type plants exhibited similar levels of bacterial growth (Figure 6C), indicating that the JA2L-mediated virulence effect of P. s. tomato is COR dependent. Together, these data support the idea that JA2L plays an important role in mediating COR-promoted virulence in the apoplast.
JA2L Suppresses SA Accumulation through Regulating SA Metabolism Genes
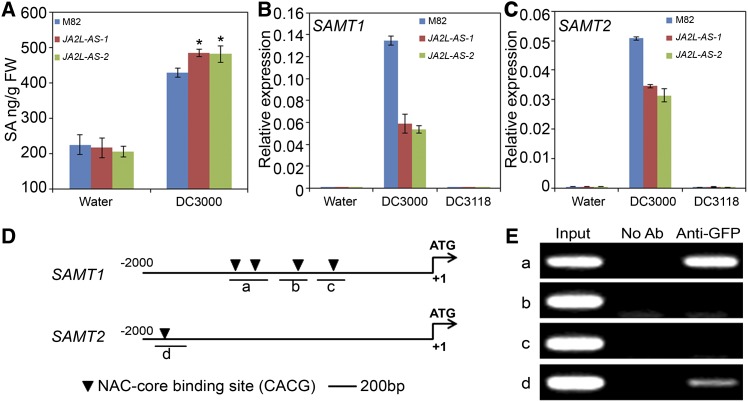
To understand how COR promotes the virulence of P. s. tomato DC3000 through induction of the TF gene JA2L, we focused on identifying JA2L downstream targets. Considering that SA is an essential signal for resistance against P. s. tomato infection and that COR suppresses SA accumulation (Kloek et al., 2001; Uppalapati et al., 2007; Zheng et al., 2012), we first measured the SA levels in wild-type and JA2L-AS plants in response to P. s. tomato DC3000 infection. As shown in Figure 7A, in JA2L-AS plants, P. s. tomato DC3000–induced accumulation of SA was substantially higher than that in wild-type plants at 24 h after inoculation (HAI), revealing that JA2L is responsible for the COR-mediated repression of SA accumulation.
Figure 7.
JA2L Suppresses SA Accumulation through Regulating the Expression of SA Metabolism Genes.
(A) Wild-type (cv M82) and JA2L-AS plants were vacuum-infiltrated with water or P. s. tomato DC3000, and SA levels were measured at 24 HAI. Student’s t test was used to compare means between the wild type and each transgenic line after incubation with P. s. tomato DC3000 (*P < 0.05). Error bars represent sd of three replicates. FW, fresh weight.
(B) and (C) Wild-type and JA2L-AS plants were vacuum-infiltrated with water, P. s. tomato DC3000, or P. s. tomato DC3118, and the expression levels of SAMT1 and SAMT2 were measured at 24 HAI. Transcript levels of SAMT genes were normalized to Actin2 expression. Error bars represent the sd of three technical replicates. This experiment was repeated three times with similar results.
(D) Schematic diagram of the promoters of SAMT1 and SAMT2. Black triangles represent NAC core binding sites. Short horizontal lines indicate DNA fragments used for ChIP-PCR experiments. Shown are 2-kb upstream sequences of each gene. The translational start site (ATG) is shown at position +1.
(E) Enrichment of the indicated DNA fragments (a and d) following ChIP using anti-GFP antibodies. Chromatin of transgenic plants expressing GFP-JA2L was immunoprecipitated with anti-GFP antibodies, and the presence of the indicated DNA in the immune complex was determined by PCR. The experiment was repeated three times with similar results. Ab, antibody.
In tomato, several genes are known to affect SA biosynthesis and metabolism (Uppalapati et al., 2007; Tieman et al., 2010). Among them, Isochorismate Synthase Gene1 encodes an enzyme involved in SA synthesis (Uppalapati et al., 2007). Salicylic Acid Methyl Transferase1 (SAMT1) encodes an enzyme that converts SA to the inactive, volatile methyl salicylate (MeSA) (Tieman et al., 2010). To test the possibility that these genes are targets of JA2L, we first examined their expression. For these experiments, tomato leaves were vacuum-infiltrated with P. s. tomato DC3000 and the expression of these genes was quantified with qRT-PCR. Interestingly, we found that SAMT1 and SAMT2, a SAMT1 homologous gene identified from the published tomato genome sequence (Tomato Genome Consortium, 2012), were induced by P. s. tomato DC3000 infection at 24 HAI (Figures 7B and 7C). Therefore, our effort to identify the transcriptional targets of JA2L was focused on SAMT1 and SAMT2. Closer observation indicated that P. s. tomato DC3118, the COR-deficient bacterial strain, failed to activate the expression of SAMT1 and SAMT2 (Figures 7B and 7C), indicating that P. s. tomato DC3000 infection activates the transcription of SAMT1 and SAMT2 in a COR-dependent manner. Importantly, while SAMT1 and SAMT2 were induced by P. s. tomato DC3000 in wild-type plants, this induction was markedly weakened in the JA2L-AS plants (Figures 7B and 7C), revealing that JA2L is essential for COR-mediated activation of SAMT expression. Together, these results support our hypothesis that COR suppresses SA accumulation through the JA2L-regulated expression of SAMT1 and SAMT2.
We then set up experiments to demonstrate that JA2L activates SAMT1 and SAMT2 expression through interaction with the NAC core binding sites identified in their promoter regions (Figure 7D). JA2L-OE plants, which express a GFP-JA2L fusion protein under the control of the 35S promoter (Figure 2D; Supplemental Figure 16), were used for ChIP assays. ChIP assays indicated that the anti-GFP antibody was able to precipitate DNA fragment a in the SAMT1 promoter and DNA fragment d in the SAMT2 promoter along with JA2L (Figure 7E; Supplemental Figure 14), revealing the direct interaction of JA2L with the NAC core binding motifs in the promoters of SAMT1 and SAMT2. In the same ChIP experiments, however, GFP-JA2L did not show any interaction with the NAC core binding motifs in the promoter of NCED1, which is a direct target of JA2 (Supplemental Figure 14).
Taken together, our results support the idea that JA2L represses SA accumulation by activating the transcription of two tomato SAMT genes, SAMT1 and SAMT2, whose protein products are able to convert SA to the inactive, volatile MeSA (Tieman et al., 2010). Given that the expression of JA2L was induced by COR and JA (Figures 2F and 2G), we propose that the JA2L-mediated transcriptional activation of SAMT genes underlies COR/JA-promoted virulence in the apoplast. Considering that JA2L is essential for COR/JA-mediated stomatal reopening (Figures 3D, 3G, and 3H) and that SA is required for stomatal defense (Melotto et al., 2006; Zeng and He, 2010; Montillet et al., 2013), we propose that the same molecular mechanism (i.e., JA2L-mediated transcriptional activation of SAMT1 and SAMT2) is responsible for JA2L-regulated stomatal reopening in response to P. s. tomato DC3000 infection.
DISCUSSION
JA2-Directed Expression of NCED1 Underlies ABA-Mediated Stomatal Closure in Response to P. s. tomato DC3000 Infection
Our results clearly demonstrate that JA2, together with its transcription target NCED1, forms a molecular module by which plants regulate P. s. tomato DC3000–triggered stomatal closure through ABA. First, both JA2 and NCED1 are guard cell–specific genes, and they show similar expression patterns in response to intrinsic and extrinsic cues. Second, genetic suppression of the activity of JA2 and NCED1 impairs ABA-mediated stomatal closure in response to P. s. tomato DC3000 infection but not JA/COR-mediated stomatal reopening. Third, JA2 regulates the transcription of NCED1, which encodes a rate-limiting enzyme of ABA biosynthesis in tomato. Fourth, genetic suppression of JA2 and NCED1 impairs ABA biosynthesis in tomato, and the resulting plants exhibit typical ABA-deficient phenotypes in terms of development and pathogen resistance. Together, these data support the notion that the JA2-NCED1 transcriptional module is involved in the regulation of ABA biosynthesis, which is required for the host to close stomata as a defense mechanism (i.e., stomatal defense) in response to P. s. tomato DC3000 infection.
Despite the importance of ABA biosynthesis in regulating plant stress responses (both abiotic stress responses and biotic stress responses), the regulatory mechanisms governing the expression of ABA biosynthetic genes remain obscure (Seo and Koshiba, 2002; Xiong and Zhu, 2003; Nambara and Marion-Poll, 2005). Previous genetic and biochemical studies in tomato and other plants found that the expression levels of the NCED gene are tightly correlated with endogenous ABA content, supporting the hypothesis that NCED is a key regulatory gene for ABA biosynthesis in response to developmental and environmental cues (Tan et al., 1997, 2003; Burbidge et al., 1999; Thompson et al., 2000a, 2000b, 2004; Iuchi et al., 2001). In this context, our results showing that JA2 regulates NCED1 expression for ABA biosynthesis reveal a molecular mechanism governing the fine-tuning of ABA accumulation in planta. Considering that the P. s. tomato DC3000–induced stomatal response deficiency of JA2-SRDX lines was weaker than that of the not mutant plants, we could not rule out the possibility that JA2 is not the only TF that can regulate NCED1 expression.
Furthermore, our results revealed the existence of an interesting regulatory loop between endogenous ABA levels and the JA2-NCED1 ABA biosynthesis module. For example, in the classical ABA-deficient sit mutant of tomato (Stubbe, 1957, 1958, 1959; Taylor et al., 2000; Thompson et al., 2000a; Harrison et al., 2011), the steady state expression levels of both JA2 and NCED1 were substantially elevated compared with those in its wild-type counterpart (Supplemental Figure 13), suggesting that the JA2-NCED1 module might involve a possible compensation mechanism for ABA biosynthesis in this ABA-deficient mutant. Not surprisingly, the dehydration-induced expression levels of both JA2 and NCED1 were markedly reduced in sit mutants as compared with those in wild-type plants, suggesting that the full activation of the JA2-NCED1 module by dehydration requires a basal level of ABA. In line with our observations, it has been reported that, in Arabidopsis, drought- and salt-activated expression of Arabidopsis NCED3 was impaired in the ABA-deficient mutants low expression of osmotically responsive gene5 (los5) and los6 (Xiong et al., 2002). Together, our results support that the JA2-NCED1 module is involved in a feedback regulatory loop for ABA homeostasis. We propose that plants might employ this feedback regulatory loop to precisely monitor the endogenous ABA status and, therefore, determine whether, when, and how to activate the JA2-NCED1 module to fine-tune the endogenous ABA levels.
Stomatal guard cells represent one of the most important cell types for ABA actions. Although vascular tissues are probably the main site of ABA biosynthesis (Nambara and Marion-Poll, 2005; Endo et al., 2008; Seo and Koshiba, 2011), several studies indicate that ABA biosynthesis is also active in guard cells. For example, the promoter activity of the Arabidopsis ABA biosynthetic gene NCED3 was detected in guard cells (Tan et al., 2003). Similarly, the mRNA and protein of AO3, another ABA biosynthetic gene of Arabidopsis, were also detected in guard cells (Koiwai et al., 2004). In line with these observations, our promoter-GUS fusion and/or GFP-protein fusion assays revealed that the tomato JA2 and NCED1 genes were richly expressed in guard cells. These results support the hypothesis that the perception of P. s. tomato DC3000 infection by the plant immune receptors may lead to increased ABA biosynthesis in guard cells. However, we failed to detect a significant increase of ABA levels from whole leaves of pathogen-infected plants (Supplemental Figure 2D). One possible explanation is that the P. s. tomato DC3000–induced increase of ABA accumulation in whole leaves is below the detection limit. Given that the JA2-NCED1 module is richly expressed in guard cells and that both JA2 and NCED1 were induced by P. s. tomato DC3000 infection, it is reasonable to speculate that the future development of cell type–specific hormone measurement techniques will be helpful to verify the role of ABA biosynthesis in guard cell immunity. It is also possible that the P. s. tomato DC3000–induced guard cell response requires a basal level of ABA.
Recently, an ABA-independent oxylipin pathway was demonstrated to play a key role in controlling stomatal defense in Arabidopsis. In their interesting study, Montillet et al. (2013) reported that the oxylipin pathway converges with the ABA pathway at the level of the anion channel SLAC1 to regulate stomatal closure.
However, in our P. s. tomato DC3000–tomato interaction system, we showed that in response to pathogen infection, while wild-type tomato plants close stomata at 1 HAI, the not mutants, which are defective in ABA accumulation (Thompson et al., 2004), were severely impaired in pathogen-induced stomatal closure (Figure 1). Importantly, we showed that JA2-SRDX plants were significantly compromised in their ability to close stomata in response to P. s. tomato DC3000 infection (Figure 3). Considering that Montillet et al. (2013) checked the stomatal response of ost1-2 and aba2-1 mutants with exogenous application of the PAMP flg22, rather than pathogen infection, we checked the stomatal behavior of wild-type, not, and LeJA2-SRDX plants in response to flg22 and found that while wild-type tomato plants closed stomata in response to flg22 application, not mutants and JA2-SRDX plants were significantly compromised in flg22-triggered stomatal closure (Supplemental Figure 7). We also examined the ABA-induced stomatal closure of these plants and found that the stomatal response of not and JA2-SRDX plants is similar to that of their wild-type counterparts (Supplemental Figure 7), indicating that exogenous ABA rescues the stomatal closure deficiency of not and JA2-SRDX plants. These results are consistent with the fact that not and JA2-SRDX plants are defective in ABA biosynthesis.
These results, together with other results described here, clearly demonstrated that JA2 and NCED1 play an important role in P. s. tomato DC3000– and flg22-triggered stomatal closure. It is most likely that the discrepancies between our observations here and those described by Montillet et al. (2013) are due to the existence of species-specific differences.
JA2L-Directed Expression of SA Metabolism Genes Was Hijacked by P. s. tomato DC3000 as a Virulence Strategy through Producing COR
We also provide evidence that the function of JA2L in regulating P. s. tomato DC3000–triggered stomatal movement is distinct from that of JA2, although they are the most closely related NAC family TF members in tomato. First, the expression of JA2L is induced by JA and COR in a Jai1/Sl-COI1–dependent manner but not by ABA. Second, similar to the JA signaling mutant jai1, JA2L-AS plants specifically impair P. s. tomato DC3000–induced stomatal reopening but not stomatal closure, and these plants are more resistant than wild-type plants to P. s. tomato DC3000, suggesting that JA2L may act in the JA/COR signaling pathway. Third, JA2L-directed expression of SA metabolism genes inhibits P. s. tomato DC3000–induced SA accumulation and, therefore, suppresses SA-mediated defense responses. These results support the notion that JA2L acts as a target of COR by which COR suppresses host immunity. Considering that JA2L is expressed in guard cells and that SA is required to trigger stomatal closure (Melotto et al., 2006; Zeng and He, 2010; Montillet et al., 2013), it is reasonable to speculate that JA2L-regulated expression of SA metabolic genes underlies the action of JA2L in promoting pathogen-induced stomatal reopening. We reasoned that the JA2L-SAMT module was hijacked by P. s. tomato DC3000 as a virulence strategy through the production of COR, which serves as a potent inducer of JA responses through targeting the SCFCOI1 receptor complex (Xie et al., 1998; Katsir et al., 2008; Fonseca et al., 2009b; Yan et al., 2009; Sheard et al., 2010). It will be important in future studies to elucidate the molecular mechanism by which COR activates the JA2L-SAMT transcriptional module.
The antagonistic interaction between JA signaling and SA signaling is believed to be involved in fine-tuning the plant defense responses depending on the lifestyle of the pathogen encountered (Spoel et al., 2007). Despite its importance, not all signaling components involved in this crosstalk have been identified. It was recently shown in Arabidopsis that the inhibition effect of SA on JA signaling occurs downstream of the SCFCOI1-JAZ complex via the TF ORA59 (Van der Does et al., 2013). Our finding here that JA2L suppresses SA accumulation represents one mechanism by which JA antagonizes SA-dependent immunity in tomato. Given that the JA/COR-induced expression of JA2L depends on the function of COI1, it is most likely that the JA2L-mediated suppression of SA signaling also occurs downstream of the SCFCOI1-JAZ complex. Besides acting as a mediator for JA–SA crosstalk and a hostage hijacked by a pathogen to promote virulence, what is the normal physiological function of JA2L? Considering that MeSA could act as an important herbivore-induced plant volatile to control damaging pests through the recruitment of natural enemies (Zhu and Park, 2005; Mallinger et al., 2011), our results showing that JA2L promotes MeSA synthesis by activating SA metabolic genes suggests that this TF may play an important role in JA-mediated indirect defense against herbivores. Our study, however, does not rule out the possibility that JA2L may target other genes for its involvement in the JA signaling pathway.
JA2 and JA2L: Structurally Similar, Functionally Distinct
Both JA2 and JA2L are typical NAC family TF members, which contain a conserved N-terminal NAC domain and a divergent C-terminal domain (Supplemental Figure 1). It is generally believed that the N-terminal NAC domain of NAC TFs is responsible for DNA binding, whereas the divergent C-terminal domain is involved in transcriptional regulation (Olsen et al., 2005; Jensen et al., 2010). JA2 and JA2L are the most closely related NAC TF members in the tomato genome; they show similar exon–intron gene structure and share 66% identity at the amino acid level. However, we show here that they function in different steps of bacteria-induced stomatal movement through distinct molecular mechanisms: whereas JA2 acts in ABA-mediated stomatal closure through regulating ABA biosynthetic genes, JA2L acts in JA/COR-mediated stomata reopening through regulating SA metabolic genes.
Recently, an elegant work demonstrated that three homologous Arabidopsis NAC TFs, ANAC019, ANAC055, and ANAC072, specifically regulate JA/COR-mediated stomatal reopening but not ABA-mediated stomatal closure (Zheng et al., 2012). The expression pattern and action mechanism of JA2L, but not those of JA2, showed high similarity to those of the three NAC TFs from Arabidopsis, which were grouped in the same clade with JA2 and JA2L in our phylogenetic analyses, raising an interesting question of how their “structure–function” relationship is determined. We reasoned that the functional divergence of JA2 and JA2L may be not only due to their promoters, which are responsible for their different expression patterns, but also due to their similar but divergent amino acid sequences. Supporting this possibility, our experiments demonstrated that JA2L and JA2 exert their regulatory effects by targeting different genes (Supplemental Figure 14), which may be achieved by recruiting or interacting with different transcriptional partners. Clearly, in-depth structure–function analyses will provide insight into the action mechanisms of these homologous NAC family TFs in regulating host immunity, pathogen virulence, and plant–pathogen coevolution.
METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) cv M82, cv Castlemart, cv Rheinlands Ruhm, and cv Ailsa Craig were used as wild types in this study. Seeds of sit (LA0574; cv Rheinlands Ruhm background) and not (LA3614; cv Ailsa Craig background) were obtained from the Tomato Genetics Resource Center at the University of California, Davis.
Tomato seeds were germinated for 48 h on moistened filter papers. Subsequently, tomato seedlings were grown in growth chambers and maintained under 16 h of light at 25°C and 8 h of dark at 18°C and 60% relative humidity. The ABA-deficient mutants (sit and not) were maintained in an unstressed state by keeping relative humidity at 80 to 90%. Homozygous jai1 plants were identified as described previously (Li et al., 2004).
DNA Constructs and Plant Transformation
DNA constructs for plant transformation were generated following standard molecular biology protocols and Gateway (Invitrogen) technology (Nakagawa et al., 2007). For JA2-SRDX plants, a 36-bp DNA sequence encoding the SRDX repression domain (LDLDLELRLGFA) was fused in frame to the 3′ end of the JA2 coding region. This fusion sequence was then cloned into the pGWB2 vector to generate the 35Spro:JA2-SRDX construct. For JA2L-AS plants, the 441-bp fragment encoding the C-terminal domain of JA2L was amplified and cloned in the antisense orientation into the pGWB2 vector to generate the JA2L-AS construct.
The above constructs were introduced into tomato cv M82 by Agrobacterium tumefaciens–mediated transformation (Yan et al., 2013). Transformants were selected based on their acquired resistance to hygromycin B. Homozygous T2 or T3 transgenic plants were used for phenotypic and molecular characterization.
Plant Treatment and Gene Expression Analyses
Methyl jasmonate (MeJA), wounding, and COR treatments were performed as described previously (Uppalapati et al., 2005; Yan et al., 2013). For ABA treatment, 4-week-old tomato plants were irrigated with water containing 100 μM ABA. For dehydration treatment, 4-week-old tomato leaves were detached and then dehydrated on filter papers at room temperature under dim light. Leaf tissues of three plants were pooled at 0, 1, 3, and 6 h after treatment for RNA extraction. For bacterial infection, 4-week-old tomato plants were vacuum-infiltrated with water, Pseudomonas syringae pv tomato DC3000, or P. s. tomato DC3118. Bacteria was inoculated at OD600nm = 0.01. Tomato leaves were harvested for RNA extraction after 24 h. RNA extraction and qRT-PCR analysis were performed as described previously (Yan et al., 2013). Expression levels of target genes were normalized to those of the tomato Actin2 gene. Primers used to quantify gene expression levels are listed in Supplemental Table 1. MeJA, COR, and ABA were purchased from Sigma-Aldrich.
For GUS staining analysis, promoters of tomato JA2, JA2L, and NCED1 were amplified by PCR and cloned into the binary vector pCambia1391Z to generate the JA2pro:GUS, JA2Lpro:GUS, and NCED1pro:GUS constructs, respectively. The resulting constructs were then introduced into tomato cv M82 by Agrobacterium-mediated transformation. The GUS staining assays were performed as described previously (Chen et al., 2011).
Stomatal Assays
Stomatal assay was performed as described previously (Melotto et al., 2006). Plants were kept under light (∼100 μmol m−2 s−1) for at least 3 h to ensure that most stomata were open before bacteria inoculation. Fully expanded healthy leaves from 5-week-old plants were immersed in water or bacterial suspension (1E + 8 colony-forming units/mL in water). At various time points, epidermis was peeled off and immediately observed with a Leica Microsystems DM5000B microscope. In the case of ABA- or flg22-induced stomatal closure, tomato leaves were immersed in MES buffer (25 mM MES-KOH, pH 6.15, and 10 mM KCl), 10 μM ABA in MES buffer, or 5 μM flg22 (Alpha Diagnostics) in MES buffer for 1 h, and epidermis was then peeled off for observation. Images of leaf peels were randomly taken, and at least 60 stomata were recorded for each sample. The width of the stomatal aperture was measured as described (Desclos-Theveniau et al., 2012). Statistical differences were analyzed with Student’s t test.
To determine dehydration-induced stomatal closure, detached leaves from wild-type and JA2-SRDX plants were dehydrated for 0 and 20 min, and the leaves were then frozen in liquid nitrogen for observing stomata with a Hitachi S-3000N scanning electron microscope. The stomatal aperture was then measured, and statistical differences were analyzed with Student’s t test.
Pathogen Infection Assays
P. s. tomato DC3000 (Melotto et al., 2006) and COR-deficient mutant P. s. tomato DC3118 (Zhao et al., 2003) were cultured at 30°C in low-salt Luria-Bertani medium (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl) with appropriate antibiotics until OD600nm of 0.8 was reached. Bacteria were collected by centrifugation and resuspended in water. For dip inoculation, 4-week-old plants were dipped into a solution of P. s. tomato (OD600nm = 0.2) with 0.02% Silwet L-77 for a few seconds and then kept under high humidity until disease symptoms developed. For vacuum infiltration, 4-week-old plants were vacuum-infiltrated with P. s. tomato (OD600nm = 0.002), and six plants were assayed for each data point. The infected leaves were surface sterilized with 15% H2O2 and washed twice before being homogenized to assay the bacterial growth. Bacterial populations were monitored by serial dilution assays (Zeng and He, 2010). Student’s t test was used to compare the means of log-transformed data.
Free SA and ABA Measurements
Free SA and ABA measurements were performed as described previously (Zhou et al., 2010; Yan et al., 2013). For free SA measurement, briefly, 4-week-old plants were vacuum-infiltrated with water or P. s. tomato DC3000 (OD600nm = 0.01). The infected leaves were harvested 24 h later, and free SA was extracted and measured by HPLC. Similarly, untreated healthy leaves from 4-week-old wild-type or transgenic plants were harvested for ABA measurement. Three replicates were taken for each data point. Statistical analyses were performed using ANOVA or Student’s t test.
Subcellular Localization
To generate the 35Spro:GFP-JA2 and 35Spro:GFP-JA2L tomatoes for subcellular localization analysis, full-length coding sequences of JA2 and JA2L, respectively, were amplified by PCR and cloned into the XhoI/SpeI sites of the binary vector GFP-pBA-C1. The resulting constructs were then transformed into tomato cv M82 for further analysis. For subcellular localization analysis, GFP fluorescence of leaves from 35Spro:GFP-JA2 and 35Spro:GFP-JA2L plants was observed and imaged with a confocal laser scanning microscope (Leica Microsystems).
EMSA
The full-length coding sequence of tomato JA2 was amplified by PCR and cloned into the pMAL-c2X vector. The recombinant MBP-JA2 protein was expressed in Escherichia coli BL21 and purified to homogeneity using an amylase resin column. Oligonucleotide probes were synthesized and labeled with biotin at their 5′ ends (Invitrogen). EMSA was performed as described previously (Chen et al., 2011; Yan et al., 2013).
ChIP-PCR Assays
ChIP assays were performed following a published protocol (Q. Chen et al., 2011; R. Chen et al., 2012; Yan et al., 2013) with minor modifications. Briefly, 1 g of 35Spro:GFP-JA2 or 35Spro:GFP-JA2L leaves was cross-linked in 1% formaldehyde and their chromatin was isolated. GFP antibody (Abcam) was used to immunoprecipitate the protein-DNA complex, and the precipitated DNA was purified using a PCR purification kit (Qiagen) for PCR analysis. Chromatin precipitated without antibody was used as a negative control, while the isolated chromatin before precipitation was used as an input control. Three independent biological repeats were performed. Primers used for ChIP-PCR are listed in Supplemental Table 1.
Accession Numbers
The accession numbers for the genes discussed in this article are as follows: JA2L (Solyc07g063410), JA2 (Solyc12g013620), SAMT1 (Solyc09g091550), SAMT2 (Solyc09g091530), NCED1 (Solyc07g056570), ZEP1 (Solyc02g090890), ABA2 (Solyc04g071960), SIT (Solyc01g009230), and FLA (Solyc07g066480).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment and Phylogenetic Analysis of JA2 and JA2L.
Supplemental Figure 2. Gene Expression and ABA Content after Pathogen Inoculation.
Supplemental Figure 3. Histochemical GUS Activity Was Detected in Various Tissues of JA2pro:GUS, JA2Lpro:GUS, and NCED1pro:GUS Plants.
Supplemental Figure 4. ABA- and MeJA-Induced Expression of JA2 and JA2L.
Supplemental Figure 5. Dehydration- and Wound-Induced Expression of JA2 and JA2L.
Supplemental Figure 6. Generation of JA2-SRDX and JA2L-AS Plants.
Supplemental Figure 7. Stomatal Aperture Response of Indicated Genotypes in Response to flg22 and ABA.
Supplemental Figure 8. ABA Contents in M82, JA2-SRDX Plants, cv Ailsa Craig, and not Mutants.
Supplemental Figure 9. Stomatal Aperture Responses of M82 and JA2-SRDX Plants to Dehydration.
Supplemental Figure 10. The ABA-Deficient Mutant not Exhibits a Dwarf Phenotype with Obviously Shorter Internodes.
Supplemental Figure 11. Dehydration-Induced Expression of ABA Biosynthetic Genes in M82 and JA2-SRDX Plants.
Supplemental Figure 12. Basal Expression Level of NCED1 in M82 and JA2-SRDX Plants.
Supplemental Figure 13. JA2 and NCED1 Show Similar Expression Patterns.
Supplemental Figure 14. JA2 and JA2L Differ from Each Other in the Ability to Bind Different Target Promoters.
Supplemental Figure 15. Growth of P. s. tomato DC3000 in M82 and JA2-SRDX Plants after Vacuum Infiltration.
Supplemental Figure 16. Molecular Identification of JA2L-OE Plants.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Jianmin Zhou for critical reading of the article. We thank the C.M. Rick Tomato Genetic Resource Center (University of California at Davis) for providing us the tomato mutant seeds of not and sit. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11030200), the Ministry of Science and Technology of China (Grant 2013AA102603), the National Natural Science Foundation of China (Grants 31030006 and 91317039), and the Postdoctoral Science Foundation of China (Grant 2013M530764).
AUTHOR CONTRIBUTIONS
M.D., Q.Z., and C.Y.L. designed the research. C.Y.L. conceived and supervised the project. M.D., Q.Z., and L.D. performed most of the experiments. S.L., H.S.L., L.Y., B.W., and H.J. generated the constructs and the transgenic plants. J.W. and Z.H. performed the hormone measurement experiments. T.H. and C.-B.L. helped grow the plants. L.K., J.L., M.D., Q.Z., and C.Y.L. analyzed the data. C.-B.L., T.H., J.L., and L.K. contributed reagents/materials/analysis tools. M.D., Q.Z., and C.Y.L. wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- ABA
abscisic acid
- SA
salicylic acid
- JA
jasmonic acid
- TF
transcription factor
- DAI
days after inoculation
- qRT-PCR
quantitative real-time PCR
- HAI
hours after inoculation
- MeSA
methyl salicylate
- ABRE
ABA-responsive element
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility-shift assay
- MeJA
methyl jasmonate
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Acharya B.R., Assmann S.M. (2009). Hormone interactions in stomatal function. Plant Mol. Biol. 69: 451–462. [DOI] [PubMed] [Google Scholar]
- Bender C.L., Alarcón-Chaidez F., Gross D.C. (1999). Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63: 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436. [DOI] [PubMed] [Google Scholar]
- Brooks D.M., Bender C.L., Kunkel B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6: 629–639. [DOI] [PubMed] [Google Scholar]
- Bu Q., Jiang H., Li C.-B., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Li C. (2008). Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18: 756–767. [DOI] [PubMed] [Google Scholar]
- Burbidge A., Grieve T.M., Jackson A., Thompson A., McCarty D.R., Taylor I.B. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 17: 427–431. [DOI] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Desclos-Theveniau M., Arnaud D., Huang T.-Y., Lin G.J.-C., Chen W.-Y., Lin Y.-C., Zimmerli L. (2012). The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 8: e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D., Zhou J.-M. (2012). Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 12: 484–495. [DOI] [PubMed] [Google Scholar]
- Endo A., et al. (2008). Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 147: 1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C., Robatzek S. (2012). Plants and pathogens: Putting infection strategies and defence mechanisms on the map. Curr. Opin. Plant Biol. 15: 699–707. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P., Browse J. (2012). The Arabidopsis JAZ2 promoter contains a G-box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 53: 330–343. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009a). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009b). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Harrison E., Burbidge A., Okyere J., Thompson A.J., Taylor I.B. (2011). Identification of the tomato ABA-deficient mutant sitiens as a member of the ABA-aldehyde oxidase gene family using genetic and genomic analysis. Plant Growth Regul. 64: 301–309. [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., De Veylder L. (2013). ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27: 325–333. [DOI] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196. [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522. [DOI] [PubMed] [Google Scholar]
- Koiwai H., Nakaminami K., Seo M., Mitsuhashi W., Toyomasu T., Koshiba T. (2004). Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 134: 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.S., Lakshmanan V., Caplan J.L., Powell D., Czymmek K.J., Levia D.F., Bais H.P. (2012). Rhizobacteria Bacillus subtilis restricts foliar pathogen entry through stomata. Plant J. 72: 694–706. [DOI] [PubMed] [Google Scholar]
- Li L., Li C., Lee G.I., Howe G.A. (2002). Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99: 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Fuglsang A.T., Palmgren M.G., Staskawicz B.J., Coaker G. (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallinger R.E., Hogg D.B., Gratton C. (2011). Methyl salicylate attracts natural enemies and reduces populations of soybean aphids (Hemiptera: Aphididae) in soybean agroecosystems. J. Econ. Entomol. 104: 115–124. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., He S.Y. (2008). Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46: 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Mittal S., Davis K.R. (1995). Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 8: 165–171. [DOI] [PubMed] [Google Scholar]
- Montillet J.-L., Hirt H. (2013). New checkpoints in stomatal defense. Trends Plant Sci. 18: 295–297. [DOI] [PubMed] [Google Scholar]
- Montillet J.-L., et al. (2013). An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel O.W., Konings H., Lambers H. (1994). Growth rate, plant development and water relations of the ABA-deficient tomato mutant sitiens. Physiol. Plant. 92: 102–108. [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakata M., Mitsuda N., Herde M., Koo A.J., Moreno J.E., Suzuki K., Howe G.A., Ohme-Takagi M. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10: 79–87. [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Sagi M., Scazzocchio C., Fluhr R. (2002). The absence of molybdenum cofactor sulfuration is the primary cause of the flacca phenotype in tomato plants. Plant J. 31: 305–317. [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Allen G.J., Hugouvieux V., Kwak J.M., Waner D. (2001a). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 627–658. [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak J.M., Allen G.J. (2001b). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330. [DOI] [PubMed] [Google Scholar]
- Schwartz S.H., Qin X., Zeevaart J.A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7: 41–48. [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2011). Transport of ABA from the site of biosynthesis to the site of action. J. Plant Res. 124: 501–507. [DOI] [PubMed] [Google Scholar]
- Sharp R.E., LeNoble M.E., Else M.A., Thorne E.T., Gherardi F. (2000). Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: Evidence for an interaction with ethylene. J. Exp. Bot. 51: 1575–1584. [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Kuo Y.-C., Mishra S., Tsai C.-H., Chien C.-C., Chen C.-W., Desclos-Theveniau M., Chu P.-W., Schulze B., Chinchilla D., Boller T., Zimmerli L. (2012). The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24: 1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351. [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104: 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe H. (1957). Mutanten der kulturtomate Lycopersicon esculentum Miller. I. Kulturpflanze 5: 190–220. [Google Scholar]
- Stubbe H. (1958). Mutanten der kulturtomate Lycopersicon esculentum Miller. II. Kulturpflanze 6: 85–115. [Google Scholar]
- Stubbe H. (1959). Mutanten der kulturtomate Lycopersicon esculentum Miller. III. Kulturpflanze 7: 82–112. [Google Scholar]
- Suhita D., Raghavendra A.S., Kwak J.M., Vavasseur A. (2004). Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol. 134: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B.C., Joseph L.M., Deng W.T., Liu L., Li Q.B., Cline K., McCarty D.R. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35: 44–56. [DOI] [PubMed] [Google Scholar]
- Tan B.C., Schwartz S.H., Zeevaart J.A., McCarty D.R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 94: 12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I.B., Burbidge A., Thompson A.J. (2000). Control of abscisic acid synthesis. J. Exp. Bot. 51: 1563–1574. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Jackson A.C., Parker R.A., Morpeth D.R., Burbidge A., Taylor I.B. (2000a). Abscisic acid biosynthesis in tomato: Regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 42: 833–845. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Jackson A.C., Symonds R.C., Mulholland B.J., Dadswell A.R., Blake P.S., Burbidge A., Taylor I.B. (2000b). Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 23: 363–374. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Thorne E., Burbidge A., Jackson A., Sharp R., Taylor I. (2004). Complementation of notabilis, an abscisic acid-deficient mutant of tomato: Importance of sequence context and utility of partial complementation. Plant Cell Environ. 27: 459–471. [Google Scholar]
- Tieman D., Zeigler M., Schmelz E., Taylor M.G., Rushing S., Jones J.B., Klee H.J. (2010). Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 62: 113–123. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S.P., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 51: 1821–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati S.R., Ayoubi P., Weng H., Palmer D.A., Mitchell R.E., Jones W., Bender C.L. (2005). The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42: 201–217. [DOI] [PubMed] [Google Scholar]
- Uppalapati S.R., Ishiga Y., Wangdi T., Kunkel B.N., Anand A., Mysore K.S., Bender C.L. (2007). The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 20: 955–965. [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M.C., Rodenburg N., Pauwels L., Goossens A., Körbes A.P., Memelink J., Ritsema T., Van Wees S.C., Pieterse C.M. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xin X.-F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Xiong L., Zhu J.-K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Zhu J.-K. (2002). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277: 8588–8596. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhai Q., Wei J., Li S., Wang B., Huang T., Du M., Sun J., Kang L., Li C.-B., Li C. (2013). Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 9: e1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., He S.Y. (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 153: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Brutus A., Kremer J.M., Withers J.C., Gao X., Jones A.D., He S.Y. (2011). A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., He S.Y., Assmann S.M. (2008). The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 56: 984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Thilmony R., Bender C.L., Schaller A., He S.Y., Howe G.A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36: 485–499. [DOI] [PubMed] [Google Scholar]
- Zheng X.-Y., Spivey N.W., Zeng W., Liu P.-P., Fu Z.Q., Klessig D.F., He S.Y., Dong X. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Wei L., Xu J., Zhai Q., Jiang H., Chen R., Chen Q., Sun J., Chu J., Zhu L., Liu C.-M., Li C. (2010). Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Park K.-C. (2005). Methyl salicylate, a soybean aphid-induced plant volatile attractive to the predator Coccinella septempunctata. J. Chem. Ecol. 31: 1733–1746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.