Abstract.
Bioaffinity conjugation between streptavidin (SA) and biotin has been widely used to link donors and acceptors for investigating the distance-dependent Förster resonance energy transfer (FRET). When studying a commonly used FRET system of (QD-SA)-(biotin-DNA-dye) [donor: quantum dot (QD); acceptor: small organic fluorescent dye; and linker: deoxyribose nucleic acid (DNA) molecule via SA-biotin conjugation], however, a contradictory finding was recently reported in the literature. It was found that the FRET lost its dependence on the number of DNA base pairs when using a phosphate-buffered saline (PBS) solution. We found that the conflicted results were caused by the ionic strength of the adopted buffer solutions. Our results suggest that the dependent FRET on the number of DNA bases is favorable in a low-ionic-strength buffer, whereas in relatively high-ionic-strength buffers, the FRET loses the DNA length dependence. We propose that the independence is mainly caused by the conformational change of DNA molecules from a stretched to a coiled mode when the cations in the high-ionic-strength buffer neutralize the negatively charged backbone of DNA molecules, thereby bringing the acceptors close to the donors.
Keywords: Förster resonance energy transfer, quantum dots, fluorophores, streptavidin and biotin conjugation, deoxyribose nucleic acid, fluorescence lifetime
1. Introduction
Förster resonance energy transfer (FRET) is an optical phenomenon in which an excited donor molecule can transfer its energy to an acceptor molecule to emit fluorescence photons at the acceptor emission wavelengths. The transfer efficiency has been found to be highly dependent upon the donor-acceptor distance. Therefore, FRET has been commonly used as a tool for investigating inter- or intramolecular distances at a level of nanometers.1,2
To efficiently observe FRET, donors and acceptors are usually linked via various conjugation structures such as amine-carboxyl reaction, thiol bonding, and bioaffinity binding.3–5 Among numerous bioconjugation structures, streptavidin (SA)-biotin binding has been widely used because of its high affinity and specificity, facile nature, and commercial availability.6–8 For example, dye-labeled biotinylated deoxyribose nucleic acid (DNA) molecules (biotin-DNA-dye) have been attached to a streptavidin-coated quantum dot (QD-SA) for FRET-based biomedical imaging9 and sensing.8,10–12 In the structure of (QD-SA)-(biotin-DNA-dye), QD and dye serve as a donor and an acceptor, respectively. Two major advantages of this system are the large quantum efficiency of the QD and the capability of controlling the donor-acceptor distance by varying the number of bases (single-stranded) or base pairs (double-stranded) of the DNA molecule (and, therefore, the length of DNA).
Studies have observed distance-dependent FRET between the QD and the dye when changing the number of the DNA bases (or base pairs),13 and have also utilized FRET data for single-molecule sensing12 and quantitative analysis of inter- or intramolecular distance.13–15 These results led to an assumed architecture for the (QD-SA)-(biotin-DNA-dye) system, which stated that “the dye acceptors on the DNA will be located at a uniform set of centrosymmetric distances from the central QD.”4,16 Based on this model, the FRET should depend on the DNA length or base number (or base pairs). However, a recent study found a contradictory result. In this study, the FRET was found to be independent of the number of DNA base pairs if the (QD-SA)-(biotin-DNA-dye) system was adopted in phosphate-buffered saline (PBS) solution.4,16 Therefore, a conclusion was drawn in the study that the (QD-SA)-(biotin-DNA-dye) system might have a very different architecture compared to the initially assumed one. A new architecture model for the (QD-SA)-(biotin-DNA-dye) system was proposed based on the fact that each SA has four biotin binding sites. This new structure proposed that some biotinylated DNA molecules might radially extend outward from the SA-coated QD surface and others could tangentially attach onto the SA-coated QD surface (see Fig. 5 in Ref. 16). The latter is the reason why some dyes might always be in close proximity to the QD surface so that the FRET becomes independent of the DNA base pairs. Thus, the authors questioned the use of an SA-biotin conjugation for linking a QD and DNAs for distance-dependent FRET applications.4,16
To further understand the (QD-SA)-(biotin-DNA-dye) system, we investigated it from a different perspective (i.e., ionic strength). Our data showed that the contradictory results in previous publications might be caused by the chemical-physical microenvironment of the DNA molecules as a result of the adoption of different buffer solutions (such as borate, Tris, PBS, and TE buffers). Therefore, we reevaluated the SA-biotin conjugation in FRET applications under varying ionic strength conditions. The finding in the current work reveals that the controversial results between length-dependent and -independent FRET are mainly attributed to the ionic strength of the adopted buffer solutions. Accordingly, we conclude that the system of (QD-SA)-(biotin-DNA-dye) is appropriate for investigating distance-dependent FRET if buffer solutions are appropriately selected.
2. Materials and Methods
2.1. Materials
2.1.1. SA-coated QDs
The SA-coated QDs (QD655-SA, peak emission: 655 nm) were purchased from Life Technologies (Grand Island, New York). The QD655-SA are expected to be 15 to 20 nm in size, with each functionalized with 5 to 10 SA molecules according to the production information provided by the vendor.17
2.1.2. Biotinylated oligonucleotides labeled with Alexa Fluor 750
In total, six strands of 3′-biotinylated oligonucleotides (10, 25, 32, 40, 50, and 70 bases) and one additional strand of nonbiotinylated oligonucleotide (50 bases) were obtained from Integrated DNA Technology Inc. (Coralville, Iowa). All oligonucleotides were labeled with Alexa Fluor 750 [N-Hydroxysuccinimide (NHS) ester] at the 5′-end. The sequences of the oligonucleotides were: (1) 5-/5Alex750N/CAA CAATAC A/3Bio/-3; (2) 5-/5Alex750N/CAA CAATAC ATC ATC TAC CAT CAT C/3Bio/-3; (3) 5-/5Alex750N/CAA CAATAC ATC ATC TAC CAT CAT CCA ACA AT/3Bio/-3; (4) 5-/5Alex750N/CAA CAATAC ATC ATC TAC CAT CAT CCA ACA ATA CAT CAT C/3Bio/-3; (5) 5-/5Alex750N/CAA CAATAC ATC ATC TAC CAT CAT CCA ACA ATA CAT CAT CTA CCATCA TC/3Bio/-3; (6) 5-/5Alex750N/CAA CAATAC ATC ATC TAC CAT CAT CCA ACA ATA CAT CAT CTA CCATCA TCC AAC AAT ACA TCA TCT ACC A/3Bio/-3; and (7) 5-/5Alex750N/CAA CAATAC ATC ATC TAC CAT CAT CCA ACA ATA CAT CAT CTA CCATCA TC/-3. Biotinylated oligonucleotides labeled with Alexa Fluor 750 were denoted as BOAF.
2.1.3. Buffer solutions
The QD’s incubation buffer [2% w/v bovine serum albumin (BSA) in 50 mM borate buffer, pH 8.3, containing 0.05% sodium azide] was purchased from Life Technologies (Grand Island, New York), and was denoted as a borate buffer. PBS packs (8 mM sodium phosphate, 2 mM potassium phosphate, 0.14 M NaCl, 10 mM KCl, pH 7.4) were purchased from Thermo Scientific (Rockford, Illinois). Tris (hydroxymethyl) aminomethane (Tris) was obtained from Bio-Rad Laboratories, Inc. (Hercules, California). Tris buffer (10 mM Tris, adjusting the pH to 8.0 with 1 M HCl), was prepared and used in the experiments. A TE buffer [10 mM Tris and 1 mM ethylenediaminetetraacetic acid (EDTA) pH 7.5] was purchased from USB Corporation (Cleveland, Ohio). Two percent BSA (weight-volume ratio) were added into PBS, Tris, and TE buffers when used for conjugation.
2.2. Methods
2.2.1. Sample preparation
Samples in the present work were prepared via three steps: conjugation between QD-SA and biotin-DNA-dye, separation of free biotin-DNA-dye from the conjugates, and dilution of the conjugates. Note that the buffer for conjugation was denoted as the “C-buffer” and the buffer used to dilute the sample (by 30-fold in volume) prior to measurement was denoted as the “D-buffer.”
A general procedure for sample preparation is described as follows. In of C-buffer, 1 pmol of QD655-SA was mixed with 10 pmol of BOAF. The sample was placed on the shaker (ThermoScientific, Asheville, North Carolina) at for 45 min at room temperature. After conjugation, the sample was filtered using a 100 K molecular weight cut-off centrifugal filter (DNA FastFlow Device, purchased from EMD Millipore Ltd., Billerica, Massachusetts) to remove the unbound DNA molecules at for 20 min. The filtered sample was collected at for 3 min according to the manual. The resulting sample was diluted to 3 mL with the D-buffer.
2.2.2. Fluorescence lifetime measurement
A custom-built fluorescence lifetime measurement system was employed to measure the change in the donor’s and acceptor’s lifetimes caused by FRET (Fig. 1). A nitrogen laser and a pumped-dye laser (nitrogen laser: OL-4300, dye laser: OL-401, both from Optical Building Blocks Corporation, Birmingham, New Jersey) were used to generate a light pulse at 490 nm with a pulse width of , using the OD 481 dye (Optical Building Blocks Corporation). Collimation of the laser beam was achieved using two lenses (L1 and L2, AC254-030, Thorlabs Inc., Newton, New Jersey). The intensity of the laser beam was controlled by a rotational neutral-density filter (Thorlabs Inc.), and a third lens (L3, A230TM-A, Thorlabs Inc.) was used to couple the beam into a multimode optical fiber (FT200EMT, Thorlabs Inc.). A laser beam from the fiber was then collimated through a lens (L4, A230TM-A, Thorlabs Inc.) to excite the sample.
Fig. 1.
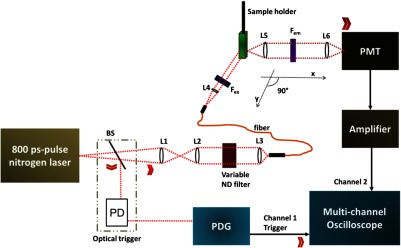
A schematic diagram showing the fluorescence lifetime measurement system. , excitation filters; , emission filters; L, lens; PMT, photomultiplier tube; BS, beam splitter; PD, photodiode; PDG, pulse-delay generator; and ND filter, natural density filter.
The spectral overlap of the donor (QD655) and the acceptor (Alexa Fluor 750) as an FRET pair is shown in Fig. 2. The overlap between the QD655 emission (solid blue line) and the AF750 excitation (the dashed red line) spectra can be clearly seen; this is not significant but is acceptable. The relatively small spectral overlap between QD655 and AF750 may lead to a relatively small FRET efficiency. However, this can be easily compensated by our sensitive detection system and the averaging method via multiple measurements. In our experiment, the FRET signals have been clearly and stably measured, while the donor’s bleed-through was kept to a the minimum. A bandpass filter with a central wavelength of 485 nm and a bandwidth of 20 nm (FF02-485/20, Semrock, Semrock, New York) was placed immediately after the lens to further clean the laser spectrum. Three neutral-density filters (OD 0.3, 0.4, and 0.6, Edmund Optics Inc., Barrington, New Jersey) were attached to the aforementioned bandpass filter only when measuring the QD655 emission to prevent the PMT from possible saturation as a result of the much stronger fluorescence emission from QD655 than from AF750. The emission filters for AF750 and QD655 are a bandpass filter with a central wavelength of 785 nm and a bandwidth of 62 nm (FF01-785/62, Semrock), and a bandpass filter with a central wavelength of 650 nm and a bandwidth of 60 nm (FF01-650/60, Semrock), respectively. A filter wheel (CFW-6, Thorlabs Inc.) was used to switch between the two emission filters. A pulse energy meter system (J-10MT-10KHZ EnergyMax sensor or J-10Si-HE Quantum EnergyMax sensor with the Labmax-Top laser power/energy meter, Santa Clara, Sunnyvale, California) was used to measure the laser pulse energy. The emitted fluorescence photons were detected by a photomultiplier tube (PMT, H10721-20, Hamamatsu, Japan). A broadband preamplifier (C5594, bandwidth from 50 kHz to 1.5 GHz, Hamamatsu, Japan) was used to convert the output of the PMT to a voltage signal and further amplify the signal. The voltage signal was then acquired by a multichannel and broadband oscilloscope (DPO 7254, 2.5 GHz, Tektronix, Beaveton, Oregon). The laser pulse was coupled into a 20-m long optical fiber (FT200EMT, Thorlabs Inc.) to generate an optical delay of about 100 ns, which was sufficient time for the electronic devices to respond for the best synchronization between the fluorescence emission pulses and data acquisitions. An optical trigger device was placed between the laser system and the fiber coupling system to split a small amount of the excitation light from the laser and generate a voltage pulse via a fast photodiode. This voltage pulse was used to trigger a digital pulse-delay generator (PDG, DG645, Stanford Research Systems, Sunnyvale, California) and further trigger the oscilloscope for data acquisition. This setup provided very high accuracy of the synchronization between the fluorescence pulse and data acquisition, therefore, a large amount of fluorescence pulses could be acquired and averaged to improve the signal-noise ratio. Each emission decay pulse recorded from the oscilloscope used for calculating fluorescence lifetime was an average of 100 times of the excitation events. For each sample, the experiment was repeated at least two times (therefore, the total number of the average is equivalent to at least 200). The mean and standard derivation (std) are calculated for each sample and are shown in the figures (i.e., ). Each sample was diluted to 3 mL and injected into a quartz cuvette (Starna Cells, Atascadero, California) that was perpendicularly fixed to the path of the laser beam.
Fig. 2.
Plot showing the spectral overlap of the donor (QD655) and the acceptor (Alexa Fluor 750) as an FRET pair.
2.2.3. Fluorescence lifetime calculation
The system impulse response function was measured and used to deconvolve the acquired fluorescence decay data. The processed data were fitted to a single exponential decay function using MATLAB (Natick, Massachusetts). The fitting was done by an iterative numerical procedure until the best agreement with the experimental decay curve was achieved.
2.2.4. AFM measurement
A aliquot of each of the conjugated 70-base B2T and B2P (B2T: the sample was incubated in borate buffer and then diluted with Tris buffer. B2P: the sample was incubated in borate buffer and then diluted with PBS buffer) samples were deposited on a biotinylated coverslip, purchased from Microsurface, Inc. (#Bio_02, Englewood, New Jersey) and kept overnight in a humidified chamber at room temperature. The resulting coverslip was rinsed three times with the same D-buffer (Tris for B2T and PBS for B2P), shaken () for 3 min, and dried in the desiccator with vacuum. The measurement was performed with a Park XE70 AFM (Santa Clara, California), using a noncontact mode with ACTA probe. At least 10 locations were selected to measure each sample. “The root-mean-square (RMS) of the image roughness was calculated since it is a commonly used parameter in AFM to quantify the height, which is more sensitive to peaks and valleys than the average roughness.”18
2.3. System of (QD-SA)-(biotin-DNA-dye)
As mentioned in Ref. 16, two possible binding modes might exist between a biotinylated DNA and an SA attached on a QD: standing-up and laying-down [Fig. 3(a)]. A significant FRET independent of the number of DNA base pairs was observed and attributed to the possibility of the laying-down binding mode. In this mode, the acceptors labeled on the DNA chains would stay within a much tighter area around the QD donor than they would in the standing-up mode [see Fig. 3(a)]. After an intensive investigation, however, we suspected that conclusion because the model did not take into consideration the steric hindrance from the adjacent SAs and the QD. The existence of the neighboring SAs on the QD might greatly reduce the possibility of the laying-down binding mode because the space between each two adjacent SA molecules may be limited for fitting of the DNA or oligonucleotide. Instead, we found that the observed base pair-independent FRET should be attributed to the buffer ionic strength which could significantly affect the FRET of the (QD-SA)-(biotin-DNA-dye) system.
Fig. 3.
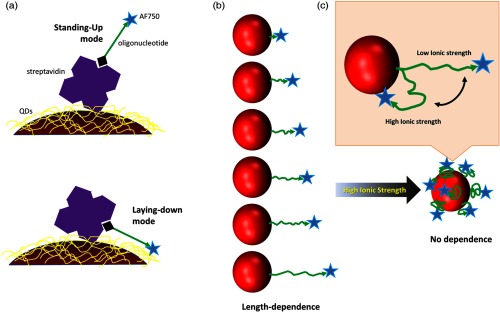
(a) Illustration of two binding modes of biotinylated oligonucleotide to streptavidin-coated quantum dots (QDs). Not to scale. (b and c) Illustration of our hypothesis in this work. At low-ionic strength, oligonucleotide stretches from the QDs, leading to length-dependent FRET. At high-ionic strength, oligonucleotide is more flexible but is prone to bending toward QDs. The separation distance between donor and acceptor becomes smaller, falling well into the FRET range, so that no length-dependent FRET was observed.
SA-coated QD655 (QD655-SA), which has a structure similar to QD609-SA used in Ref. 16 with a longer peak emission wavelength (655 nm), was selected as a donor in this study. A near-infrared fluorophore, Alexa Fluor 750 (AF750), was adopted as the acceptor based on the reasonable spectral overlap with the QD655-SA donor (see Fig. 2). Although a biotin was attached to the 3′-end of an oligonucleotide (a single-stranded DNA), the fluorophore of AF750 was labeled at the 5′-end. The AF750-labeled biotinylated oligonucleotide was denoted as BOAF. Six oligonucleotides with base numbers of 10, 25, 32, 41, 50, and 70 were adopted (see details in Sec. 2). After conjugation, a QD655–SA∼BOAF structure was formed. While the bindings of biotin to SA occur in the C-buffer, the oligonucleotide conformation (stretched or coiled) depends on the D-buffer. It is reasonable to hypothesize that the donor-acceptor distance will correlate with the base number of BOAF if the standing-up binding mode is dominant and the oligonucleotide is in the stretched conformation [see Fig. 3(b)]. Therefore, the measured FRET (indicated by the AF750 lifetime in this study) will depend on the base number of the oligonucleotide. Whenever the oligonucleotide transits from the stretched to the coiled structure as a result of the change of the D-buffer, the donor-acceptor distance and thus the FRET will lose correlation with the base number [see Fig. 3(c)]. By contrast, if the laying-down mode is dominant, the FRET will not (or will very weakly) correlate with the base number, no matter the conformation of the oligonucleotide and the composition of the buffers. To characterize the FRET event, the fluorescence lifetimes of both the acceptor AF750 and the donor QD655 were measured with a customized instrument (see Sec. 2.2.2 for details). Compared with intensity, the fluorescence lifetime is insensitive to the errors caused by unknown fluorophore concentration variation, and is, therefore, more reliable. A molar ratio of QD655-SA to BOAF at was employed in the present study. Since each QD has 6 to 10 SA molecules according to the provided information from the vendor, Life Technologies, the molar amount of SA is 6 to 10 pmol. Consequently, the biotin-SA ratio in this study was to 1.67. Such a relatively low ratio has the following advantages: (1) it can reduce the possibility of multiple BOAFs binding on one SA; and (2) the effect of acceptor concentration on the FRET is minimized and the donor-acceptor distance is the major factor affecting the FRET.
3. Results and Discussion
3.1. Fluorescence Lifetime Calculation
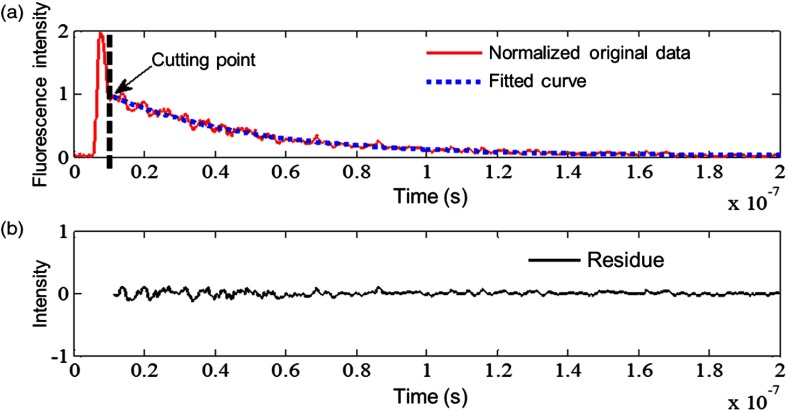
A tail fitting strategy was adopted to maximally remove two possible noises: (1) the laser leakage and (2) the emission from the acceptors (AF750) that are directly excited by the laser. The raw decay curve was truncated after an inflexion point and then data-fitting via a single exponential function was performed. As an example, Fig. 4 shows the step-by-step process for the 25 base borate (C-buffer) to the Tris (D-buffer) sample. The cutting point was selected based on the original decay curve. The lifetime was fitted by using the data after the cutting point. Remember that both the laser leakage and the direct excitation of the acceptors decay much faster than the FRET signal. Therefore, the head part of the decay curve is the data that can possibly be contaminated by the two types of noise.
Fig. 4.
Fluorescence lifetime calculation method. (a) Illustration of the decay curve for 25-base BOAF with borate as C-buffer and Tris as D-buffer and a single exponential tail fitting by setting a cut-off point. (b) Residues are shown versus time.
3.2. Effect of Buffers (ionic strength) on FRET
When borate was used as a C-buffer and Tris as a D-buffer, for oligonucleotides with 10 bases, the fluorescence lifetime of the acceptor AF750 was found to be [Fig. 5(a)], which was significantly longer than the natural fluorescence lifetime of AF750 ().19 There exists documented evidence that the lifetime of the acceptor increases in an FRET system,20,21 although it has been less rigorously studied than the increase of the donor’s lifetime. On the other hand, the average lifetime of the donor QD655 for all the bases was measured to be 38.8 ns [Fig. 5(b)], consistent with the value given by the manufacturer ( to 40 ns). However, it should be noticed that the donor’s lifetime decreased when the acceptor (AF750)-labeled 10-base DNA linker was used. The lifetime of QD is decreased from 38.8 to 34 ns in Tris, while decreased from 38.8 to 32 ns in PBS [Fig. 5(b)]. The result indicated an obvious FRET occurrence between QD655 and AF750. The lifetime reduction of the donor (from 38.8 to 32 or 34 ns) is not as significant as the lifetime lengthening of the acceptor (from to ). This is mainly attributed to the following factors: (1) the relatively low () acceptor per donor (APD) ratio in the conjugation step; (2) significantly higher extinction coefficient and quantum yield of the donor (QD655) compared to the acceptor (AF750). The above factors contribute to the observation that the donor’s lifetime had not been significantly affected by the acceptors.
Fig. 5.
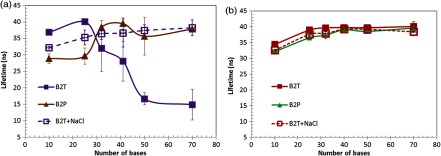
Fluorescence lifetimes of (a) the acceptor AF750 and (b) the donor QD655-SA vary as the base number of oligonucleotide increases. Excitation: 490 nm; emission: bandpass filter for (a) and bandpass filter for (b). B2T: the sample was incubated in borate buffer and then diluted with Tris buffer. B2P: the sample was incubated in borate buffer and then diluted with PBS buffer. B2T + NaCl: 140 mM NaCl was added into the B2T sample.
Apparently, the lifetime of AF750 in QD655–SA∼BOAF was found to gradually decrease as the base number increased, as shown by the purple curve with filled squares in Fig. 5(a). When the base number is greater than 50, no significant change in the lifetime of AF750 is observed, which indicates that acceptors may be out of the FRET range and/or the background fluorescence is dominant. Therefore, the length-dependent FRET was observed in our system of QD655–SA∼BOAF. This result provides strong evidence for our hypothesis of the dominant standing-up mode in Fig. 3(a). The observed lifetimes of and for the 50 and 70 bases samples could be partially attributed to the background fluorescence noise (see Sec. 3.3). On the other hand, it has been observed that when some gold nanoparticles22 or QDs23 are adopted as donors (and dyes as acceptors), the FRET distance is much longer than the conventional FRET distance between an organic dye donor-acceptor pair. However, it is hard to quantify which mechanisms are dominant. This change may be caused by either one or both. Figure 5(b) shows that the lifetime of the donor QD655 is relatively stable for all oligonucleotide sequences (with an average lifetime 38.8 ns and varying within a range of ). This is mainly attributed to the fact that the ratio of acceptor-donor was relatively low in the conjugation and that the donor’s lifetime had not been significantly affected by the acceptors.
Interestingly, the length-dependent FRET disappears when the Tris is replaced with PBS as the D-buffer [the brown curve with solid triangles in Fig. 6(a)]. The lifetimes of AF750 at 50 and 70 bases significantly increase compared with those measured in the Tris buffer. The lifetime fluctuates when the base increases (in a range of with an average of 35 ns), suggesting a large FRET occurrence. This result implies that the length-dependent FRET is favorable in a relatively low-ionic-strength buffer (borate and Tris, 10 mM) as opposed to the length independence in a relatively high-ionic-strength buffer (PBS, 162.7 mM). To further investigate the effect of ionic strength on the length-dependent or -independent FRET, we implemented a straightforward experiment of adding ca. 140 mM NaCl to the B2T samples (C-buffer: borate; D-buffer: Tris) and measuring the lifetime of AF750 again. As shown by the dashed purple curve with open squares in Fig. 5(a), the lifetime of AF750 loses the length-dependence that is originally observed in the Tris buffer and remains a relative stable value for all oligonucleotide sequences (with an average of 36.5 ns and varies within a range of ). This length-independent FRET resembles the result observed in the PBS buffer. Accordingly, we conclude that the length-dependent FRET is favorable in low-ionic-strength buffers, such as 10 mM Tris, and unfavorable in relatively high-ionic-strength buffers, such as PBS or Tris + NaCl. It has been reported that oligonucleotide becomes more flexible when the ionic strength increases.14,24 The reason for this is that excess cations screen the negatively charged backbone of the oligonucleotide. At a low-ionic strength, oligonucleotide stretches outward from the QD655-SA, leading to the length-dependent FRET [Fig. 3(b)]. At a high-ionic strength, oligonucleotide is of high flexibility, making it prone to bend toward QD655-SA [Fig. 3(c)] and lose the distance dependence. We also excluded the effect of pH on FRET by adjusting the pH from 6.0 to 9.0 (data not shown), although the difference in pH between the Tris buffer (8.0) and the PBS buffer (7.4) used in this study was not significant.
Fig. 6.

Fluorescence lifetimes of the acceptor AF750 in QD655–SA∼BOAF with varying base numbers in different buffer solutions. T2T: the samples were conjugated in Tris buffer and then diluted with Tris buffer. P2P: the samples were conjugated in PBS buffer and then diluted with PBS buffer. T2P: the samples were conjugated in Tris buffer and then diluted with PBS buffer. P2T: the samples were conjugated in PBS buffer and then diluted with Tris buffer. The excitation light and emission filters are the same as in Fig. 5.
While the conformation of oligonucleotide can be altered at a high-ionic strength, another question is whether such a conformation change will hinder the binding between BOAF and QD655-SA in the initiate conjugation step (in C-buffer). To address this question, we investigated the lifetime change of AF750 in different combinations of Tris and PBS buffer solutions (as C- or D-buffer, or both). Note that the total volume for conjugation is while the final diluted solution is (30-fold dilution). As shown in Fig. 6(a), a length-dependence of AF750 lifetime was observed for the T2T sample (Tris as both C- and D-buffers), while no dependence for the P2P sample (PBS as both C- and D-buffers) was found. When the sample was conjugated in Tris and then diluted with PBS buffer (T2P), the dependence disappeared [Fig. 6(b) brown curve with solid circles]. By contrast, when the sample was conjugated in PBS and diluted with the Tris buffer (P2T), the length-dependence of the AF750 lifetime reappeared [the purple curve with open circles in Fig. 6(b)]. Taken together, the use of PBS as a C-buffer is not able to significantly affect the binding between BOAF and QD655-SA. It is worth noting that adding 2% (weight-volume ratio) BSA into the C-buffer is necessary to maintain good stability of the QD655-SA and a relatively high viscosity of the solution for the centrifugal filtering process (see Sec. 2 for details).
As long as the D-buffer (or the final solution) is Tris with an ionic strength of , a length-dependent FRET is observable. In fact, such a low-ionic-strength Tris buffer has been widely used in the literature for investigating the DNA linked FRET system.4,8,10,11,15 Like Tris, TE is another commonly used buffer with a lower ionic strength compared to PBS. We also obtained similar results when replacing Tris with the TE buffer (Figs. 7 and 8).
Fig. 7.
Fluorescence lifetime of the acceptor AF750 in QD655–SA∼BOAF with varying base numbers in buffer solutions of B2Te and B2Te + NaCl. B2Te: the sample was incubated in borate buffer and then diluted with TE buffer. B2Te + NaCl: 140 mM NaCl was added into the B2Te sample. Excitation: 490 nm; emission: bandpass filter.
Fig. 8.

Fluorescence lifetime of the acceptor AF750 in QD655–SA∼BOAF with varying base numbers in buffer solutions of (a) Te2Te and (b) Te2P and P2Te. Te2Te: the sample was incubated in TE buffer and then diluted with TE buffer. Te2P: the sample was incubated in TE buffer and then diluted with PBS buffer. P2Te: the sample was incubated in PBS buffer and then diluted with TE buffer. Excitation: 490 nm; emission: bandpass filter.
Currently, it is unclear why the samples with 10 bases do not show the highest value in the acceptor’s lifetime in Figs. 5(a) and 8(a). Possible reasons may include: (1) self-quenching of acceptors may happen because the average distance between two adjacent acceptors becomes short when the DNA base is small; (2) FRET may become saturated if the donor-acceptor distance is so short; and (3) some system measurement errors may exist.
3.3. Background Noise of Fluorescence (Measured From Control Samples)
The lifetime of the AF750 with all the different lengths of the biotinylated DNA molecule along with the nonbiotinylated DNA was measured in PBS, Tris, and TE as a control of the acceptor’s lifetime (Fig. 9). The calculated lifetimes are , are independent of the number of bases, and are close to the natural lifetime of AF at 0.7 ns.
Fig. 9.
Lifetime of Alexa Fluor 750 in Tris, TE, and PBS versus the number of DNA bases for (a) biotinylated DNA and (b) 50 bases nonbiotinylated DNA.
To further evaluate the background fluorescent noise, another two control samples were adopted: (1) QD655-SA alone and (2) QD655-SA with nonbiotinylated AF750-labeled oligonucleotides (50 bases). In Fig. 10(a), the lifetimes of the QD655-SA alone samples in B2T and B2P are shown to be and , respectively, when measured using the AF750 emission filter (). They are much shorter than the lifetime of the QD655-SA alone samples ( or ) in Fig. 10(b) when measured using the QD emission filter (). However, they are much longer than the laser pulse width (). Therefore, these results indicate that a small amount of fluorescence can be emitted from the QD655-SA itself within the spectrum band of the AF750 emission filter (so called bleed-through from the donor to the acceptor channel). However, we found that the intensity of the bleed-through is usually much weaker compared with the FRET signal from the BOAF with bases (remember that the fluorescence lifetime is calculated based on the emission decay curve). Accordingly, the effect of these fluorescence noises on the FRET can be ignored. Therefore, we conclude that the direct emission from QDs may be one of the dominant sources of the received photons when the measured lifetime is around or below 7.3 ns.
Fig. 10.
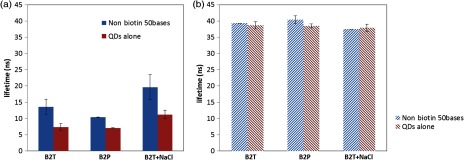
Fluorescence lifetime at the acceptor’s (a) and the donor’s emission channels (b). Blue: 50-base oligonucleotide without biotin; red: QD655-SA alone. Excitation: 490 nm; emission: bandpass filter for (a) and bandpass filter for (b). Samples of B2T, B2P, and B2T + NaCl are the same as in Fig. 5.
After the QD655-SA was mixed with the nonbiotinylated AF750-labeled oligonucleotides, the same procedures as those used for processing the biotinylated samples were followed. Compared with the QD655-SA alone samples, the lifetimes in B2T and B2P increase [ and , respectively, see Fig. 10(a)]. However, these two values are much larger than the natural lifetime of AF750 (). This is mainly attributed to the FRET between the QD655-SA and the remaining AF750 in the solution (although the same filtering steps have adopted, a small amount of AF750-labeled oligonucleotides may remain in the solution). This type of FRET is strongly dependent on the concentration of AF750 instead of the base numbers of the oligonucleotides. After the filtering, the amount of the remaining AF750-labeled oligonucleotide is usually small, and the intensity of this type of FRET is also much weaker than the FRET intensity from the SA-biotin conjugated samples with bases . Consequently, this type of FRET is not dominant until the bases are close to or above 50. Similarly, we conclude that this type of FRET may be another dominant source of the received photons when the measured lifetime is around or below 13.6 ns. It is worth pointing out that the required concentration to generate this type FRET should be significantly smaller than that of the FRET occurring between a dye-dye pair considering the large extinction coefficient, quantum yield, the long FRET distance,23 the lifetime of the QDs, and the sensitive measurement system.
On the other hand, the QD655-SA lifetime measured in the spectrum of QD655 emission filter remains stable [ in B2T and in B2P, see Fig. 10(b)], which indicates that the remaining AF750 does not significantly change the QD655 fluorescence emission properties. Similar results were found in samples where NaCl was added [see Figs. 10(a) and 10(b)].
3.4. Discussion About Using the Acceptor Lifetime as FRET Indicator
Conventionally, when lifetime is used for FRET studies, the shortening of the donor’s lifetime (instead of the lengthening of the acceptor’s lifetime) is used as the FRET indictor if the donor and acceptor have comparable lifetimes, extinction coefficients, quantum yields, and widths of the emission spectra. This is mainly due to two reasons. (1) Usually, the donor channel has a relatively weaker bleed-through from the acceptor emission than the acceptor channel has from the donor emission. This is because the wavelength of the laser is usually close to the excitation peak wavelength of the donor, but is far away from that of the acceptor, therefore, the donor emission is usually stronger than the acceptor emission. (2) The donor is less likely to be excited (and further emit light) by the emission light of the acceptor because there is usually none or an ignorable spectral overlap between the acceptor emission and donor excitation spectra. In contrast, it is possible that some acceptors are directly excited (and further emit light) by the emission light of the donors because of the spectral overlap between the donor emission and acceptor excitation spectra. This type of phenomenon can be called reabsorption-and-reemission and is different from FRET (FRET does not involve donor emission).
However, compared with the acceptor, if the donor has a much narrower emission spectrum, a much longer lifetime, a much larger extinction coefficient, and a much higher quantum yield, the lengthening of the acceptor’s lifetime can be used and may be even better as an FRET indicator.20,21 This is true for the FRET system adopted in this study (QD655-DNA-AF750). The above parameters for QD655 and AF750 are listed in Table 1 for comparison.
Table 1.
Comparison of fluorescence parameters of the donor and acceptor studied in the current work.a
| Width of the emission spectrum (nm) | Lifetime (ns) | Quantum yield | Extinction coefficient () | ||
|---|---|---|---|---|---|
| FWHMb | FWTMc | ||||
| QD655 (donor) | 0.6 | 2,900,000 at 488 nm | |||
| AF750 (acceptor) | 0.7 | 0.12 | 240,000 at 749 nm | ||
See the data from the manufacturer.17
Full width at the half maximum.
Full width at one-tenth of the maximum.
The concern of the donor’s (QD655) bleed-through to the acceptor (AF750) channel can be efficiently minimized because QD655 has a narrow emission spectrum and almost no overlap with the pass band of the emission filter of the acceptor channel. This has been validated by the control sample (i.e., the sample with only QDs) in which the contribution of this type of noise is weak. In addition, the effect of the fluorescence emission caused by the direct excitation of the acceptor AF750 on the FRET signal can be very easily eliminated because its lifetime and, therefore, the emission decay is much shorter than the FRET based emission pulse. This type of fluorescence noise can be easily eliminated by using a time gating method to get rid of the head part of the raw data. Regarding the effect of the reabsorption-and-reemission on FRET, our data clearly showed that it is ignorable in the adopted QD655-DNA-AF750 system. This can be seen from the following facts. (1) If the effect of the reabsorption-and-reemission was dominant, the acceptor lifetime should be . However, when QD655 is mixed with nonbiotin DNA-dye, the measured acceptor lifetime is between 10 and 20 ns (see the data in Fig. 10), which is much shorter than the QD655 lifetime (). Therefore, the measured signal should not be caused by the reabsorption-and-reemission effect. Instead, it is mainly caused by the slight bleed-through from the QD655. In Figs. 5 and 7, the dependence of the acceptor lifetime on the DNA length is eliminated when adding NaCl into the sample. This fact indicates that the measured signal from the acceptor channel should not be caused by the reabsorption-and-reemission phenomenon because this phenomenon is independent of NaCl concentration. Therefore, the effect of reabsorption-and-reemission is ignorable. The following reasons may explain why the effect of the reabsorption-and-reemission is ignorable in the adopted QD655-DNA-AF750: (a) both the donor and acceptor have a very low concentration (1 pmol donor and 10 pmol acceptor) in our sample; (b) the acceptor has a low quantum yield and small extinction coefficient; and (c) the spectral overlap between the donor and acceptor is small.
Here, we summarize and compare the possible signal components in the acceptor channel of the adopted QD655-DNA-AF750: (a) when the acceptors (AF750) are well attached on the donor (QD655) and they are within the FRET distance range, the FRET is the dominant effect; (b) when the acceptors are attached on the donor but are separated so far that they are out of the FRET distance, two possible effects may be dominant: the bleed-through from donor (QD655) to acceptor (AF750) and the FRET between the donors and those unattached free acceptors (i.e., the residue of the free acceptors, because they are free and have a small possibility of reaching the vicinity of the donor); (c) the reabsorption-and-reemission may be the weakest effect, and it does not show an observable effect in our results.
In contrast, using the donor’s lifetime shortening as the FRET indicator has some disadvantages. Specifically, to shorten a bright QD’s long lifetime (QD655 ), a large amount of acceptors (AF750) is usually needed. This is because: (a) the donor (QDs) usually has a much larger quantum yield and extinction coefficient, and a much longer lifetime than the acceptor (AF750); (b) all or the majority of donors (QDs) should have enough acceptors (AF750) to generate an obvious FRET (in contrast, if only a small portion of QDs are quenched by the AF750, the lifetime will not significantly decrease).
When a large amount of acceptor (AF750) is used, it can generate some problems. (a) More residue of the free AF750 (unattached DNA-AF750) may exist in the sample (although a centrifugal filter was always used to get rid of the free DNA-AF750 as much as possible). These free dyes in the sample would also shorten the lifetime of the donor (QDs) (but not the FRET that we are investigating). Thus, it is difficult to differentiate the effect of the free dye from the effect of the attached dye. (b) Therefore, the donor’s lifetime will also depend on the acceptor concentration (the more AF750 residue in the sample gives the shorter of the donor’s lifetime), which can be confused with the FRET we are investigating via the QD-DNA-dye system.
More importantly, it may not be practical to attach a large amount of acceptors on one QD (donor). This is because the number of the SA on each QD655 is limited to 10 (based on manufacturer provided data). Although each SA has four biotin-binding sites, in practice it is highly possible that to 2 binding sites on each SA are not available for biotin-DNA-AF750 because they may be blocked by the QD and/or the surrounding other SAs due to steric hindrance. Thus, the number of the acceptors that can be attached on one donor is (). Therefore, achieving a large number of the APD may be impractical. Thus, if a significant reduction of the donor’s lifetime is found, it is highly possible that it is caused by the residue of the free acceptors, which has nothing to do with the FRET that we are investigating via the QD-DNA-dye system.
However, if one is using the lengthening of the acceptor’s lifetime as the FRET indicator, the FRET efficiency is very high because of the high quantum yield and extinction coefficient of the QDs. Therefore, we can maintain a small number of acceptors per donor (), which is practically possible, and then detect the lifetime change of the acceptor for FRET study. Thus, the residue of free acceptors (AF750) is small and the FRET is mainly depends on the donor-acceptor separation distance, which is what we want to investigate.
3.5. Further Evidence From Atomic Force Microscopy
To further verify our hypothesis, AFM was employed to investigate the attachment of the QD655–SA∼BOAF (70 bases) on the surface of a biotinylated coverslip (Bio_02, MicroSurfaces Inc., Englewood, New Jersey). Since the 70 bases linker caused no FRET in low salt solutions but did cause a dramatic FRET in high salt solutions, it was an all-or-nothing condition. Furthermore, it has a relatively long chain so that its conformation change could be possibly be detected by AFM. Compared with the biotinylated oligonucleotides, the biotins on the coverslip have much smaller dimensions. Thus, the steric hindrance from the surrounding SAs on the QD655 can be dramatically reduced. In addition, because of the low ratio of BOAF to SA used in the present study, there are unoccupied binding sites of SA in QD655–SA∼BOAF. They will possibly be available for the biotin moiety on the surface of the coverslip so that QD655–SA∼BOAF may be immobilized on the surface.
As shown in Fig. 11(a), few QD655–SA∼BOAF were observed on the surface when the biotinylated coverslip was incubated with the B2T conjugated sample (10 locations were measured and compared). The RMS of the B2T sample was estimated to be 0.765 nm, which does not significantly differ from that of the original biotinylated coverslip sample [0.619 nm, Fig. 11(c)]. On the contrary, the RMS value was found to increase remarkably to 1.988 nm when incubating with the B2P sample [Fig. 11(b)]. Furthermore, the observed peak heights of the biotinylated chains on the coverslip (coated by the manufacturer) were found to be 10 to 18 nm [see Fig. 11(c)]. After conjugation with QD655–SA∼BOAF, we found that the height values of the peaks for the B2P sample increased by 15 to 20 nm [Fig. 11(b)], which fits well in the diameter range of QD655-SA according to the vendor’s manual. Because the sample was measured after the buffer solution had dried, the height contributed by oligonucleotide molecules could hardly be observed. Consequently, we conclude that many more QD655–SA∼BOAFs are attached on the coverslip surface in the B2P buffer than in the B2T buffer. This result indicates that the binding between the SA on QD655 and the biotin on the coverslip was more feasible in the B2P sample (with high-ionic strength) than in the B2T sample (with high-ionic strength). Figure 11(d) shows a possible mechanism to explain the above result. For the B2T sample, the oligonucleotide molecules may be stretched-out because of the low-ionic strength of the buffers [see the left panel in Fig. 11(d)]. These stretched oligonucleotides may significantly reduce the possibility of the SAs on the QD655 conjugating with the biotins on the coverslip. In contrast, for the B2P sample, the oligonucleotide molecules may be significantly coiled because of the high-ionic strength of the buffers [see the right panel in Fig. 11(d)]. Thus, the possibility of the SAs on the QD655 being exposed to the biotins on the coverslip can dramatically increase, which leads to the immobilization of the QD655. The above result may be considered an additional evidence for verifying our hypothesis about the effect of the buffer ionic strength on the conformation change of the oligonucleotides.
Fig. 11.

Representative AFM images of biotinylated coverslips incubated with the conjugated solution of QD655–SA∼BOAF (70 bases). (a) B2T, (b) B2P samples (note that the peak heights are not limited by the maximum scale of 30 nm), (c) biotinylated coverslip alone. (d) Illustration of the attachment of QD655–SA∼BOAF (70 bases) on the biotinylated coverslip surface. At low-ionic strength (in Tris buffer), the available binding sites of streptavidin were blocked by stretched-out oligonucleotide molecules (left), leading to difficulty in attaching. At high-ionic strength (in PBS buffer), the binding sites were exposed (right), which facilitates the attachment.
4. Conclusions
In the buffers with low-ionic strength (such as borate, Tris, and TE buffer), length-dependent FRET between QD655-SA and BOAF was confirmed by the observation of the acceptor AF750s fluorescence lifetime. In the buffers with high-ionic strength (such as PBS and NaCl-added Tris), a strong length-independent FRET was observed. The independence was likely attributed to the increased flexibility of the oligonucleotide chain when the cations screen the negatively charged backbone of the oligonucleotide. This flexibility increases the possibility of the terminal-attached acceptors (AF750) approaching the vicinity of the donor. If the buffer solutions and the SA-biotin molecular ratio are appropriately selected, we draw the following conclusions based on the above data: (1) the system of (QD-SA)-(biotin-DNA-dye) is appropriate for investigating the distance-dependent FRET between the QD and the dye if they are linked by a single-stranded DNA, and (2) the effect of the SAs multiple binding modes on the FRET (proposed in Ref. 16) may not be dominant compared with the effect of the buffer ionic strength.
Acknowledgments
This work was supported in part by funding from the NIH/NIBIB 7R15EB012312-02 (B. Yuan), the CPRIT RP120052 (B. Yuan), and the NSF CBET-1253199 (B. Yuan). The authors are grateful to Dr. K. Nguyen and Dr. Y. Hong for sharing lab equipment for this study, and to Dr. Y. Pei for constructive discussion.
Biographies
Bahar Saremi received her BS degree from Amirkabir University of Technology in Iran and is currently working toward the PhD degree in bioengineering at the University of Texas at Arlington, Texas, USA. Her current research has been about developing contrast agents and investigating the properties of dyes with potential for further applications in fluorescence imaging systems.
Ming-Yuan Wei received the PhD degree in environmental science from the Chinese Academy of Science, Beijing, China, in 2009. He was a postdoctoral research associate at West Virginia University. In June 2012, he joined with Professor Baohong Yuan’s group in the Department of Bioengineering, University of Texas at Arlington, Arlington, Texas, USA. His current research interests include the development of contrast agents for ultrasound-switchable fluorescence imaging with thermoresponsive polymer or nanoparticles and environment-sensitive fluorophores.
Yuan Liu received the BS degree in biomedical engineering from Huazhong University of Science and Technology, Wuhan, China, in 2008, and the MS degree in biomedical engineering from The Catholic University of America, Washington, D.C., USA, in 2010. She is finishing her PhD degree in bioengineering at the University of Texas at Arlington, USA. Her research interests include developing ultrasound-mediated fluorescence imaging techniques, optical and ultrasound contrast agents, and imaging systems for fast microbubble motion detection.
Bingbing Cheng received the bachelor of engineering degree in biomedical engineering from Beijing Jiaotong University, Beijing, China, in 2011. He is currently working toward the PhD degree with Professor Baohong Yuan’s group in the Ultrasound and Optical Imaging Laboratory, Department of Biomedical Engineering, University of Texas at Arlington, USA. His current research interests include the development of ultrasound-switchable fluorescence imaging contrast agents based on thermosensitive polymer or nanoparticles and environment-sensitive fluorophores.
Baohong Yuan received the BS degree in microelectronics from the Harbin Institute of Technology, Harbin, China, in 1997, and the PhD degree in biomedical engineering from the University of Connecticut, USA, in 2006. He is currently an associate professor of biomedical engineering at the University of Texas at Arlington, USA. His research interest is to explore and develop new imaging technology, including contrast agents and instruments, for understanding cancer mechanisms, early detecting and diagnosing cancers, and monitoring cancer treatment efficiency.
References
- 1.Stryer L., “Fluorescence energy-transfer as a spectroscopic ruler,” Annu. Rev. Biochem. 47, 819–846 (1978). 10.1146/annurev.bi.47.070178.004131 [DOI] [PubMed] [Google Scholar]
- 2.Sahoo H., “Forster resonance energy transfer—a spectroscopic nanoruler: principle and applications,” J. Photochem. Photobiol. C 12(1), 20–30 (2011). 10.1016/j.jphotochemrev.2011.05.001 [DOI] [Google Scholar]
- 3.Algar W. R., Tavares A. J., Krull U. J., “Beyond labels: a review of the application of quantum dots as integrated components of assays, bioprobes, and biosensors utilizing optical transduction,” Anal. Chim. Acta 673(1), 1–25 (2010). 10.1016/j.aca.2010.05.026 [DOI] [PubMed] [Google Scholar]
- 4.Sapsford K. E., et al. , “Analyzing nanomaterial bioconjugates: a review of current and emerging purification and characterization techniques,” Anal. Chem. 83(12), 4453–4488 (2011). 10.1021/ac200853a [DOI] [PubMed] [Google Scholar]
- 5.Medintz I. L., et al. , “Quantum dot bioconjugates for imaging, labelling and sensing,” Nat. Mater. 4(6), 435–446 (2005). 10.1038/nmat1390 [DOI] [PubMed] [Google Scholar]
- 6.Goldman E. R., et al. , “Avidin: a natural bridge for quantum dot-antibody conjugates,” J. Am. Chem. Soc. 124(22), 6378–6382 (2002). 10.1021/ja0125570 [DOI] [PubMed] [Google Scholar]
- 7.Lidke D. S., et al. , “Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction,” Nat. Biotechnol. 22(2), 198–203 (2004). 10.1038/nbt929 [DOI] [PubMed] [Google Scholar]
- 8.Zhang C. Y., et al. , “Single-quantum-dot-based DNA nanosensor,” Nat. Mater. 4(11), 826–831 (2005). 10.1038/nmat1508 [DOI] [PubMed] [Google Scholar]
- 9.Roy R., Hohng S., Ha T., “A practical guide to single-molecule FRET,” Nat. Methods 5(6), 507–516 (2008). 10.1038/nmeth.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C. Y., Johnson L. W., “Microfluidic control of fluorescence resonance energy transfer: breaking the FRET limit,” Angew. Chem. Int. Ed. 46(19), 3482–3485 (2007). 10.1002/(ISSN)1521-3773 [DOI] [PubMed] [Google Scholar]
- 11.Levy M., Cater S. F., Ellington A. D., “Quantum-dot aptamer beacons for the detection of proteins,” Chembiochem 6(12), 2163–2166 (2005). 10.1002/cbic.v6:12 [DOI] [PubMed] [Google Scholar]
- 12.Zhang C. Y., Johnson L. W., “Quantum-dot-based nanosensor for RRE IIB RNA-Rev peptide interaction assay,” J. Am. Chem. Soc. 128(16), 5324–5325 (2006). 10.1021/ja060537y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy M. C., et al. , “Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy,” Biophys. J. 86(4), 2530–2537 (2004). 10.1016/S0006-3495(04)74308-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H., et al. , “Ionic strength-dependent persistence lengths of single-stranded RNA and DNA,” Proc. Natl. Acad. Sci. U. S. A. 109(3), 799–804 (2012). 10.1073/pnas.1119057109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohng S., Ha T., “Single-molecule quantum-dot fluorescence resonance energy transfer,” Chemphyschem 6(5), 956–960 (2005). 10.1002/(ISSN)1439-7641 [DOI] [PubMed] [Google Scholar]
- 16.Boeneman K., et al. , “Quantum dot DNA bioconjugates: attachment chemistry strongly influences the resulting composite architecture,” ACS Nano 4(12), 7253–7266 (2010). 10.1021/nn1021346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson I., Spence M. T. Z., “Molecular probes handbook, a guide to fluorescent probes and labeling technologies,” http://www.lifetechnologies.com/us/en/home/references/molecular-probes-the-handbook.html (28 July 2014).
- 18.Gadelmawla E. S., et al. , “Roughness parameters,” J. Mater. Process Technol. 123(1), 133–145 (2002). 10.1016/S0924-0136(02)00060-2 [DOI] [Google Scholar]
- 19.Hassan M., et al. , “Fluorescence lifetime imaging system for in vivo studies,” Mol. Imaging 6(4), 229–236 (2007). [PMC free article] [PubMed] [Google Scholar]
- 20.Lakowicz J. R., Principles of Fluorescence Spectroscopy, Springer, New York: (2006). [Google Scholar]
- 21.Shorte S. L., Frischknecht F., Imaging Cellular and Molecular Biological Functions, Springer, Berlin: (2007). [Google Scholar]
- 22.Mayilo S., et al. , “Long-range fluorescence quenching by gold nanoparticles in a sandwich immunoassay for cardiac troponin T,” Nano Lett. 9(12), 4558–4563 (2009). 10.1021/nl903178n [DOI] [PubMed] [Google Scholar]
- 23.Grecco H. E., et al. , “Ensemble and single particle photophysical proper-ties (two-photon excitation, anisotropy, FRET, lifetime, spectral conversion) of commercial quantum dots in solution and in live cells,” Microsc. Res. Tech. 65(4–5), 169–179 (2004). 10.1002/(ISSN)1097-0029 [DOI] [PubMed] [Google Scholar]
- 24.Ambia-Garrido J., Vainrub A., Pettitt B. M., “A model for structure and thermodynamics of ssDNA and dsDNA near a surface: a coarse grained approach,” Comput. Phys. Commun. 181(12), 2001–2007 (2010). 10.1016/j.cpc.2010.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]