Abstract
Our previous study reported that galaxamide, which is a cyclo-pentapeptide containing five leucines that was extracted from Galaxaura filamentosa, displayed remarkable anticancer cytotoxicity. This novel cyclo-peptide provided a new skeleton for the structural modifications used in finding new drugs with better anticancer properties. In this study, five analogues were synthesized based on changing the number of d/l amino acids by adding a new amino acid, phenylalanine. Galaxamide and five of its analogues were evaluated through MTT assays to examine their cytotoxic activities. We found that modified analogue 5, which is referred to as A5, displayed broad spectrum cytotoxic activity toward every cell line tested; in addition, the IC50 of A5 was lower than that of galaxamide and the other analogues. Furthermore, we used flow cytometry and western blot assays to investigate whether galaxamide and A5 could induce cancer cell apoptosis. The flow cytometric studies showed that HepG2 cells treated with different concentrations of galaxamide or A5 over 72 h displayed significant and dose-dependent increases in the percentages of early-stage apoptotic cells. Western blotting revealed that both compounds induce caspase-dependent apoptosis in HepG2 cells through a mitochondria-mediated pathway. The results demonstrate that galaxamide and its analogues have potential applications as clinical anticancer drugs.
Keywords: galaxamide, peptide, synthesis, anticancer, apoptosis
1. Introduction
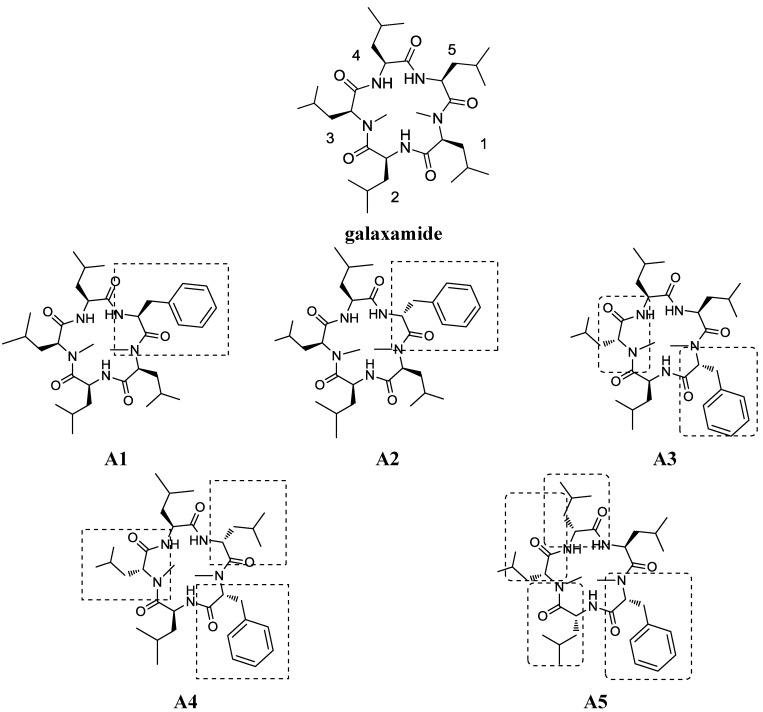
The ocean has more species diversity than land, supplying numerous organisms with chemical properties that are different from their terrestrial counterparts [1]. Cyclic peptides are an important class of marine compounds that may exhibit strong antitumor, antiviral and antibiotic activities [2]. Many structurally novel bioactive cyclopeptides have been isolated from marine cyanobacteria in recent decades [3]. Cyclic peptides tend to have a greater binding affinity for protein targets than their linear counterparts or small molecules, because their bond rotation is restricted and their structures are conformationally constrained [4]. Galaxamide is a novel cyclic pentapeptide isolated from Galaxaura filamentosa in the South China Sea, Xisha Islands. Our earlier research resulted in the isolation, structural determination and total synthesis of galaxamide, revealing significant antitumor activity against the human renal cell carcinoma GRC-1 and hepatocellular carcinoma HepG2 cell lines [5]. The pharmacological and toxicological profiles of galaxamide strongly suggested that galaxamide might be a promising anticancer drug candidate. Therefore, we modified galaxamide and investigated its structure-activity relationship to discover novel antitumor agents among the galaxamide analogues. In this work, we changed the number of d-amino acids in galaxamide [6] and different residues, including leucine and phenylalanine, which is considered an active group in cyclopentapeptide [7], were replaced to synthesize analogues 1–5 (referred to as A1–A5, Figure 1). All of the compounds were evaluated for their cytotoxicity against four human cancer cell lines, HepG2, MCF-7, SW480 and U87, as well as normal human liver cells (L02). As expected, several modified compounds showed strong antitumor activities; in particular, A5 demonstrated stronger cytotoxic activities than galaxamide against all of the tested cancer cell lines. Furthermore, we demonstrated that galaxamide and A5 could activate caspase-3, caspase-9 and PARP, inducing apoptosis in HepG2 and elucidating the mechanism of its anti-tumor activity.
Figure 1.
Structure of galaxamide and galaxamides A1–A5.
2. Results
2.1. Chemistry
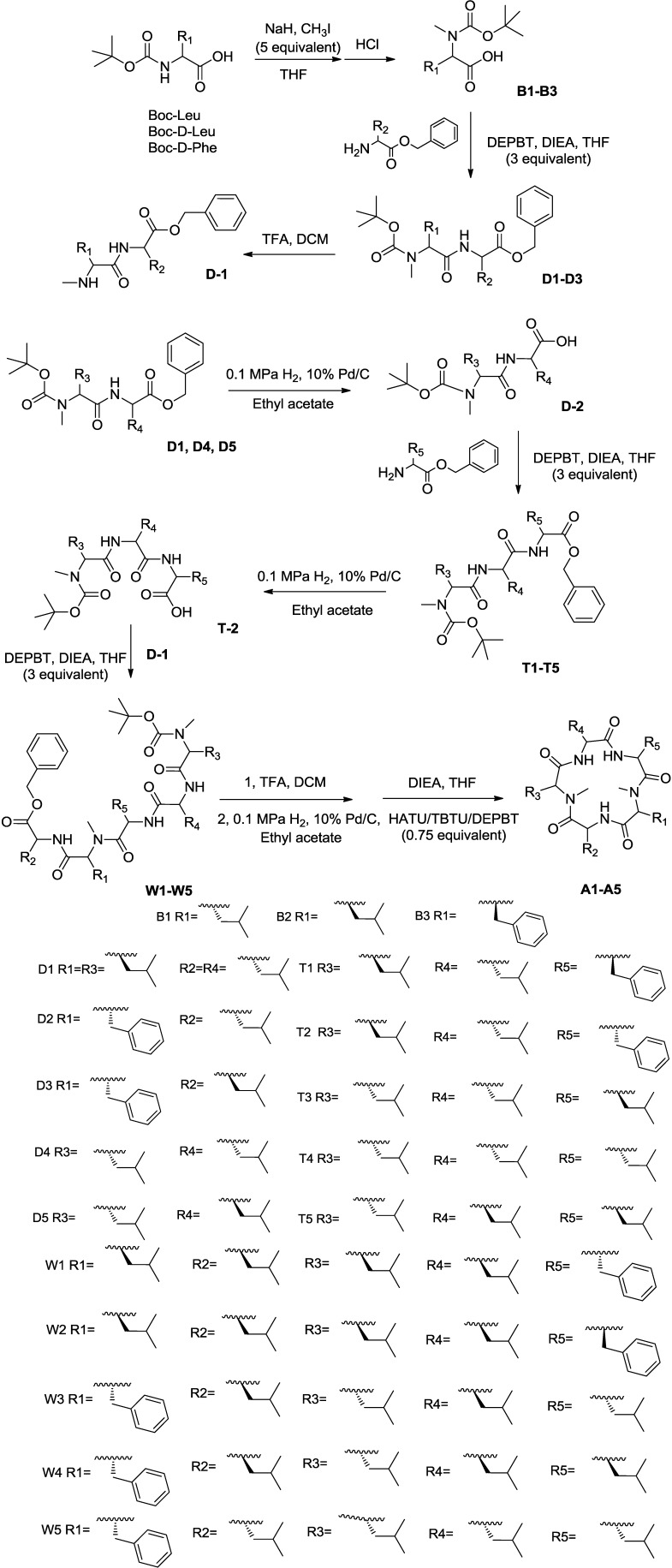
In our previous paper, we described the synthesis of galaxamide, which is a cyclic peptide [5]. We employed the same solution phase method to synthesize galaxamides A1–A5, as shown here (Scheme 1) for A5.
Scheme 1.
The synthesis of galaxamides A1–A5. DEPBT, 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one; DIEA, diisopropylethylamine; DCM, dichloromethane.
Following the strategy as Sakurai [8] described, our synthesis began with the commercially available N-(tert-butoxycarbonyl)-leucine, N-(tert-butoxycarbonyl)-d-leucine and N-(tert-butoxycarbonyl)-d-phenylalanine, reacting with CH3I/NaH [9] in THF, generating Compounds B1–B3, respectively. A reaction between B1–B3 and amino acids protected by benzyl ester in the presence of a coupling reagent, specifically 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one (DEPBT) and diisopropylethylamine (DIEA), generated dipeptides D1–D5 in an 85.0%–92.0% yield [10]. Additionally, we removed the t-butyl carbamate (referred to as Boc) group from D1–D3 using TFA to produce D-1. A hydrogen reduction over Pd/C [11] removed the benzyl group from D1, D4 and D5 before amino acids protected by benzyl ester were coupled using DEPBT, generating tripeptides T1–T5 in an 83%–88% yield. Subsequently, the benzyl group was removed to obtain T-2. C-1 coupled with T-2 via DEPBT, generating pentapeptides W1–W5. Each coupling reaction was concentrated upon completion and subjected directly to silica gel column chromatography using n-hexane/acetone (20:1).
The cyclization is a key step during the synthesis of cyclic peptides. After removing the Boc group and benzyl groups from pentapeptides W1–W5 using TFA and a hydrogen reduction over Pd/C, respectively, the dried, unpurified free amine/free acid linear pentapeptide was dissolved in 2:2:1 THF/CH3CN/CH2Cl2 (0.003 M). Afterwards, three coupling agents, specifically DEPBT, 2-(1H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) (0.75 equivalent each) with DIEA (six equivalent) were added to the reaction [12,13]. The reaction was usually completed within approximately four days. The final product was generated after removing the solvent by distillation under reduced pressure. The mixture was dissolved in EtOAc and washed with saturated ammonium chloride solution [5]. The organic layers were dried, filtered and concentrated. The unpurified mixture was subjected to reverse phase HPLC to obtain A1–A5 in a 44.0%–47.0% yield.
2.2. Biological Activity
2.2.1. Galaxamide and Its Analogues Inhibit the Proliferation of Different Cancer Cells
To examine the effect of galaxamide and its five analogues on cell proliferation, four cancer cell lines and one normal cell line, including human hepatocellular carcinoma HepG2, human breast carcinoma MCF-7, human colon carcinoma SW480 and human glioma cell line U87, as well as human normal liver cell line L02, were exposed to all of the compounds at different concentrations before being subjected to an MTT assay. Galaxamide and A1–A5 were cytotoxic toward different cancer cell lines while exhibiting low cytotoxicity toward normal liver cell line L02. All of the tested compounds exhibited strong cytotoxic activity against HepG2 cells. When comparing galaxamide and its five analogues, A5 demonstrated the highest cytotoxicity against every cell line. The results are summarized in Table 1 and discussed below.
Table 1.
Cytotoxicity of compounds galaxamide and A1–A5 against HepG2, L02, SW480, U87 and MCF-7 cells (IC50, μg/mL) a.
| Compound/Cell Line | HepG2 b | L02 b | SW480 b | U87 b | MCF-7 b |
|---|---|---|---|---|---|
| Galaxamide | 4.63 ± 0.14 | >40 | >40 | 10.61 ± 0.24 | 14.09 ± 1.35 |
| A1 | 4.11 ± 0.16 | >40 | 12.37 ± 1.01 | >40 | 9.40 ± 0.43 |
| A2 | 4.56 ± 0.20 | >40 | 5.75 ± 0.19 | >40 | 9.64 ± 0.35 |
| A3 | 5.58 ± 0.11 | >40 | 10.20 ± 0.15 | 20.16 ± 0.30 | 7.23 ± 0.27 |
| A4 | 6.27 ± 0.13 | >40 | 8.60 ± 0.08 | 2.82 ± 0.10 | 6.56 ± 0.18 |
| A5 | 1.46 ± 0.09 | 10.55 ± 0.60 | 5.10 ± 0.12 | 1.85 ± 0.11 | 4.18 ± 0.15 |
a The IC50 values are reported as the means ± the standard error from three independent experiments; b HepG2, human hepatoma cancer cell line; L02, human normal liver cell line; SW480, human colon carcinoma cell line; U87, human Glioma cell line; MCF-7, human breast cancer cell line.
2.2.2. Effect of Galaxamide and A5 on the Cell Morphology and Nuclear Integrity
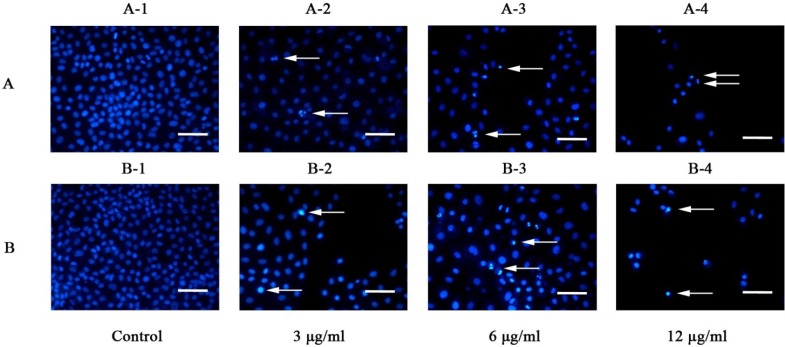
Galaxamide and A5 induced morphological changes in HepG2 cells. Treating HepG2 cells with 3, 6, 12 μg/mL of galaxamide and A5 for 72 h resulted in the detachment of most cells from the tissue culture plates and cell death. These behaviors might be caused by cytotoxicity toward the cells. The number of cells detached from the culture plates increased when increasing the concentrations of galaxamide and A5 in the plates. To examine whether the cells died through apoptotic mechanisms, the control and the galaxamide- and A5-treated cells were stained with Hoechst33342. The resulting morphological changes were observed under a fluorescence microscope (Figure 2). The control cells exhibited normal morphologies with round nuclei. However, the galaxamide- and A5-treated cells showed chromatin condensation at the edges of the nuclear membrane and nuclear fragmentation. As the concentration of galaxamide and A5 was increased to 12 μg/mL, most of them were stimulated to death, but morphological changes still could be observed.
Figure 2.
Morphology observation of galaxamide and A5-induced apoptosis, respectively, in HepG2 cells as indicated by Hoechst33342 staining. Cells were incubated in the absence or presence of 3, 6 and 12 μg/mL galaxamide (A) and A5 (B). HepG2 cells were fixed and stained with Hoechst33342 to visualize the nucleus (blue) under a fluorescent microscope at a magnification of 200×; scale bar, 50 μm. Arrows indicate apoptotic features (condensed chromatin and nuclear fragmentation). Results are representative of triplicate experiments.
2.2.3. Galaxamide and A5 Induce HepG2 Apoptosis
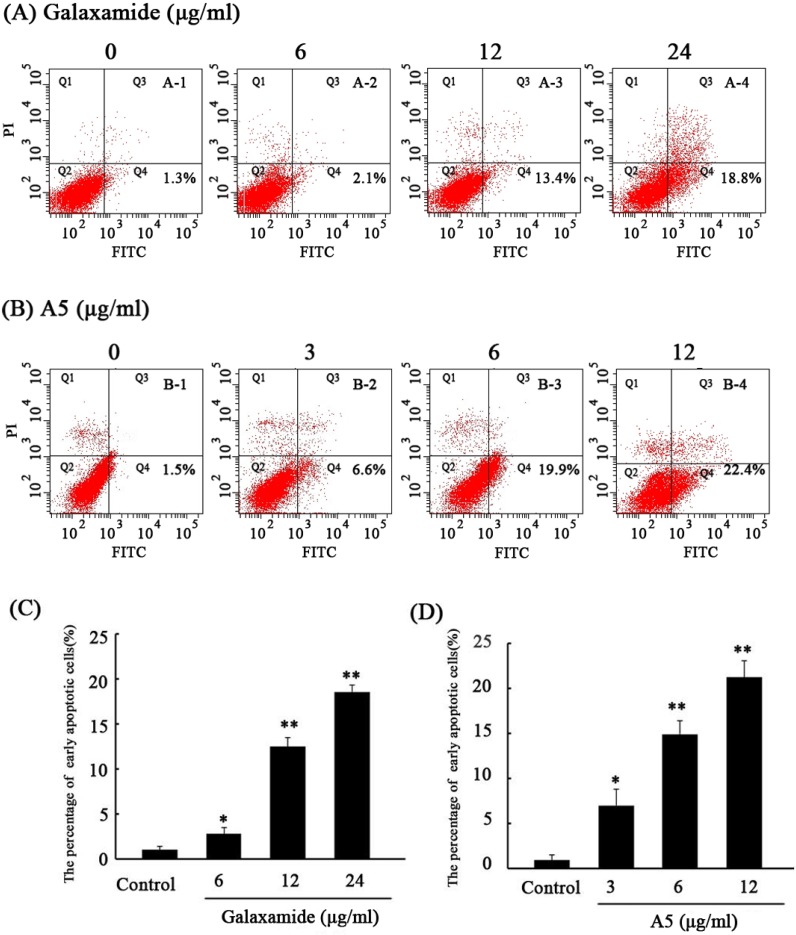
To study the mechanism of the anti-tumor effect, HepG2 was stained with Annexin V/PI and subjected to flow cytometry after treatment with galaxamide and A5 (MW = 627.5 g/mol) at 6, 12 and 24 μg/mL for 72 h. As shown in Figure 3, the lower right quadrants represent the early apoptotic cells. After 72 h of treatment with galaxamide (MW = 593.4 g/mol) at different concentrations (6, 12 and 24 μg/mL), the percentage of early apoptotic cells increased from 1.06% ± 1.0% to 2.3% ± 0.3%, 4.3% ± 1.1% and 18.8% ± 1.9%, respectively (p < 0.05 Figure 3A,C). Lower concentrations of A5 (3, 6 and 12 μg/mL) increased the percentage of early apoptotic cells from 1.5% ± 0.4% to 6.6% ± 1.7%, 16.6% ± 1.5% and 22.4% ± 2.4%, respectively (p < 0.05, Figure 3B,D). In addition, galaxamide and A5 induced early apoptosis in HepG2 dose-dependently. Therefore, galaxamide and A5 induced apoptosis in HepG2 cells.
Figure 3.
Galaxamide and A5 affect the apoptosis of HepG2. HepG2 cells were treated with galaxamide or A5 at different concentrations for 72 h. The control and treated HepG2 cells were assessed using Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) dual parameter flow cytometry. A dual parameter dot plot of FITC-fluorescence (x-axis) vs. PI-fluorescence (y-axis) shows a logarithmic intensity. A dual parameter dot plot of FITC-fluorescence (x-axis) vs. PI-fluorescence (y-axis) shows a logarithmic intensity. Quadrants: lower left, the live cells; lower right, the early apoptotic cells; upper left, the necrotic cells; and upper right, late apoptotic cells. (A,B) Representative Annexin V/PI double staining in the galaxamide (A) or A5 treatment group (B); and (C,D) a comparison of the percentages of early apoptosis in the galaxamide (C) or A5 treatment group (D). * p < 0.05 or ** p < 0.01 vs. the control groups.
2.2.4. Galaxamide and A5 Activate Caspase-3, Caspase-9 and PARP in HepG2
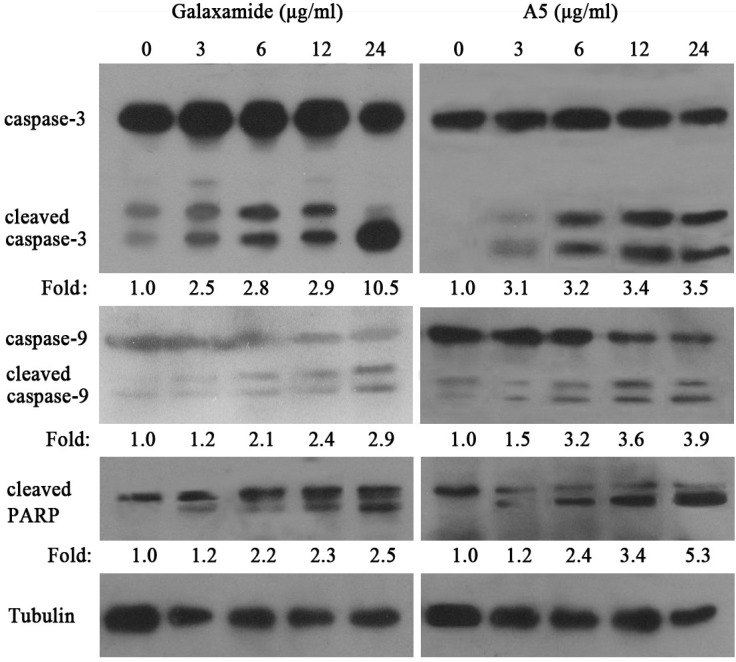
Based on the increased apoptosis of galaxamide- and A5-treated cells, we next examined whether caspase-3, caspase-9 and PARP activities were changed after treatment with galaxamide and A5. As shown in Figure 4, the cleaved caspase-3, caspase-9 and PARP expressions were significantly increased after their treatment (3, 6, 12, and 24 μg/mL) for 72 h. These findings suggest that galaxamide and A5 might induce apoptosis by regulating mitochondria-mediated pathways.
Figure 4.
The expression of apoptosis-associated proteins in HepG2 cell treated with galaxamide and A5 for 72 h. A western blot was used to analyze whole cell lysates for caspase-3, caspase-9 and cleaved PARP expression after treatment with galaxamide and A5, and equal amounts of the protein were loaded onto each lane. One representative immunoblot from three independent experiments is presented. The fold change was calculated as the ratio of cleaved caspase-3, cleaved caspasse-9 and cleaved PARP in the different concentration of Galaxamide or A5 to their expression in the control cells, after normalization to tubulin in each lane. All data shown are representative of at least three separate studies.
3. Discussion
In our previous paper [5], we described the isolation, structural determination and synthesis of galaxamide. In the present study, galaxamide and analogues A1–A5 were synthesized; they all exhibited significant anti-tumor activity against different cancer cells. Furthermore, our data demonstrated that galaxamide and the best cytotoxic analogue, A5, could induce cell apoptosis through a mitochondria-mediated apoptosis pathway. Galaxamide and its analogues might be anti-tumor therapeutic agents ready for further study.
Galaxamide and A1–A5 were tested for inhibitory activity against the growth of several tumor cell lines and one normal liver cell line. As shown in Table 1, some modified compounds exhibited much better activity than galaxamide. Notably, A5 showed broad-spectrum antitumor activity against HepG2, SW480, U87 against MCF-7 cell lines with IC50 values of 1.46, 5.1, 1.85 and 4.18 μg/mL, which were 3.17-, 8.59-, 5.73- and 3.37-fold more active than galaxamide (with IC50 value of 4.63, 43.82, 10.61 and 14.09 μg/mL), respectively. Galaxamide and A1–A4 exhibited low cytotoxic activity toward normal liver cell line L02 (IC50 > 40 μg/mL). Although A5 achieved an IC50 value of 10.55 μg/mL, this value was about tenfold higher than that of HepG2. Galaxamide and its analogues affect cancer cells differently than normal cells. Moreover, all of the cyclopeptide exhibited strong antiproliferative activity against HepG2 cell, revealing that galaxamide and its analogues might be more effective against HepG2 than other types of tumor cells. Both A1 and A2 displayed better cytotoxic activities against HepG2, MCF-7 and SW480. A3 and A4 did not exhibited equal cytotoxic activity against HepG2 compared to galaxamide, but they performed better than galaxamide against SW480 and MCF-7; in addition, A4 had an IC50 value of 2.82 μg/mL and was more active than galaxamide.
Relative to the structure, A5 has four d-amino acids in Positions 2–5. In comparison, A1 has phenylalanine, while A2 is changed into a d-amino acid at Position 1. A3 has two d-amino acids in Positions 2 and 4, and A4 has three d-amino acids in Positions 1, 2 and 4. The structural analysis revealed that: (1) A1 and A2, which contain d/l-phenylalanine, exhibited favorable activity; (2) A5 was more potent than the other analogues, suggesting that the compounds with four d-amino acids, including a d-N-Me-phenylalanine, exhibit better activity than analogues bearing fewer d-amino acids than A2–A4. Future analogues that incorporate additional d-amino acids at the other positions will be synthesized and tested.
Apoptosis is an important homeostatic mechanism that balances cell division and death while maintaining the appropriate number of cells in the body [14]. Many therapeutic peptides, such as aplidine and didemnin B, which both were isolated from marine animals, could induce apoptosis in cancer cells [15,16]. Cyclopentapeptide sansalvamide A is a derivative of sansalvamide A, which is a depsipeptide with antitumor activity that promotes apoptosis in cancer cells [17]. Galaxamide and its analogues were also peptide compounds. In the present study, Annexin V/PI double-staining assays revealed that galaxamides and A5 induced apoptosis in HepG2 cells. Therefore, inducing apoptosis might be the primary mechanism for the anti-cancer activity of galaxamides. In addition, a lower concentration of A5 induced a better apoptosis effect than galaxamides. Similar to the results of the cytotoxicity experiments, we believe that A5 has stronger anti-cancer activity.
Apoptosis can be induced through two distinct pathways [18]: the ligation of the TNF/Fas-receptor with its ligand followed by caspase-8 activation to activate caspase-3 directly and the mitochondria-mediated caspase-9 activation pathway. Initiator caspases cleave procaspases are able to activate effector caspases (caspase-3, -6 and -7) or amplify the caspase cascade by increasing the activation of the initiator caspases. Both pathways converge at caspase-3, leading to cell death. In the present study, caspase-3, caspase-9 and PARP were all activated in HepG2 after being treated with galaxamide or A5. Therefore, the apoptosis of HepG2 induced by galaxamide and A5 might be related to a mitochondria-mediated pathway.
In summary, galaxamide and its analogues have potent antitumor effects and cause cell apoptosis in vitro. Future analogues that incorporate additional d-amino acids at other positions will be synthesized and tested. In addition, improving the yield of the cyclization is also necessary.
4. Experimental Section
4.1. Chemistry
The reagents and solvents were commercially available. The solvents were dried and purified through standard procedures before use. The NMR spectra were obtained on a Varian Inova-500 NB spectrometer in CDCl3, while using TMS as an internal standard; the coupling constants (J) are in Hz. The ESI mass spectra were obtained on a LCQ DECA XP LC-MS mass spectrometer. Column chromatography was performed using silica gel (Qingdao Ocean Chemical Factory, Qingdao, China, 200–300 mesh) eluted with petroleum ether/acetone or n-hexane/acetone; C18 reversed phase silica gel (Welch Material Inc., Shanghai, China, 45 μm) was eluted with methanol-water.
4.1.1. N-Boc-Me-Leu-Leu-OBn (D1)
Yield: 90.0%; colorless crystal (CH3OH); 1H NMR (500 MHz, CDCl3): 7.36–7.40 (m, 5H), 6.51 (s, 1H), 5.16–5.20 (m, 2H), 4.65 (s, 2H), 2.75 (s, 3H), 1.63–1.68 (m, 4H), 1.50–1.54 (m, 2H), 1.48 (s, 9H), 0.90–0.96 (m, 12H); MS (ESI) m/z: 449.3 [M + H]+, 466.5 [M + NH4]+, 471.6 [M + Na]+.
4.1.2. N-Boc-Me-d-Phe-Leu-OBn (D2)
Yield: 86.8%; colorless crystal (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.33–7.44 (m, 5H), 7.21–7.24 (m, 5H), 5.40 (d, J = 10.0 Hz, 1H), 5.22–5.25 (m, 1H), 5.10 (q, 2H), 4.92–4.97 (m, 1H), 2.97–3.01 (m, 2H), 2.70 (s, 3H), 1.63–1.69 (m, 1H), 1.54–1.58 (m, 1H), 1.48 (s, 9H), 0.93–0.96 (m, 1H), 0.84–0.90 (m, 6H); MS (ESI) m/z: 483.7 [M + H]+, 500.6 [M + NH4]+, 505.6 [M + Na]+.
4.1.3. N-Boc-Me-d-Phe-d-Leu-OBn (D3)
Yield: 85.6%; colorless crystal (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.35–7.40 (m, 5H), 7.26–7.30 (m, 5H), 5.41 (d, J = 10.0 Hz, 1H), 5.24–5.30 (m, 1H), 5.11 (q, 2H), 4.94–4.97 (m, 1H), 3.01 (m, 2H), 2.70 (s, 3H), 1.64–1.69 (m, 1H), 1.57–1.60 (m, 1H), 1.48 (s, 9H), 0.93–0.96 (m, 1H), 0.84–0.88 (m, 6H); MS(ESI) m/z: 483.7 [M + H]+, 500.6 [M + NH4]+, 505.6 [M + Na]+.
4.1.4. N-Boc-Me-d-Leu-Leu-OBn (D4)
Yield: 91.3%; colorless crystal (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.35–7.44 (m, 5H), 6.52 (s, 1H), 5.14–5.20 (m, 2H), 5.02 (d, 1H, J = 10.0 Hz), 4.61 (s, 2H), 3.04 (d, 2H, J = 5.0 Hz), 1.62–1.70 (m, 4H), 1.50–1.55 (m, 2H), 1.48 (s, 9H), 0.91–0.93 (m, 12H); MS (ESI) m/z: 449.3 [M + H]+, 466.5 [M + NH4]+, 471.6 [M + Na]+.
4.1.5. N-Boc-Me-d-Leu-d-Leu-OBn (D5)
Yield: 92.0%; colorless crystal (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.37–7.47 (m, 5H), 6.50 (s, 1H), 5.14–5.22 (m, 2H), 5.02 (d, 1H, J = 10.0 Hz), 4.51 (s, 2H), 3.04 (d, 2H, J = 5.0 Hz), 1.66–1.72 (m, 4H), 1.49–1.55 (m, 2H), 1.38 (s, 9H), 0.91–0.93 (m, 12H); MS (ESI) m/z: 449.3 [M + H]+, 466.5 [M + NH4]+, 471.6 [M + Na]+.
4.1.6. N-Boc-Me-Leu-Leu-Phe-OBn (T1)
Yield: 90.7%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.32–7.37 (m, 5H), 7.22–7.26 (m, 5H), 7.14 (d, 2H, J = 6.8 Hz), 5.12 (s, 2H), 4.65–4.68 (m, 2H), 4.53– 4.56 (m, 1H), 2.98–3.01 (m, 2H), 2.42 (s, 3H), 1.59–1.63 (m, 2H), 1.47–1.53 (m, 2H), 1.43 (s, 9H), 1.38–1.42 (m, 2H), 0.86–0.91 (m,12H); MS (ESI) m/z: 595.7 [M + H]+, 612.7 [M + NH4]+, 617.9 [M + Na]+.
4.1.7. N-Boc-Me-Leu-Leu-d-Phe-OBn (T2)
Yield: 91.5%; colorless crystal (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.32–7.40 (m, 5H), 7.24–7.30 (m, 5H), 6.78 (d, 1H, J = 10.0 Hz), 5.19 (d, 1H, J = 5.0 Hz), 4.73 (q, 1H), 4.49 (q, 1H), 2.95 (d, 2H, J = 10.0 Hz), 2.61 (s, 3H), 1.63–1.70 (m, 4H), 1.35 (s, 9H), 1.25–1.30 (m, 2H), 0.86–0.92 (m, 12H); MS (ESI) m/z: 595.7 [M + H]+, 612.7 [M + NH4]+, 617.9 [M + Na]+.
4.1.8. N-Boc-Me-d-Leu-Leu-Leu-OBn (T3)
Yield: 87.1%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.32–7.36 (m, 5H), 6.64 (d, J = 5.0 Hz, 1H), 6.56 (d, J = 5.0 Hz, 1H), 5.23 (d, J = 10.0 Hz, 1H), 5.16–5.19 (m, 2H), 4.69–4.73 (m, 1H), 4.53–4.56 (m, 1H), 2.97 (s, 3H), 1.87–1.93 (m, 2H), 1.53–1.56 (m, 4H), 1.42–1.46 (m, 9H), 1.36–1.39 (m, 3H), 0.92–0.96 (m, 18H); MS (ESI) m/z: 562.8 [M + H]+, 579.8 [M + NH4]+, 584.9 [M + Na]+.
4.1.9. N-Boc-Me-d-Leu-Leu-d-Leu-OBn (T4)
Yield: 86.6%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.37–7.47 (m, 5H), 6.65 (d, J = 5.0 Hz, 1H), 6.56 (d, J = 5.0 Hz, 1H), 5.25 (d, J = 10.0 Hz, 1H), 5.17–5.22 (m, 2H), 4.75–4.77 (m, 1H), 4.53–4.56 (m, 1H), 2.98 (s, 3H), 1.88–1.92 (m, 2H), 1.52–1.56 (m, 4H), 1.39–1.44 (m, 9H), 1.34–1.39 (m, 3H), 0.92–0.97 (m, 18H); MS (ESI) m/z: 562.8 [M + H]+, 579.8 [M + NH4]+, 584.9 [M + Na]+.
4.1.10. N-Boc-Me-d-Leu-d-Leu-Leu-OBn (T5)
Yield: 87.2%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm):7.35–7.40 (m, 5H), 6.60 (d, J = 5.0 Hz, 1H), 6.51 (d, J = 5.0 Hz, 1H), 5.19 (d, J = 10.0 Hz, 1H), 5.06–5.11 (m, 2H), 4.59–4.63 (m, 1H), 4.43–4.46 (m, 1H), 2.87 (s, 3H), 1.77–1.81(m, 2H), 1.43–1.52 (m, 4H), 1.32–1.37 (m, 9H), 1.24–1.27 (m, 3H), 0.88–0.96 (m, 18H); MS (ESI) m/z: 562.8 [M + H]+, 579.8 [M + NH4]+, 584.9 [M + Na]+.
4.1.11. N-Boc-Me-Leu-Leu-Phe-Me-Leu-LeuOBn (W1)
Yield: 78.6%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.33–7.41 (m, 7H), 7.20–7.24 (m, 3H) ,7.18 (d, 1H, J = 10.0 Hz), 6.68 (d, 1H, J = 10.0 Hz), 6.61 (d, 1H, J = 10.0 Hz), 5.24–5.30 (m, 2H), 5.13 (s, 2H), 5.06–5.11 (m, 1H), 4.93–4.99 (m, 2H), 2.91–2.95 (m, 2H), 2.83 (s, 3H), 2.73 (s, 3H), 1.77–1.80 (m, 2H), 1.65–1.68 (m, 6H), 1.53–1.59 (m, 4H), 1.45 (s, 9H), 0.93–0.99 (m, 14H), 0.84–0.88 (m, 10H); MS (ESI) m/z: 837.2 [M + H]+, 854.1 [M + NH4]+, 859.1 [M + Na]+.
4.1.12. N-Boc-Me-Leu-Leu-d-Phe-Me-Leu-Leu-OBn (W2)
Yield: 81.0%; colorless oil CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.31 (m, 7H), 7.23–7.26 (m, 3H) , 6.76 (d, 1H, J = 10.0 Hz), 6.36 (d, 1H, J = 10.0 Hz), 5.30 (d, 1H, J = 10.0 Hz), 5.13 (s, 2H), 5.01–5.05 (m, 1H), 4.83–4.90 (m, 2H), 4.70–4.73 (m, 1H), 4.56–4.59 (m, 1H), 2.96 (s, 3H), 2.73–2.77 (m, 2H), 2.67 (s, 3H), 1.64–1.67 (m, 2H), 1.59–1.62 (m, 6H), 1.46–1.50 (m, 4H), 1.41 (s, 9H), 0.88–098 (m, 24H), MS (ESI) m/z: 837.0 [M + H]+, 854.0 [M + NH4]+, 859.0 [M + Na]+.
4.1.13. N-Boc-Me-d-Leu-Leu-Leu-d-Phe-Leu-OBn (W3)
Yield: 82.5%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.35–7.45 (m, 7H), 7.26–7.30 (m, 3H), 6.76 (d, 1H, J = 10.0 Hz), 6.65 (d, 1H, J = 10.0 Hz), 6.37 (d, 1H, J = 10.0 Hz), 5.37 (s, 2H), 5.31–5.35 (m, 1H), 5.03–5.09 (m, 2H), 4.90–4.95 (m, 1H), 4.68–4.72 (m, 2H), 3.47 (s, 3H), 2.95 (s, 3H), 2.72–2.76 (m, 2H), 1.76–1.82 (m, 2H), 1.59–1.68 (m, 6H), 1.51–1.57 (m, 4H), 1.44 (s, 9H), 0.92–0.98 (m, 24H); MS (ESI) m/z: 837.0 [M + H]+, 854.0 [M + NH4]+, 859.0 [M + Na]+.
4.1.14. N-Boc-Me-d-Leu-Leu-d-Leu-d-Phe-Leu-OBn (W4)
Yield: 81.5%; colorless oil (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.33–7.43 (m, 7H), 7.23–7.28 (m, 3H), 6.75 (d, 1H, J = 10.0 Hz), 6.64 (d, 1H, J = 10.0 Hz), 6.36 (d, 1H, J = 10.0 Hz), 5.35 (s, 2H), 5.31–5.33 (m, 1H), 5.03–5.10 (m, 2H), 4.88–4.92 (m, 1H), 4.68–4.75 (m, 2H), 3.57 (s, 3H), 2.97 (s, 3H), 2.74–2.78 (m, 2H), 1.76–1.81 (m, 2H), 1.57–1.66 (m, 6H), 1.53–1.55 (m, 4H), 1.45 (s, 9H), 0.91–0.95 (m, 24H); MS (ESI) m/z: 837.0 [M + H]+, 854.0 [M + NH4]+, 859.0 [M + Na]+.
4.1.15. N-Boc-Me-d-Leu-d-Leu-Leu-d-Phe-d-Leu-OBn (W5)
Yield: 82.5%; colorless oil(CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.35–7.45 (m, 7H), 7.26–7.30 (m, 3H), 6.76 (d, 1H, J = 10.0 Hz), 6.65 (d, 1H, J = 10.0 Hz), 6.37 (d, 1H, J = 10.0 Hz), 5.37 (s, 2H), 5.31–5.35 (m, 1H), 5.03–5.09 (m, 2H), 4.90–4.95 (m, 1H), 4.68–4.72 (m, 2H), 3.47 (s, 3H), 2.95 (s, 3H), 2.72–2.76 (m, 2H), 1.76–1.82 (m, 2H), 1.59–1.68 (m, 6H), 1.51–1.57 (m, 4H), 1.44 (s, 9H), 0.92–0.98 (m, 24H); MS (ESI) m/z: 837.0 [M + H]+, 854.0 [M + NH4]+, 859.0 [M + Na]+.
4.1.16. Cyclo(Phe-N-Me-Leu-Leu-N-Me-Leu-Leu) (A1)
Yield: 42.0%; white powder (CH3OH); 1H-NMR (500 MHz, CDCl3), δ(ppm): 7.27–7.31 (m, 5H), 7.04 (d, J = 10.0 Hz, 1H), 6.93 (d, J = 10.0 Hz, 1H), 6.02 (d, J = 10.0 Hz, 1H), 5.17 (t, J = 15.0 Hz, 1H), 4.80–4.87 (m, 3H), 3.34 (dd, J = 5.0 Hz, 5.0 Hz, 1H), 3.20–3.27 (m, 6H), 2.65 (s, 3H), 1.94–2.00 (m, 1H), 1.66–1.75 (m, 2H), 1.47–1.59 (m, 7H), 1.17–1.22 (m, 1H), 0.90–0.98 (m, 24H); 13C-NMR (125 MHz, CDCl3), δ (ppm): 173.1, 172.3, 170.4, 170.2, 169.3, 137.4, 129.4(2C), 128.5(2C), 126.9, 64.2, 53.9, 53.2, 48.2, 47.8, 40.8, 40.6, 38.8, 36.9, 36.4, 34.6, 29.6, 25.2(2C), 24.7, 24.6, 23.3, 23.2, 22.8, 22.6(2C); MS (ESI) m/z/: 628.9 [M + H]+, 645.8 [M + NH4]+, 650.8 [M + Na]+.
4.1.17. Cyclo(d-Phe-N-Me-Leu-Leu-N-Me-Leu-d-Leu) (A2)
Yield: 42.5%; white powder (CH3OH); 1H-NMR (500 MHz, CDCl3), δ(ppm): 8.13(s, 1H), 7.71–7.74 (m, 1H), 7.19–7.29 (m, 5H), 6.91 (d, J = 10.0Hz, 1H), 5.18 (t, J = 10.0Hz, 1H), 4.87–4.91 (m, 1H), 4.74–4.79 (m, 3H), 3.05–3.09 (m, 2H), 2.91–2.94 (m, 1H), 2.84 (s, 1H), 2.80 (s, 2H), 2.62 (s, 3H), 1.88–1.93 (m, 1H), 1.77–1.82 (m, 1H), 1.50–1.61 (m, 6H), 1.37–1.43 (m, 1H), 1.06–1.12 (m, 1H), 0.89–0.97 (m, 18H), 0.66–0.73 (m, 6H); 13C-NMR (125 MHz, CDCl3), δ (ppm): 175.9, 171.7, 171.5, 170.1, 170.0, 136.2, 129.1(2C), 128.8(2C), 127.2, 57.8, 53.7(2C), 48.7, 47.8, 41.5, 41.1, 37.7, 37.5, 34.5, 31.4, 29.5, 24.9(2C), 24.8, 24.5, 23.2, 23.1, 22.9(2C), 22.2, 21.9, 21.8, 21.6; MS (ESI) m/z: 628.8 [M + H]+, 645.9 [M + NH4]+, 650.8 [M + Na]+.
4.1.18. Cyclo(Me-d-Leu-Leu-Leu-Me-d-Phe-Leu) (A3)
Yield: 44.5% ; white powder (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 8.17(d, J = 10.0 Hz, 1H), 7.07–7.23 (m, 5H), 6.85 (d, J = 5.0 Hz, 1H), 6.69 (d, J = 10.0 Hz, 1H), 5.23 (dd, J = 5.0 Hz, J = 5.0 Hz, 1H), 4.83 (dd, J = 5.0 Hz, J = 5.0 Hz, 1H), 4.68 (dd, J = 5.0 Hz, J = 5.0 Hz, 1H), 4.50 (dd, J = 10.0 Hz, J = 10.0 Hz, 1H), 3.47 (t, J = 20.0 Hz, 1H), 3.2 (s, 3H), 3.11 (s, 1H), 3.03 (d, J = 10.0 Hz, 2H), 2.97 (s, 3H), 2.12 (s, 1H), 1.74–1.84 (m, 4H), 1.49–1.52 (m, 2H), 1.32–1.39 (m, 4H), 0.76–1.00 (m, 24H); 13C-NMR (125 MHz, CDCl3), δ (ppm): 173.6, 173.2, 171.5, 170.1, 169.7, 136.3, 129.6(2C), 128.5(2C), 126.9, 57.5, 54.3, 52.6, 47.8, 46.8, 40.7(2C), 39.2, 37.5, 37.1, 31.3, 31.2, 29.6, 24.8, 24.4, 23.6, 23.3(2C), 23.0, 22.6(2C), 22.2, 22.0, 21.8; MS (ESI) m/z: 628.6 [M+H]+, 645.7 [M + NH4]+, 650.4 [M + Na]+.
4.1.19. Cyclo(Me-d-Leu-Leu-d-Leu-Me-d-Phe-Leu) (A4)
Yield: 43.3%; white powder (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.20–7.27 (m, 4H), 7.06 (d, J = 10.0 Hz, 1H), 6.85 (d, J = 10.0 Hz, 1H), 6.40 (m, 1H), 6.17 (t, 1H), 5.45 (dd, J = 5.0, 5.0 Hz, 1H), 4.99 (td, 1H), 4.68 (td, 1H), 4.41–4.49 (m, 2H), 3.20–3.27 (m, 4H), 3.07 (d, J = 5.0 Hz, 1H), 3.01–3.07 (m, 1H), 2.96–2.99 (m, 1H), 2.78 (d, J = 5.0 Hz, 3H), 1.81 (dd, J = 5.0, 5.0 Hz, 2H), 1.59–1.62 (m, 4H), 1.43–1.48 (m, 2H), 1.30 (dd, J = 10.0, 10.0 Hz, 2H), 0.98–1.03 (m, 6H), 0.93 (dd, J = 10.0, 10.0 Hz, 12H), 0.84 (d, J = 5.0 Hz, 3H), 0.76 (d, J = 5.0 Hz, 3H); 13C-NMR (125 MHz, CDCl3), δ (ppm): 175.7, 172.3, 171.4(2C), 169.5, 136.8(3C), 128.8(2C), 126.8, 57.2, 56.2(2C), 52.6(3C), 48.4(2C), 48.0(2C), 41.3, 40.9, 37.5(3C), 33.9, 30.9(2C), 29.9(2C), 25.2, 23.1, 22.8, 22.7; MS (ESI) m/z: 628.7 [M + H]+, 645.8 [M + NH4]+, 650.6 [M + Na]+.
4.1.20. Cyclo(Leu-d-N-Me-Phe-d-Leu-d-N-Me-Leu-d-Leu) (A5)
Yield: 47.0%; white powder (CH3OH); 1H-NMR (500 MHz, CDCl3), δ (ppm): 7.22–7.31 (m, 5H), 7.06 (d, J = 10.0 Hz, 1H), 6.95 (d, J = 10.0 Hz, 1H), 6.07 (d, J = 10.0 Hz, 1H), 5.17 (t, J = 10.0 Hz, 1H), 4.82 (m, 3H), 3.33 (m, 1H), 3.20 (m, 5H), 3.02 (d, J = 10.0 Hz, 1H), 2.66 (s, 3H), 1.95–2.00 (m, 3H), 1.77–1.85 (m, 2H), 1.66–1.75 (m, 2H), 1.47–1.52 (m, 3H), 1.2 (dd, J = 5.0, 5.0 Hz, 1H), 0.81–0.95 (m, 24H); 13C-NMR (125 MHz, CDCl3), δ (ppm): 173.4, 172.9, 172.1, 171.4, 168.5, 138.8, 129.5(2C), 128.8, 128.7, 127, 68.36(2C), 59.37, 55.37, 47.66, 47.3, 41.7, 36.0(3C), 30.8(2C), 25.4(3C), 23.0(3C), 22.6(3C), 22.03(2C); MS (ESI) m/z: 628.6[M + H]+, 645.7 [M + NH4]+, 650.6 [M + Na]+.
4.2. Antitumor Activity in Vitro
4.2.1. Cell Culture
Human hepatocellular carcinoma HepG2, liver L02, colon carcinoma SW480, breast cancer MCF-7 and glioma U87 cells were obtained from the China cell bank of the Institute of Biochemistry and Cell Biology and maintained in Dulbecco’s modification Eagle’s medium (DMEM, Corning, NY, USA) with 10% fetal bovine serum (FBS, Hyclone, Logan, UT, USA), 1% penicillin-streptomycin and an antifungal agent. All of the cells were incubated at 37 °C in a humidified atmosphere composed of 95% air and 5% CO2.
4.2.2. Anti-Proliferative Activity Using MTT Assays
The cell viability was determined by measuring the ability of cells to transform MTT (Genview, Houston, TX, USA) to a purple formazan dye. The cells were seeded in 96-well tissue culture plates at 2.5 × 103 cells/well for 24 h. The cells were then incubated with galaxamide and its analogues at different concentrations for 72 h. After incubation, 20 μL/well of MTT solution (5 mg/mL phosphate buffered saline) were added and incubated for 5 h. The medium was aspirated and replaced with 100 μL/well of DMSO to dissolve the formazan salt. The color intensity of the formazan solution, which reflects the cell growth condition, was measured at 570 nm using a microplate spectrophotometer (SpectroAma™ 250, Winooski, VT, USA).
4.2.3. Cell Apoptosis Analysis
The apoptotic cells were quantified using an Annexin V-FITC (Sigma-Aldrich, USA) Apoptosis Detection Kit according to the manufacturer’s instructions. Briefly, the cells were plated in a 6-well plates and treated with galaxamide and A5 (6 μg/mL, 12 μg/mL, 24 μg/mL) for 72 h before being collected and resuspended in 200 μL of binding buffer. Five μL of Annexin V-fluorescein isothiocyanate (FITC) and 5 μL of PI were added. The analysis was performed with a FACScan flow cytometer (Coulter Epics Elite, Miami, FL, USA). The cells in the FITC-positive and PI-negative fraction were regarded as apoptotic cells.
4.2.4. Observations of Nuclear Damage
Exponentially growing HepG2 cells (1 × 105 cells/mL) were incubated for 72 h with 3, 6, 12 μg/mL of galaxamide and A5, respectively. Apoptotic nuclear morphology was visualized by the Hoechsst33342 staining technique. Cells were fixed with 3.7% of paraformaldehyde for 10 min at room temperature and washed three times with PBS. Thus, paraformaldehyde-fixed cells were stained using Hoechsst33342 (10 μg/mL) in the dark for 10 min. After washing three times with PBS, cells were visualized using fluorescence microscope.
4.2.5. Western Blot Analysis
The protein was extracted with RIPA buffer and quantified using the Bradford method. Equal amounts of protein were loaded into each well. After SDS-PAGE, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% non-fat milk for 1 h and incubated with a primary antibody (Cell Signaling Technology, Danvers, MA, USA, Rabbit monoclonal antibody, 1:1000) at 4 °C overnight. After washing two times with TBST (Tris-buffered saline with 0.1% Tween-20) buffer, the membrane was incubated with a secondary antibody (goat-anti-rabbit horseradish peroxidase (HRP)-conjugated 1:15,000) at room temperature for 1 h. The membranes were washed three times (8 min each time) with TBST. The intensity of the specific immunoreactive bands was detected through enhanced chemiluminescence (ECL).
4.2.6. Statistical Analysis
The data are expressed as the mean ± standard error of mean (SEM). The statistical comparisons were performed using the Student’s t-test, and the differences between groups were considered significant at p < 0.05.
5. Conclusions
This paper has reported the synthesis and the biological evaluation of novel cyclopeptides, including galaxamide and its analogues A1–A5, against a series of cancer cells and normal liver cells. Some modified analogues exhibited strong cytotoxicity against cancer cells. Meanwhile, galaxamide and A5 activated an apoptotic pathway involving the sequential activation of caspases 9, 3 and PARP in the HepG2 cells. Interest in exploring the chemical structure relationship of galaxamide and analogues with anti-tumor properties for new drug design has grown.
Acknowledgments
This research was supported by the General projects of the National Natural Science Foundation (Grant Nos. 21172094, 41376155, 21372100, 31201028, U1301131), a major program of the technology office in Guangzhou (Grant No. 2013000000163), the emerging industrial strategy plan of Shenzhen City (Grant Nos. JCYJ20120615172324607) and the National High Technology Research and Development Program (“863” Program) of China (Grant No. 2013AA092902).
Author Contributions
Bin Du and Shihai Xu designed the research and wrote the manuscript; Xi Xiao, Xiaojian Liao and Shaoling Qiu performed the chemistry experiments; Xi Xiao and Zihao Liu performed the biology experiments; all authors approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Donia M., Hamann M.T. Marine natural products and their potential applications as anti-infective agents. Lancet Infect. Dis. 2003;3:338–348. doi: 10.1016/S1473-3099(03)00655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J.S. Marine drug research and development ramble. Chem. Rev. 1995;95:2115–2134. doi: 10.1021/cr00038a013. [DOI] [Google Scholar]
- 3.Blunt J.W., Copp B.R., Munro M.H.G., Northcote P.T., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2011;28:196–268. doi: 10.1039/c005001f. [DOI] [PubMed] [Google Scholar]
- 4.Davies J.S. The cyclization of peptides and depsipeptides. J. Peptide. Sci. 2003;9:471–501. doi: 10.1002/psc.491. [DOI] [PubMed] [Google Scholar]
- 5.Xu W.J., Liao X.J., Xu S.H., Diao J.Z. Isolation, structure determination, and synthesis of Galaxamide, a rare cytotoxic cyclic pentapeptide from a marine algae Galaxaura filamentosa. Org. Lett. 2008;10:4569–4572. doi: 10.1021/ol801799d. [DOI] [PubMed] [Google Scholar]
- 6.Davis M.R., Styers T.J., Rodriguez R.A., Pan P.S., Vasko R.C., McAlpine S.R. Synthesis and cytotoxicity of a new class of potent decapeptide macrocycles. Org. Lett. 2008;10:177–180. doi: 10.1021/ol702403r. [DOI] [PubMed] [Google Scholar]
- 7.Katerina O., Gerald L., David V. Comprehensive study of Sansalvamide A analogues and their structure–activity relationships against drug-resistant colon cancer cell lines. J. Med. Chem. 2007;51:530–544. doi: 10.1021/jm070731a. [DOI] [PubMed] [Google Scholar]
- 8.Sakurai A., Okumura Y. Synthesis of viscumamide and its analogs. Chem. Soc. Jpn. 1979;52:540–543. doi: 10.1246/bcsj.52.540. [DOI] [Google Scholar]
- 9.Cheung S.T., Benoiton N.L. N-Methylamino acids in peptides synthesis. V. The synthesis of N-tert-butyloxycarbony1, N-methylamino acids by N-methylation. Can. J. Chem. 1977;55:906–910. doi: 10.1139/v77-125. [DOI] [Google Scholar]
- 10.Li H.T., Jiang X.H., Ye Y.H., Fan C.X., Romoff T., Goodman M. 3-(Diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one(DEPBT): A new coupling reagent with remarkable resistance to racemization. Org. Lett. 1999;1:91–93. doi: 10.1021/ol990573k. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J.D., Ma D.W. Total synthesis of microsclerodermin E. Angew. Chem. Int. Ed. Engl. 2003;42:5348–5351. doi: 10.1002/anie.200352423. [DOI] [PubMed] [Google Scholar]
- 12.Belofsky G.N., Jensen P.R., Fenical W. A new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999;40:2913–2916. doi: 10.1016/S0040-4039(99)00393-7. [DOI] [Google Scholar]
- 13.Rodriguez R.A., Pan P.S., Pan C.M., Ravula S., Lapera S., Singh E.K., Styers T.J., Brown J.D., Cajica J., Parry E., et al. Synthesis of second-generation Sansalvamide A derivatives: Novel templates as potential antitumor agents. J. Org. Chem. 2007;72:1980–2002. doi: 10.1021/jo061830j. [DOI] [PubMed] [Google Scholar]
- 14.Martin S.J., Green D.R. Apoptosis and cancer: The failure of controls on cell death and cell survival. Crit. Rev. Oncol. Hematol. 1995;8:137–153. doi: 10.1016/1040-8428(94)00124-C. [DOI] [PubMed] [Google Scholar]
- 15.Gajate C., An F., Mollinedo F. Rapid and selective apoptosis in human leukemic cells induced by aplidine through a Fas/CD95- and mitochondrial-mediated mechanism. Clin. Cancer Res. 2003;9:1535–1545. [PubMed] [Google Scholar]
- 16.Baker M.A., Grubb D.R., Lawen A. Didemnin B induces apoptosis in proliferating but not resting peripheral blood mononuclear cells. Apoptosis. 2002;7:407–412. doi: 10.1023/A:1020078907108. [DOI] [PubMed] [Google Scholar]
- 17.Joseph B.K., Mark N.P., Leslie D.A., Veronica C.A., Jeanette R.M., Shelli R.M. Synthesis and evaluation of biotinylated sansalvamide A analogs and their modulation of Hsp90. Bioorg. Med. Chem. Lett. 2011;21:4716–4719. doi: 10.1016/j.bmcl.2011.06.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan G., O’Rourke K., Chinnaiyan A.M., Gentz R., Ebner R., Ni J., Dixit V.M. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]