Abstract
A cell-based high-throughput screen that assessed the cellular stability of a tumor suppressor protein PDCD4 (Programmed cell death 4) was used to identify a new guanidine-containing marine alkaloid mirabilin K (3), as well as the known compounds mirabilin G (1) and netamine M (2). The structures of these tricyclic guanidine alkaloids were established from extensive spectroscopic analyses. Compounds 1 and 2 inhibited cellular degradation of PDCD4 with EC50 values of 1.8 μg/mL and 2.8 μg/mL, respectively. Mirabilin G (1) and netamine M (2) are the first marine natural products reported to stabilize PDCD4 under tumor promoting conditions.
Keywords: natural product, guanidine alkaloid, PDCD4, tumor suppressor
1. Introduction
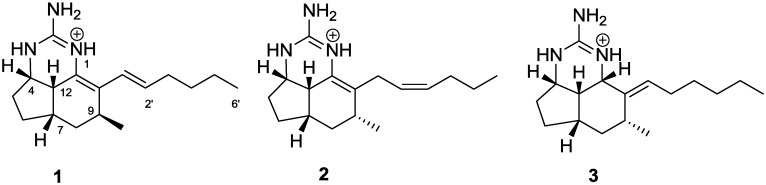
Tumor suppressor proteins can prevent or repress malignant cell growth by regulating progression of the cell cycle or by promoting apoptosis via alteration in gene expression patterns. Rather than directly affecting transcription, PDCD4 (programmed cell death 4) is a novel tumor suppressor that inhibits protein translation through interaction with the eukaryotic translation initiation factors eIF4A and eIF4G [1]. PDCD4 can inhibit the transformation, migration, and invasion of cancer cells in vitro [2,3,4], and overexpression of PDCD4 in transgenic mice was found to significantly suppress tumorigenesis [5]. Down regulation of PDCD4 expression has been associated with the onset of a number of human tumors including colorectal [6], brain [7], ovarian [8], and liver carcinomas [9]. Since cellular PDCD4 levels are in part regulated by phosphorylation-dependent proteasomal degradation in response to tumor promoters [10,11,12], stabilizing PDCD4 provides an attractive potential therapeutic target. With the aim to identify small molecule stabilizers of PDCD4, a high throughput cell-based reporter screen was developed where the stability of PDCD4 was assessed under tumor promoting conditions [13]. The assay was designed to monitor tetradecanoylphorbol-13-acetate (TPA)-induced degradation of a PDCD4-luciferase construct, and a 50% or greater recovery of the luciferase signal was defined as a hit. The screening assay was used to test natural product extracts sourced from a diverse collection of marine invertebrates, terrestrial plants, and microbial isolates from the Natural Products Repository of the US National Cancer Institute. Testing of 135,678 extracts yielded 42 confirmed hits to date. Previously we reported on the PDCD4 stabilizing activity of tubercidin from a halophilic actinomycete Actinopolyspora erythrea [14], as well as terrestrial plant metabolites including the cryptocaryols, a series of α-pyrone-containing 1,3 polyols obtained from Cryptocarya sp. [15]; erioflorin, a sesquiterpene lactone isolated from the wooly sunflower Eriophyllum lanatum [16], and the isoflavone pomiferin triacetate [17]. Herein we report on the activity of compounds isolated from the organic solvent extract of the marine sponge Acanthella cavernosa (NSC # C005465), collected in Southwestern Australia. Bioassay-guided fractionation resulted in the purification of two known compounds, mirabilin G (1) [18,19] and netamine M (2) [20], along with a new analogue, mirabilin K (3), as shown in Figure 1. These metabolites belong to a class of tricyclic guanidine alkaloids exemplified by the ptilocaulins, [21,22,23] netamines, [20,24] and mirabilins [18,19,25].
Figure 1.
Structures of the tricyclic guanidine alkaloids mirabilin G (1), netamine M (2), and mirabilin K (3).
2. Results and Discussion
An aliquot of the crude CH2Cl2/MeOH extract (170 mg) of the sponge was subjected to diol flash chromatography eluting sequentially with hexanes, CH2Cl2, EtOAc, and MeOH. The PDCD4 stabilizing activity concentrated in the MeOH fraction, which was further subjected to C18, C8, and C2 reversed-phase chromatography. The trifluoroacetate salt of mirabilin G (1, 2.4 mg, 1.41% crude weight) was purified using a flash C8 reversed-phase column, while the trifluoroacetate salt of mirabilin K (3, 0.8 mg, 0.47% crude weight) was purified by C2 semipreparative HPLC. Each chromatography fraction was monitored by LCMS and 1H NMR, and it was apparent that signals corresponding to an additional analogue were lost with the addition of aqueous TFA to the chromatography solvents. The isolation was subsequently repeated without TFA using 373 mg of extract, with the PDCD4 stabilizing activity again concentrated in the MeOH eluent from the diol flash column. This fraction was then further purified by flash C8 reversed-phase chromatography to yield mirabilin G (1, 7.4 mg, 1.98% crude weight), netamine M (2, 4.8 mg, 1.29% crude weight) and an impure fraction that by MS and NMR analysis contained mirabilin K (3).
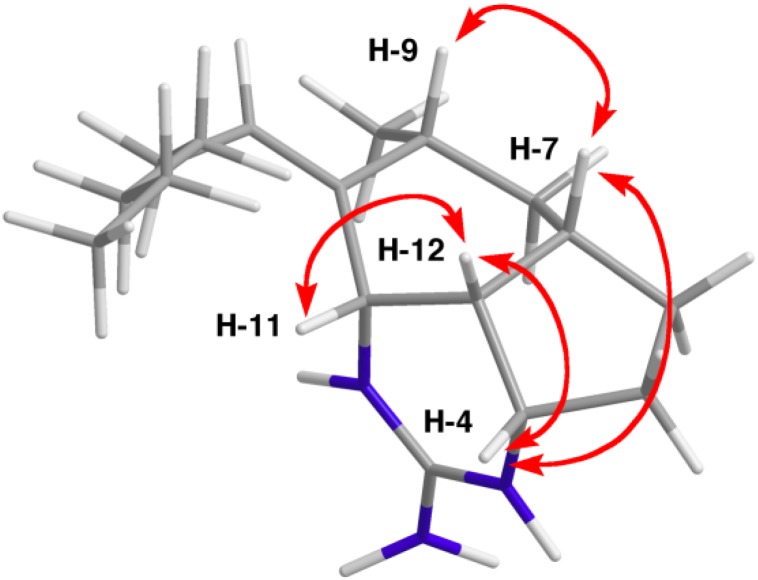
Mirabilin G trifluoroacetate salt (1) and netamine M naturally occurring counterion (2) were identified by comparison of their spectroscopic and chirooptical data with published values [18,19,20]. HRESIMS data for compound 3 established a molecular formula of C17H30N3, which required 5 double bond equivalents. Examination of the 1H and 13C NMR data for 3 revealed close correspondence to the NMR data for compound 1 (see Supplementary Table S1). However, compound 3 had only 3 sp2 hybridized carbon resonances which were characteristic of a guanidine group (δC 154.0) and a trisubstituted olefin (δC 138.1 and δC 132.2/δH 5.42). Another significant difference between 1 and 3 was the presence of a second nitrogen-substituted methine group (δH 3.98, d, J = 6.3 Hz, δC 54.9). This doublet methine showed HMBC correlations to C-2 (δC 154.0), C-7 (δC 34.6), C-9 (δC 30.0), C-12 (δC 39.6), and the two olefinic carbons, which placed it at C-11. The double bond was positioned between C-10 and C-1′ to complete the structure of mirabilin K (3). The relative configuration of 3 was assigned from ROESY data and coupling constant analysis. ROESY correlations from H-4 to both H-7 and H-12, H-7 to H-9, and H-11 to H-12 (see Table 1 and Figure 2) positioned H-4, H-7, H-9, H-11 and H-12 on a common face of the molecule. The 9-Me-anti assignment was further confirmed by analysis of the coupling constants of the resonances assigned to H-8α and β. Observation of a strong ROESY correlation from H-7 to H-8β (δH 1.84, ddd, J = 13.5, 5.0, 5.0 Hz) placed this resonance on the β-face of the molecule, while the coupling constant values observed for H-8α (δH 1.10, ddd, J = 13.5, 12.6, 12.6 Hz) confirmed it had a trans-di-axial relationship with both the H-7 and H-9 protons. The relative configuration of mirabilin K (3) was therefore proposed to be (4S*,7S*,9R*,11S*12R*).
Table 1.
NMR data for mirabilin K (3) trifluoroacetate salt in CDCl3.
| Position | δC a, Type | δH, Mult. (J in Hz) b | HMBC c | ROESY |
|---|---|---|---|---|
| 1-N | 7.71, br s | |||
| 2 | 154.0, C | |||
| C2-NH2 | 7.00, br s | |||
| 3-N | 7.05, br s | |||
| 4 | 52.9, CH | 3.80, br t (3.9) | 2, 6, 7 | 5b, 7, 12 |
| 5a | 32.9, CH2 | 1.89, m | 4, 7, 12 | |
| 5b | 1.64, m | 4, 7, 12 | 4 | |
| 6a | 29.6, CH2 | 1.99, m | 4, 5, 12 | |
| 6b | 1.43, m | 4, 5, 12 | ||
| 7 | 34.6, CH | 2.05, m | 4,6,12 | 4, 8a, 9 |
| 8a | 37.2, CH2 | 1.84, ddd (13.5, 5.0, 5.0) | 7, 9, 9Me, 12 | 7, 8b, 9 |
| 8b | 1.10, ddd (13.5, 12.6, 12.6) | 6, 9, 9Me, 12 | 8a | |
| 9 | 30.0, CH | 2.50, m | 8, 10, 11, 1′ | 7 |
| 9-Me | 22.4, CH3 | 1.06 3H, d (6.8) | 8, 9, 10 | 8a |
| 10 | 138.1, C | |||
| 11 | 54.9, CH | 3.98, d (6.3) | 7, 9, 10, 12, 1′ | 1′, 12 |
| 12 | 39.6, CH | 2.31, m | 4, 7, 8, 11 | 4, 11 |
| 1′ | 132.2, CH | 5.42, br t (7.1) | 9, 11, 3′ | 11, 2′, 3′ |
| 2′ | 27.8 CH2 | 2.02, m | 1, 10, 3′ | |
| 3′ | 29.6, CH2 | 1.24, m | 4′, 5′ | |
| 4′ | 31.5, CH2 | 1.26, m | 3′, 5′ | |
| 5′ | 22.5, CH2 | 1.28, m | 6′, 4′ | |
| 6′ | 14.0, CH3 | 0.88, t (6.8) | 4′, 5′ |
a Recorded at 150 MHz; b Recorded at 600 MHz; c Optimized for 8.3 Hz, correlations are from the proton(s) stated to the indicated carbon.
Figure 2.
Chem3D minimized core structure of mirabilin K (3) showing crucial ROESY correlations used to assign the relative configuration (4S*,7S*,9R*,11S*12R*).
Mirabilin G (1) and netamine M (2) were found to inhibit TPA-induced degradation of PDCD4 with EC50 values of 1.8 and 2.8 μg/mL, respectively, while mirabilin K (3) was inactive at the highest test concentration of 20 μg/mL (Table 2). Compound 2 was found to have a broader PDCD4 stabilizing range, exhibiting approximately 90% recovery of the luciferase signal at 5.5 μg/mL and cytotoxicity observed at 17 μg/mL. In contrast, mirabilin G (1) showed a maximum of 60% luciferase recovery and it was found to be cytotoxic at a significantly lower concentration of 4.2 μg/mL.
Table 2.
Biological activities of compounds 1–3.
| Compound | PDCD4 EC50 a (μg/mL) | PDCD4 EC90 a (μg/mL) | Cytotoxicity b (μg/mL) |
|---|---|---|---|
| 1 | 1.8 | - | 4.2 |
| 2 | 2.8 | 5.5 | 17.0 |
| 3 | n.a. c | - | 10.6 |
| Rapamycin d | 0.02 | 0.1 | >1.0 |
a EC50 and EC90 values represent the minimum concentration required for 50% and 90% recovery of the PDCD4-luciferase signal from TPA-induced degradation; b Values presented as the minimum concentration required for total luciferase signal loss, relative to TPA treated control cells; c Not active; d Positive control, concentration in μM.
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were recorded on a PerkinElmer 241 Polarimeter (PerkinElmer, Waltham, MA, USA) using a 1 dm cell in the solvent indicated. Ultraviolet-visible spectra were run as methanol solutions on a Varian Cary 50-Bio UV-Vis scanning spectrophotometer (Varian, Palo Alto, CA, USA). NMR spectra were recorded on a Bruker Avance DRX-600 spectrometer (Bruker BioSpin, Billerica, MA, USA) operating at 600 MHz for 1H nuclei and 150 MHz for 13C nuclei. Residual solvent signals were used as reference: CDCl3 δH 7.25, δC 77.0. Standard Bruker pulse sequences were utilized. High-resolution mass spectra were recorded using the Agilent 1100 Series LC/MSD model G1946D, with an API-electrospray source (Agilent Technologies, Santa Clara, CA, USA). Semi-preparative reversed-phase HPLC was run on a Varian ProStar 215 HPLC system (Varian, Palo Alto, CA, USA) using a Chromanetics Lichosorb C2 column (310 Å; 250 × 10 mm, Chromanetics Scientific Products, Trenton, NJ, USA) and eluting with a linear gradient of H2O (0.05% TFA) to MeCN at 2.0 mL/min. Reversed-phase flash column chromatography was carried out on C8 YMC stationary support with a pore size of 40–63 μm. Normal phase flash chromatography was carried out on Applied Separations Spe-ed SPE 2g diol cartridges (Applied Separations, Allentown, PA, USA).
3.2. Extraction and Isolation
The sponge sample was repeatedly extracted with CH2Cl2–MeOH (1:1) and 100% MeOH according to the methodology described by McCloud [26] to give the organic solvent crude extract. A portion of the crude organic extract (170 mg) was subjected to diol flash chromatography eluting with hexanes, CH2Cl2, EtOAc and MeOH. Mirabilin G (1) trifluoroacetate salt (2.4 mg, 1.41% crude weight) was purified from the MeOH fraction using a flash C8 reversed-phase column, eluting with MeOH–aqueous TFA (0.05%) (7:3). Mirabilin K (3) trifluoroacetate salt (0.8 mg, 0.47% crude weight) was purified from the MeOH diol fraction by semipreparative HPLC using a C2 reversed-phase column, eluting with MeCN–aqueous TFA (0.05%) (7:3), at 2 mL/min. Another portion of the crude extract (373 mg) was subjected to diol flash chromatography eluting with hexanes, CH2Cl2, EtOAc and MeOH, with the activity again concentrated in the MeOH fraction. The active fraction was further subjected to flash C8 reversed-phase chromatography eluting with MeOH-H2O (6:4) to yield mirabilin G (1) (7.4 mg, 1.98% crude weight) and mirabilin K (2) (4.8 mg, 1.29% crude weight).
Mirabilin G (1) naturally-occurring counterion: clear oil; 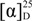 = +170 (c 0.1, CHCl3); UV (MeOH) λ (log ε) 202 (4.09), 260 (4.10) nm; 1H NMR (CHCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Supplementary Table S1; HRESIMS m/z [M + H]+ 274.2377 (calcd for C17H28N3, 274.2278). Trifluoroacetate salt: yellow oil;
= +170 (c 0.1, CHCl3); UV (MeOH) λ (log ε) 202 (4.09), 260 (4.10) nm; 1H NMR (CHCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Supplementary Table S1; HRESIMS m/z [M + H]+ 274.2377 (calcd for C17H28N3, 274.2278). Trifluoroacetate salt: yellow oil; 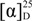 = +64 (c 0.325, CHCl3); HRESIMS m/z [M]+ 274.2270 (calcd for C17H28N3, 274.2278).
= +64 (c 0.325, CHCl3); HRESIMS m/z [M]+ 274.2270 (calcd for C17H28N3, 274.2278).
Netamine M (2) naturally-occurring counterion: clear oil; 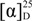 = +60 (c 0.1, CHCl3); UV (MeOH) λ (log ε) 202 (4.20), 230 (3.89) nm; ECD (MeOH) λmax (Δε) 204 (−2.9), 207 (0), 253 (0), 231 (+8.2), 288 (0) nm; 1H NMR (CHCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Supplementary Table S1; HRESIMS m/z [M]+ 274.2274 (calcd for C17H28N3, 274.2278).
= +60 (c 0.1, CHCl3); UV (MeOH) λ (log ε) 202 (4.20), 230 (3.89) nm; ECD (MeOH) λmax (Δε) 204 (−2.9), 207 (0), 253 (0), 231 (+8.2), 288 (0) nm; 1H NMR (CHCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Supplementary Table S1; HRESIMS m/z [M]+ 274.2274 (calcd for C17H28N3, 274.2278).
Mirabilin K (3) trifluoroacetate salt: yellow oil; 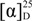 = +40 (c 0.1, CHCl3); UV (MeOH) λ (log ε) 202 (4.07), 240 (3.54) nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Table 1; HRESIMS m/z [M]+ 276.2439 (calcd for C17H30N3, 276.2434).
= +40 (c 0.1, CHCl3); UV (MeOH) λ (log ε) 202 (4.07), 240 (3.54) nm; 1H NMR (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data, see Table 1; HRESIMS m/z [M]+ 276.2439 (calcd for C17H30N3, 276.2434).
3.3. PDCD4 Assay
Stabilization of PDCD4 was assessed as previously described [14], using HEK-293 cells that could be readily transfected with appropriate PDCD4-luciferase plasmids. In brief, HEK-293 renal cells expressing a fusion protein comprised of a fragment of PDCD4 containing the stability regulatory region (amino acids 39–91) and luciferase were plated (2000 cells/well, 40 μL/well) in 384-well opaque white plates and allowed to attach overnight (18 h). TPA (final concentration 10 nM) was added followed (within 15 min) by test samples or controls. Following an 8 h incubation, luciferase activity was measured 10–15 min after the addition of Steadylite Plus (Perkin-Elmer) reagent. Controls were DMSO only (no TPA), TPA only, and TPA + rapamycin (100 nM final concentration). The activities of compounds were calculated using the following formula: activity (%) = (RLUcompound+TPA − RLUTPA)/(RLUDMSO − RLUTPA) × 100. Cytotoxicity was estimated based on the loss of luciferase activity in cells treated with TPA and the test compound relative to control cells treated only with TPA.
4. Conclusions
Using a high-throughput screen to identify natural products with the ability to stabilize the tumor suppressor protein PDCD4, we isolated three tricyclic guanidine alkaloids from an extract of the Australian marine sponge Acanthella cavernosa. The known compounds, mirabilin G (1) and netamine M (2), were found to be the active PDCD4 stabilizing compounds in the extract, while the structural analogue, mirabilin K (3), had a new structure but was inactive. Compounds that prevent PDCD4 degradation have attracted considerable attention recently as evidenced by efforts directed toward the total synthesis and SAR studies of the cryptocaryols [27,28,29]. To the best of our knowledge, this is the first report of a structural class of marine alkaloids that can rescue cellular PDCD4 levels under tumor-promoting conditions.
Acknowledgments
We thank David Newman (NCI) and Thomas McCloud (SAIC-Frederick) for the sponge extracts and Marzena Dyba and Sergey Tarasov (Biophysics Resource, SBL, NCI-Frederick) for assistance with the HRLCMS studies. This research was supported in part by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research. This project was also funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This work was further supported by the Deutsche Forschungsgemeinschaft (DFG) (GRK1172).
Supplementary Files
Author Contributions
J.S.B., M.M.B., and C.L.T. performed the screening assays. T.G. and K.R.G isolated and identified the structure of the natural products. T.S., N.H.C., C.J.H., M.L.P, and J.B.M. designed experiments and analyzed data. T.G. and K.R.G wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yang H.S., Jansen A.P., Komar A.A., Zheng X., Merrick W.C., Costes S., Lockett S.J., Sonenberg N., Colburn N.H. The Transformation Suppressor PDCD4 is a Novel Eukaryotic Translation Initiation Factor 4A Binding Protein that Inhibits Translation. Mol. Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang H.S., Jansen A.P., Nair R., Shibahara K., Verma A.K., Cmarik J.L., Colburn N.H. A Novel Transformation Suppressor, PDCD4, Inhibits AP-1 Transactivation but not NF-κB or ODC Transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 3.Leupold J.H., Yang H.S., Colburn N.H., Asangani I., Post S., Allgayer H. Tumor Suppressor PDCD4 Inhibits Invasion/Intravasation and Regulates Urokinase Receptor (u-PAR) Gene Expression via Sp-transcription Factors. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- 4.Nieves-Alicea R., Colburn N.H., Simeone A.M., Tari A.M. Programmed Cell Death 4 Inhibits Breast Cancer Cell Invasion by Increasing Tissue Inhibitor of Metalloproteinases-2 Expression. Breast Cancer Res. Treat. 2009;114:203–209. doi: 10.1007/s10549-008-9993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen A.P., Camalier C.E., Colburn N.H. Epidermal Expression of the Translation Inhibitor Programmed Cell Death 4 Suppresses Tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 6.Asangani I.A., Rasheed S.A., Nikolova D.A., Leupold J.H., Colburn N.H., Post S. MicroRNA-21 (miR-21) Post-transcriptionally Downregulates Tumor Suppressor PDCD4 and Stimulates Invasion, Intravasation and Metastasis in Colorectal Cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y., Liu W., Chao T., Zhang Y., Yan X., Gong Y., Qiang B., Yuan J., Sun M., Peng X., et al. MicroRNA-21 Down-regulates the Expression of Tumor Suppressor PDCD4 in Human Glioblastoma Cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 8.Wang X., Wei Z., Gao F., Zhang X., Zhou C., Zhu F., Wang Q., Gao Q., Ma C., Sun W., et al. Expression and Prognostic Significance of PDCD4 in Human Epithelial Ovarian Carcinoma. Anticancer Res. 2008;28:2991–2996. [PubMed] [Google Scholar]
- 9.Zhang H., Ozaki I., Mizuta T., Hamajima H., Yasutake T., Eguchi Y., Ideguchi H., Yamamoto K., Matsuhashi S. Involvement of Programmed Cell Death 4 in Transforming Growth Factor-β 1-Induced Apoptosis in Human Hepatocellular Carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 10.Dorrello N.V., Peschiaroli A., Guardavaccaro D., Colburn N.H., Sherman N.E., Pagano M. S6K1- and βTRCP-Mediated Degradation of PDCD4 Promotes Protein Translation and Cell Growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 11.Schmid T., Jansen A.P., Baker A.R., Hegamyer G., Hagan J.P., Colburn N.H. Translation Inhibitor PDCD4 is Targeted for Degradation during Tumor Promotion. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 12.Schmid T., Bajer M.M., Blees J.S., Eifler L.K., Milke L., Rübsamen D., Schulz K., Weigert A., Baker A.R., Colburn N.H., et al. Inflammation-induced Loss of PDCD4 is Mediated by Phosphorylation-Dependent Degradation. Carcinogenesis. 2011;32:1427–1433. doi: 10.1093/carcin/bgr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L.-X., Huang S.-X., Tang S.-K., Jiang C.-L., Duan Y., Beutler J.A., Henrich C.J., McMahon J.B., Schmid T., Blees J.S., et al. Actinopolysporins A-C and Tubercidin as a PDCD4 Stabilizer from the Halophilic Actinomycete Actinopolyspora erythraea YIM 90600. J. Nat. Prod. 2011;74:1990–1995. doi: 10.1021/np200603g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blees J.S., Schmid T., Thomas C.L., Baker A.R., Benson L., Evans J.R., Goncharova E.I., Colburn N.H., McMahon J.B., Henrich C.J., et al. Development of a High-throughput Cell-based Reporter Assay to Identify Stabilizers of Tumor Suppressor PDCD4. J. Biomol. Screen. 2010;15:21–29. doi: 10.1177/1087057109351028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grkovic T., Blees J.S., Colburn N.H., Schmid T., Thomas C.L., Henrich C.J., McMahon J.B., Gustafson K.R. Cryptocaryols A–H, A-Pyrone-Containing 1,3-Polyols from Cryptocarya Sp. Implicated in Stabilizing The Tumor Suppressor PDCD4. J. Nat. Prod. 2011;74:1015–1020. doi: 10.1021/np100918z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blees J.S., Bokesch H.R., Rübsamen D., Schulz K., Milke L., Bajer M.M., Gustafson K.R., Henrich C.J., McMahon J.B., Colburn N.H., et al. Erioflorin Stabilizes the Tumor Suppressor PDCD4 by Inhibiting its Interaction with the E3-ligase -TrCP1. PLoS One. 2012;7:e46567. doi: 10.1371/journal.pone.0046567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajer M.M., Kunze M.M., Blees J.S., Bokesch H.R., Chen H., Dong Z., Gustafson K.R., Biondi R.M., Henrich C.J., McMahon J.B., et al. Characterization of Pomiferin Triacetate as a Novel mTOR and Translation Inhibitor. Biochem. Pharmacol. 2014;88:313–321. doi: 10.1016/j.bcp.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capon R.J., Miller M., Rooney F. Mirabilin G: A New Alkaloid from a Southern Australian Marine Sponge, Clathria species. J. Nat. Prod. 2001;64:643–644. doi: 10.1021/np000564g. [DOI] [PubMed] [Google Scholar]
- 19.El-Naggar M., Conte M., Capon R.J. Mirabilins Revisited: Polyketide Alkaloids from a Southern Australian Marine Sponge, Clathria sp. Org. Biomol. Chem. 2010;8:407–412. doi: 10.1039/b915624k. [DOI] [PubMed] [Google Scholar]
- 20.Gros E., Al-Mourabit A., Martin M.-T., Sorres J., Vacelet J., Frederich M., Aknin M., Kashman Y., Gauvin-Bialecki A. Netamines H-N, Tricyclic Alkaloids from the Marine Sponge Biemna laboutei and their Antimalarial Activity. J. Nat. Prod. 2014 doi: 10.1021/np4009283. [DOI] [PubMed] [Google Scholar]
- 21.Harbour G.C., Tymiak A.A., Rinehart K.L., Shaw P.D., Hughes R.G., Mizsak S.A., Coats J.H., Zurenko G.E., Li L.H., Kuentzel S.L., et al. Ptilocaulin and Isoptilocaulin, Antimicrobial and Cytotoxic Cyclic Guanidines from the Caribbean Sponge Ptilocaulis aff. P. spiculifer (Lamarck, 1814) J. Am. Chem. Soc. 1981;103:5604–5606. doi: 10.1021/ja00408a071. [DOI] [Google Scholar]
- 22.Tavares R., Daloze D., Braekman J.C., Hajdu E., van Soest R.W.M. 8β-Hydroxyptilocaulin, a New Guanidine Alkaloid from the Sponge Monfloata arbuscula. J. Nat. Prod. 1995;58:1139–1142. doi: 10.1021/np50121a029. [DOI] [Google Scholar]
- 23.Patil A.D., Freyer A.J., Offen P., Bean M.F., Johnson R.K. Three New Tricyclic Guanidine Alkaloids from the Sponge Batzella sp. J. Nat. Prod. 1997;60:704–707. doi: 10.1021/np970014r. [DOI] [Google Scholar]
- 24.Sorek H., Rudi A., Gueta S., Reyes F., Martin M.J., Aknin M., Gaydou E., Vacelet J., Kashman Y. Netamines A–G: Seven New Tricyclic Guanidine Alkaloids from the Marine Sponge Biemna laboutei. Tetrahedron. 2006;62:8838–8843. doi: 10.1016/j.tet.2006.06.063. [DOI] [Google Scholar]
- 25.Barrow R.A., Murray L.M., Lim T.K., Capon R.J. Mirabilins (A–F): New Alkaloids from a Southern Australian Marine Sponge, Arenochalina mirabilis. Aust. J. Chem. 1996;49:767–773. [Google Scholar]
- 26.McCloud T.G. High Throughput Extraction of Plant, Marine and Fungal Specimens for Preservation of Biologically Active Molecules. Molecules. 2010;15:4526–4563. doi: 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., O’Doherty G.A. Cryptocaryols A and B: Total Synthesis, Stereochemical Revision, Structural Elucidation, and Structure-Activity Relationship. J. Am. Chem. Soc. 2013;135:9334–9337. doi: 10.1021/ja404401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy D.S., Mohapatra D.K. Total Synthesis and Structure Confirmation of Cryptocaryol A. Eur. J. Org. Chem. 2013;2013:1051–1057. doi: 10.1002/ejoc.201201309. [DOI] [Google Scholar]
- 29.Cuccarese M.F., Wanf Y., Beuning P.J., O’Doherty G.A. Cryptocaryol Structure-activity Relationship Study of Cancer Cell Cytotoxicity and Ability to Stabilize PDCD4. ACS Med. Chem. Lett. 2014;5:522–526. doi: 10.1021/ml4005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.