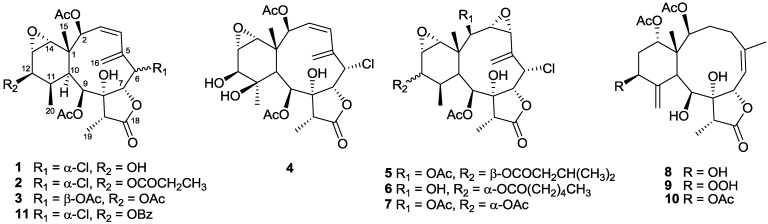
Abstract
Ten new briarane diterpenoids, briaviolides A–J (1–10), together with six known briaranes, solenolides A and D, excavatolide A, briaexcavatolide I, 4β-acetoxy-9-deacetystylatulide lactone and 9-deacetylstylatulide lactone, were isolated from the Taiwanese soft coral, Briareum violacea. Their structures were determined on the basis of spectroscopic data (1H- and 13C-NMR, 1H–1H COSY, HSQC, HMBC and NOESY), HR-MS and chemical methods. The absolute configuration of briaviolide A (1) was determined by X-ray crystallographic analysis. Compounds 5, 9 and derivative 11 showed moderate inhibitory activities on superoxide-anion generation and elastase release by human neutrophils in response to N-formyl-methionyl-leucyl-phenylalanine/Cytochalasin B (fMLP/CB).
Keywords: Briareum violacea, briarane diterpenoids, briaviolides, anti-inflammatory activities
1. Introduction
The briarane diterpenoids [1] continue to attract the attention of natural product chemists because of their structural complexity and interesting biological activities, such as anti-inflammatory [2], antiviral [3], cytotoxic [4,5,6], antifouling [7,8], immuno-modulatory [9], insecticidal [10] and reversal of multidrug resistance [11]. The structures of these diterpenoids are characterized by a highly oxygenated bicyclo[8.4.0]tetradecane skeleton that is frequently attached with a γ-lactone moiety. Since the first structural elucidation of briarein A isolated from Briareum asbestinum in 1977 [12], more than 450 briarane-type diterpenoids have been reported from Octocorallia, including Gorgonacea, Pennatulacea, Alcyonacea and Stolonifera [13,14,15]. In serial studies of the Taiwanese gorgonian corals, many new briaranes have been isolated, including juncenolides A–G from Junceella juncea [16,17,18], frajunolides A–K from J. fragilis [19,20] and briaviodiol A from B. violacea [21].
In this paper, we report the investigation of Taiwanese soft coral Briareum violacea that provided ten new briarane-type diterpenoids, briaviolides A–J (1–10) (Figure 1), along with six known analogues, solenolides A and D, excavatolide A, briaexcavatolide I, 4β-acetoxy-9-deacetystylatulide lactone and 9-deacetylstylatulide lactone. The structures of new compounds were established by spectroscopic and chemical methods. Among them, the structure of 1 was further confirmed by single-crystal X-ray analysis. The in vitro anti-inflammatory activities of new compounds (1–10) and new derivative 11 were also tested for their inhibition of elastase release and superoxide-anion generation from human neutrophils.
Figure 1.
Briaviolides A–J (1–10) isolated from Briareum violacea.
2. Results and Discussion
Briaviolide A (1) was isolated as colorless prisms. Its ESIMS revealed two isotopic [M + Na] + and [M + Na + 2]+ peaks for pseudo-molecular ions at m/z 521 and 523 (3:1) and HRESIMS at m/z 521.1554 [M + Na]+, indicating a molecular formula C24H31O9Cl, which contains one chlorine atom and accounts for nine degrees of unsaturation. The IR spectrum of 1 showed absorption frequencies at 3524, 1767 and 1737 cm−1, indicating the presence of hydroxyl, γ-lactone and carbonyl ester functionalities, respectively. The 1H- and 13C-NMR spectroscopic data (Table 1 and Table 2) exhibited signals of two acetate methyl singlets at δH 2.09 (δC 21.1) and 2.17 (δC 22.1), with corresponding carbonyl signals at δC 170.3, 170.2, respectively. The carbonyls showed respective HMBC connectivity (Figure 2) with two methine doublets at δH 6.09 and 5.30, revealing the positions of each acetate group (C-2 and C-9, respectively). The carbonyl signal of a γ-lactone (δC 174.9) was connected with a secondary methyl doublet at δH 1.17 (δC 6.3) by the HMBC (H-19/C-18) and COSY (H-19/H-17) correlations. The connection between C-17 and C-8 (δC 83.9) was elucidated by the HMBC correlation from H-19 to C-8. The hydroxyl proton (δH 3.52) of C-8 was correlated with the adjacent lactonide carbon C-7 and C-9 by virtue of the HMBC correlations from 8-OH to C-7 and C-9. These correlations along with the H-6/H-7 (COSY) correlation fixed the position of the lactone ring. The signals of an exocyclic double bond were observed at δH 6.24 (br s) and 5.95 (d, J = 2.1 Hz) (δC 118.7) and were conjugated with an endocyclic double bond at δH 5.63 (dd, J = 11.4, 9.0 Hz; δC 131.0), 5.90 (d, J = 11.4 Hz; δC 128.0) by related HMBC and COSY correlations. The correlations of H-6/H-7 (COSY) and H-16/C-6 (HMBC) thus connected the conjugated double bond with the γ-lactone ring. The signals of a pair of methine protons at δH 3.23 (H-13) and δH 2.99 (H-14) were assigned for an epoxide ring. This moiety was neighbored on a quaternary carbon (δC 40.6) and a hydroxylated carbon (δC 70.5; δH 3.71) by the HMBC correlation of H-12/C-13 and H-14/C-1. The COSY correlations of H-12/H-11/H-20 and H-9/H-10 indicated the presence of two proton sequences. These two sequences were connected by the HMBC correlations of H-20/C-10 and H-9/C-11. By deducing the unsaturation of two acetates, conjugated diene, a γ-lactone ring and the epoxide ring, the remaining two degrees of unsaturation strongly suggested that Compound 1 possesses two additional rings belong to briarane-type diterpenoids.
Table 1.
1H-NMR spectroscopic data (in ppm, J in Hz) of briaviolides A–J (1–10) and Compound 11.
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 a | 9 b | 10 c | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 6.09 (d, 9.0) | 6.12 (d, 9.2) | 6.10 (d, 9.5) | 6.21 (d, 10.0) | 5.13 (d, 9.3) | 3.66 (d, 8.0) | 5.23 (d, 9.5) | 5.34 (d, 9.2) | 5.16 (d, 7.6) | 5.76 (d, 8.8) | 6.19 (d, 9.0) |
| 3 | 5.63 (dd, 11.4, 9.0) | 5.16 (dd, 11.2, 9.2) | 5.61 (dd, 11.5, 9.5) | 5.60 (dd, 11.6, 9.6) | 3.39 (dd, 9.3, 3.9) | 3.39 (dd, 8.0, 3.6) | 3.41 (dd, 9.5, 4.4) | 2.90 (m) | 2.90 (td, 14.8, 5.2) | 3.32 (m) | 5.65 (dd, 12.0, 9.0) |
| 1.45 (m) | 1.49 (m) | 1.67 (m) | |||||||||
| 4 | 5.90 (d, 11.4) | 5.89 (d, 11.2) | 6.15 (d, 11.5) | 5.93 (d, 10.0) | 3.60 (d, 4.0) | 3.72 (d, 3.6) | 3.66 (d, 4.0) | 2.47 (br d, 10.0) | 2.48 (br d, 14.0) | 2.53 (br d, 15.6) | 5.93 (d, 12.0) |
| 1.73 (td, 15.2, 4.4) | 1.83 (dd, 14.0, 4.4) | 1.80 (m) | |||||||||
| 6 | 5.21 (br s) | 5.20 (br s) | 5.75 (d, 10.0) | 5.08 (br d, 2.8) | 5.37 (d, 2.0) | 5.40 (br s) | 5.43 (d, 3.5) | 5.31 (d, 11.6) | 5.41 (br d, 9.6) | 5.88 (d, 8.8) | 5.23 (br s) |
| 7 | 4.94 (d, 3.6) | 4.93 (d, 3.2) | 4.66 (d, 10.0) | 4.97 (d, 4.4) | 5.03 (d, 2.0) | 4.68 (br s) | 5.02 (d, 3.0) | 5.52 (d, 9.6) | 5.51 (d, 10.0) | 6.01 (d, 9.6) | 4.79 (d, 3.6) |
| 9 | 5.30 (d, 7.8) | 5.24 (d, 7.6) | 5.29 (d, 8.0) | 5.76 (d, 6.0) | 5.32 (m) | 5.30 (d, 9.0) | 5.33 (d, 9.0) | 4.68 (br s) | 4.40 (m) | 5.10 (m) | 5.30 (d, 7.5) |
| 10 | 1.95 (dd, 7.8, 2.0) | 2.01 (dd, 7.2, 2.0) | 2.03 (m) | 2.22 (d, 6.0) | 1.74 (dd, 8.8, 2.4) | 1.83 (m) | 2.04 (dd, 9.0, 2.5) | 3.24 (s) | 3.18 (br s) | 3.89 (s) | 1.84 (d, 7.2) |
| 11 | 2.07 (m) | 2.05 (m) | 2.22 (m) | 2.30 (m) | 2.30 (m) | 2.35 (m) | 2.10 (m) | ||||
| 12 | 3.71 (d, 4.2) | 4.62 (d, 4.4) | 4.63 (d, 5.0) | 3.56 (s) | 4.64 (d, 4.8) | 4.68 (br s) | 4.70 (dd, 5.0, 2.5) | 4.13 (dd, 10.8, 5.2) | 4.40 (m) | 5.50 (m) | 4.99 (d, 4.5) |
| 13 | 3.23 (d, 3.3) | 3.15 (d, 3.2) | 3.18 (br d, 1.5) | 3.24 (d, 4.0) | 3.12 (br s) | 3.60 (m) | 3.53 (dd, 3.5, 5.5) | 2.01 (m) | 2.09 (ddd, 14.4, 6.0, 3.2) | 2.38 (m) | 3.32 (d, 3.0) |
| 1.50 (m) | 1.67 (ddd, 14.4, 9.2, 3.2) | 1.86 (m) | |||||||||
| 14 | 2.99 (d, 3.6) | 2.97 (d, 3.6) | 2.99 (d, 3.0) | 2.94 (d, 4.0) | 2.90 (d, 3.6) | 3.24 (d, 3.0) | 2.87 (d, 3.0) | 4.68 (br s) | 4.73 (t, 3.2) | 5.10 (m) | 3.04 (d 3.3) |
| 15 | 1.13 (s) | 1.12 (s) | 1.14 (s) | 1.21 (s) | 1.23 (s) | 1.11 (s) | 1.19 (s) | 1.08 (s) | 1.17 (s) | 1.42 (s) | 1.17 (s) |
| 16 | 6.24 (br s) | 6.26 (br s) | 6.10 (s) | 6.23 (br s) | 6.13 (d, 2.0) | 5.61 (br s) | 6.11 (d, 2.5) | 1.85 (s) | 1.91 (s) | 2.04 (s) | 6.31 (br s) |
| 5.95 (d, 2.1) | 6.00 (s) | 5.72 (s) | 5.94 (s) | 6.04 (d, 2.0) | 5.98 (d, 1.8) | 6.07 (d, 2.5) | 6.04 (d, 1.8) | ||||
| 17 | 2.37 (q, 6.9) | 2.35 (q, 7.6) | 2.48 (q, 7.0) | 2.37 (q, 7.6) | 2.45 (q, 6.9) | 2.43 (q, 7.2) | 2.41 (q, 7.0) | 3.05 (q, 7.2) | 3.09 (q, 7.2) | 3.63 (q, 7.6) | 2.34 (q, 7.5) |
| 19 | 1.17 (d, 6.9) | 1.14 (d, 7.6) | 1.16 (d, 7.0) | 1.18 (d, 7.6) | 1.18 (d, 6.9) | 1.16 (d, 7.2) | 1.19 (d, 6.5) | 1.02 (d, 7.2) | 1.06 (d, 7.2) | 1.43 (d, 7.6) | 1.18 (d, 7.5) |
| 20 | 1.04 (d, 6.9) | 1.05 (d, 6.8) | 1.05 (d, 7.0) | 1.29 (s) | 1.03 (d, 6.9) | 1.05 (d, 7.2) | 1.06 (d, 7.5) | 5.45 (s) | 5.32 (br s) | 5.65 (s) | 1.16 (d, 7.2) |
| 5.22 (s) | 5.25 (br s) | 5.48 (s) | |||||||||
| 2' | 2.38 (m) | 2.20 (m) | 2.28 (m) | ||||||||
| 3' | 1.15 (t, 8.0) | 2.10 (m) | 1.61 (m) | 7.46 (m) | |||||||
| 4' | 0.96 (d, 6.3) | 1.31 (m) | 7.59 (t, 7.2) | ||||||||
| 5' | 0.96 (d, 6.3) | 1.31 (m) | 7.47 (m) | ||||||||
| 6' | 0.90 (t, 7.0) | 7.59 (t, 7.2) | |||||||||
| 7' | 7.46 (m) | ||||||||||
| 2-OAc | 2.09 (s) | 2.07 (s) | 2.08 (s) | 2.18 (s) | 2.12 (s) | 2.20 (s) | 2.14 (s) | 1.90 (s) | 1.96 (s) | 2.06 (s) | 2.09 (s) |
| 6-OAc | 2.07 (s) | ||||||||||
| 9-OAc | 2.17 (s) | 2.15 (s) | 2.13 (s) | 2.08 (s) | 2.19 (s) | 2.20 (s) | 2.22 (s) | 2.19 (s) | |||
| 12-OAc | 2.19 (s) | 2.06 (s) | 2.05 (s) | ||||||||
| 14-OAc | 1.90 (s) | 1.94 (s) | 1.85 (s) | ||||||||
| 8-OH | 3.52 (s) | 3.48 (s) | 7.38 (s) | 3.49 (s) | |||||||
| 9-OH | 7.79 (d, 7.6) |
a Recorded in d6-acetone at 400 MHz; b Recorded in pyridine-d5 at 400 MHz; c Recorded in CD3OD at 400 MHz.
Table 2.
13C-NMR spectroscopic data (δ in ppm, mult.) of briaviolides A–J (1–10) and Compound 11 a.
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 b | 9 c | 10 d | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 40.6 (s) | 41.9 (s) | 40.8 (s) | 42.9 (s) | 38.3 (s) | 38.8 (s) | 38.5 (s) | 49.5 (s) | 48.9 (s) | 47.7 (s) | 41.9 (s) |
| 2 | 75.5 (d) | 75.8 (d) | 75.3 (d) | 75.6 (d) | 74.9 (d) | 74.0 (d) | 75.5 (d) | 74.8 (d) | 76.6 (d) | 73.4 (d) | 76.0 (d) |
| 3 | 131.0 (d) | 130.4 (d) | 130.4 (d) | 129.1 (d) | 60.0 (d) | 62.3 (d) | 59.9 (d) | 32.3 (t) | 32.4 (t) | 30.1 (t) | 130.6 (d) |
| 4 | 128.0 (d) | 127.7 (d) | 128.0 (d) | 129.1 (d) | 57.0 (d) | 57.8 (d) | 56.9 (d) | 29.5 (t) | 29.4 (t) | 27.3 (t) | 127.9 (d) |
| 5 | 136.6 (s) | 135.8 (s) | 137.2 (s) | 136.3 (s) | 133.3 (s) | 135.1 (s) | 133.9 (s) | 141.3 (s) | 144.0 (s) | 141.0 (s) | 135.8 (s) |
| 6 | 62.4 (d) | 63.2 (d) | 75.3 (d) | 61.6 (d) | 61.1 (d) | 61.2 (d) | 61.1 (d) | 120.4 (d) | 121.7 (d) | 120.3 (d) | 62.9 (d) |
| 7 | 78.3 (d) | 78.3 (d) | 81.5 (d) | 78.9 (d) | 76.4 (d) | 76.5 (d) | 76.2 (d) | 79.9 (d) | 81.3 (d) | 79.4 (d) | 78.4 (d) |
| 8 | 83.9 (s) | 84.4 (s) | 80.4 (s) | 82.5 (s) | 83.9 (s) | 84.9 (s) | 84.8 (s) | 83.6 (s) | 85.1 (s) | 82.9 (s) | 84.4 (s) |
| 9 | 69.9 (d) | 70.5 (d) | 69.9 (d) | 68.6 (d) | 69.4 (d) | 68.7 (d) | 68.6 (d) | 75.7 (d) | 75.0 (d) | 73.9 (d) | 70.6 (d) |
| 10 | 38.1 (d) | 38.9 (d) | 37.2 (d) | 41.5 (d) | 37.4 (d) | 32.9 (d) | 33.0 (d) | 41.8 (d) | 42.2 (d) | 40.5 (d) | 38.9 (d) |
| 11 | 40.1 (d) | 39.1 (d) | 36.6 (d) | 77.0 (d) | 36.0 (d) | 35.9 (d) | 35.9 (d) | 156.2 (s) | 152.1 (s) | 151.1 (s) | 38.6 (d) |
| 12 | 70.5 (d) | 73.7 (d) | 72.4 (d) | 74.9 (d) | 71.4 (d) | 69.7 (d) | 69.2 (d) | 69.4 (d) | 83.8 (d) | 70.5 (d) | 73.3 (d) |
| 13 | 59.6 (d) | 58.3 (d) | 57.1 (d) | 59.5 (d) | 56.7 (d) | 53.0 (d) | 52.2 (d) | 38.7 (t) | 33.8 (t) | 33.4 (t) | 58.3 (d) |
| 14 | 62.9 (d) | 62.9 (d) | 62.0 (d) | 62.1 (d) | 61.4 (d) | 62.4 (d) | 61.3 (d) | 75.7 (d) | 76.3 (d) | 73.8 (d) | 63.2 (d) |
| 15 | 16.0 (q) | 17.7 (q) | 15.6 (q) | 15.5 (q) | 16.4 (q) | 15.7 (q) | 16.4 (q) | 15.2 (q) | 14.9 (q) | 12.9 (q) | 17.6 (q) |
| 16 | 118.7 (d) | 119.1 (d) | 123.1 (d) | 117.6 (d) | 121.0 (d) | 118.8 (d) | 120.6 (d) | 28.4 (q) | 27.9 (q) | 26.4 (q) | 119.3 (d) |
| 17 | 45.0 (d) | 46.3 (d) | 45.1 (d) | 45.6 (d) | 45.5 (d) | 45.2 (d) | 45.2 (d) | 45.8 (d) | 45.6 (d) | 44.2 (d) | 46.3 (d) |
| 18 | 174.9 (s) | 173.3 (s) | 174.7 (s) | 175.3 (s) | 174.1 (s) | 174.5 (s) | 174.0 (s) | 175.1 (s) | 180.4 (s) | 176.4 (s) | 173.5 (s) |
| 19 | 6.3 (q) | 8.2 (q) | 6.2 (q) | 6.9 (q) | 6.1 (q) | 6.2 (q) | 6.1 (q) | 8.3 (q) | 7.0 (q) | 6.2 (q) | 8.1 (q) |
| 20 | 8.8 (q) | 11.2 (q) | 9.3 (q) | 18.2 (q) | 9.7 (q) | 13.1 (q) | 12.9 (q) | 106.2 (t) | 111.5 (t) | 106.8 (t) | 11.4 (q) |
| 2-OCOCH3 | 170.3 (s) | 169.0 (s) | 169.7 (s) | 170.2 (s) | 169.0 (s) | 169.8 (s) | 170.0 (s) | 168.6 (s) | 172.3 (s) | 169.4 (s) | 169.3 (s) |
| 2-OCOCH3 | 21.1 (q) | 23.6 (q) | 21.4 (q) | 22.0 (q) | 20.9 (q) | 22.0 (q) | 20.5 (q) | 22.4 (q) | 21.5 (q) | 19.8 (q) | 22.6 (q) |
| 6-OCOCH3 | 169.6 (s) | ||||||||||
| 6-OCOCH3 | 21.0 (q) | ||||||||||
| 9-OCOCH3 | 170.2 (s) | 168.7 (s) | 170.2 (s) | 169.8 (s) | 169.5 (s) | 169.4 (s) | 168.9 (s) | ||||
| 9-OCOCH3 | 22.1 (q) | 22.7 (q) | 21.0 (q) | 21.1 (q) | 21.9 (q) | 21.9 (q) | 22.6 (q) | ||||
| 12-OCOCH3 | 170.3 (s) | 169.2 (s) | 168.0 (s) | ||||||||
| 12-OCOCH3 | 21.9 (q) | 20.9 (q) | 19.5 (q) | ||||||||
| 14-OCOCH3 | 169.0 (s) | 172.7 (s) | 168.9 (s) | ||||||||
| 14-OCOCH3 | 22.1 (q) | 21.3 (q) | 19.8 (q) | ||||||||
| 1′ | 171.2 (s) | 171.9 (s) | 173.1 (s) | 164.7 (s) | |||||||
| 2′ | 29.1 (t) | 43.2 (t) | 34.0 (t) | 129.7 (s) | |||||||
| 3′ | 10.9 (q) | 25.7 (d) | 24.8 (t) | 129.5 (d) | |||||||
| 4′ | 22.3 (q) | 31.4 (t) | 128.5 (d) | ||||||||
| 5′ | 22.3 (t) | 22.4 (t) | 133.0 (d) | ||||||||
| 6′ | 14.0 (q) | 128.5 (d) | |||||||||
| 7′ | 129.5 (d) |
a Assignments made using the HSQC and HMBC techniques; b Recorded in Acetone-d6 at 100 MHz; c Recorded in pyridine-d5 at 100 MHz; d Recorded in CD3OD at 100 MHz.
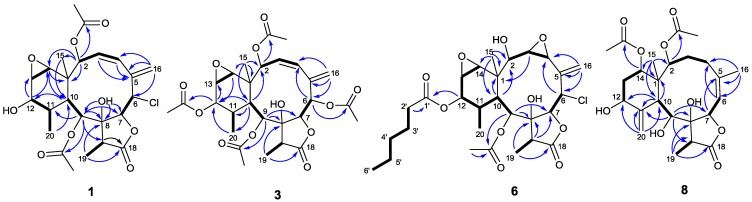
Figure 2.
Key HMBC (arrows) and COSY (bold lines) correlations of 1, 3, 6 and 8.
A literature survey revealed that the 1H- and 13C-NMR spectroscopic data of 1 showed similarity with those of briaexcavatolide I [22]. The only difference between them was 1 having two acetyl groups, while there are three in briaexcavatolide I. Acetylation of 1 gave a triacetyl product that was identical to briaexcavatolide I after comparing with their 1H and 13C NMR data. Benzoylation of 1 yielded a monobenzoyl derivative 11 that confirmed the secondary hydroxyl group at C-12.
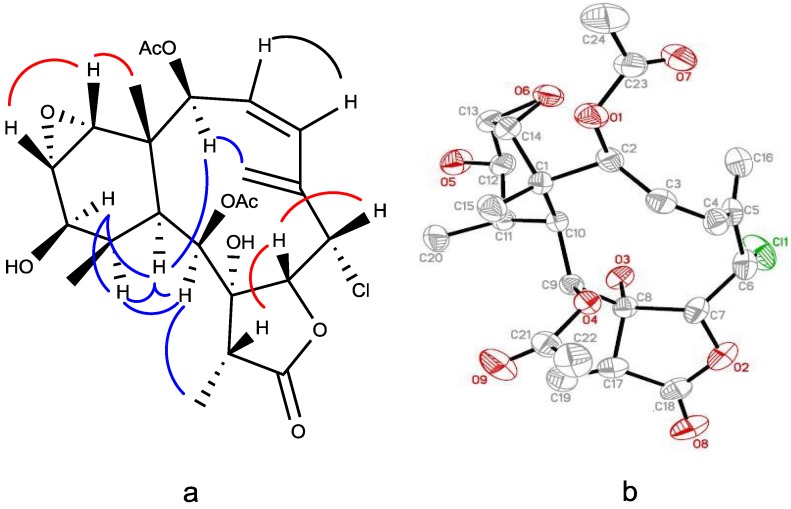
The relative configuration of 1 was determined by NOESY (Figure 3a) and X-ray diffraction analysis (Figure 3b). Naturally occurring briaranes have β-face of Me-15 and α-orientation of H-10. The NOESY correlations between H-2/H-16, H-2/H-10, H-10/H-12, H-12/H-11, H-11/H-9 and H-9/Me-19 required that all of these groups were in α-face, and correlations of H-6/H-7, H-7/H-17, Me-15/H-14 and H-14/H-13 indicated β-disposition for these groups. The correlations between H-16/H-2 and H-2/H-10 suggested that the conjugated diene had a s-cis geometry and forced the ten-membered ring to adopt a boat-like conformation with C-1 and C-5 at the bow and stern positions. An X-ray crystallographic analysis established the complete structure and stereochemistry of 1 as shown by the Oak Ridge Thermal Ellipsoid Plot (ORTEP) diagram stereo-drawing in Figure 3b. The negative optical rotation value of 1 was similar to that of briaexcavatolide I [23] in direction and magnitude, suggesting that 1 and briaexcavatolide I had 1S,10S-configurations in the ring junction. Thus, the structure of briaviolide A (1) was determined as (1S,2S,3Z,6S,7R,8R,9S,10S,11R,12R,13S,14R,17R)-6-chloro-13,14-epoxy-2,9-diacetoxy-8,12-dihydroxybriaran-3(4),5(16)-dien-18,7-olide.
Figure 3.
(a) Key NOESY correlations of Compound 1; (b) ORTEP (Oak Ridge Thermal Ellipsoid Plot) diagram showing the crystallographic atom-numbering scheme and solid state conformation of 1.
Briaviolide B (2) was isolated as a colorless gum, having a molecular formula of C27H35O10Cl as deduced from the high-resolution ESIMS. Similar to those of 1, Compound 2 showed IR bands at 3453, 1783, 1738 and 1732 cm−1, indicating hydroxyl, γ-lactone and ester carbonyl functionalities, respectively. Comparisons of its 1H- and 13C-NMR data (Table 1 and Table 2) with those of 1 revealed strong resemblance in all signals, except that the C-12 and H-12 signals in the NMR spectra of 2 were shifted downfield to δC 73.7 and δH 4.62 (d, J = 4.4 Hz), respectively, suggesting that Compound 2 had an ester group at position C-12. This ester group was revealed to be a propionyloxy group (δH 2.38, m; δH 1.15, t, J = 8.0 Hz). The structure of 2 was further supported by COSY, HSQC and HMBC experiments. The NOESY cross-peaks of 2 and 1 were quite similar, suggesting that they have the same relative configuration. Thus, briaviolide B (2) was established to be a 12-propionyloxy derivative of 1.
The HRESIMS and 13C-NMR data (Table 2) of Compound 3 suggested a molecular formula of C28H36O12 that contains eleven degrees of unsaturation. It was found that the 1H-, 13C-NMR and IR spectroscopic data of 3 were very similar to those of Compound 1, except for the signals of two more acetyl groups, including two methyl singlets, δH 2.07 (δC 21.0), δH 2.19 (δC 21.9), and the respective two carbonyls, δC 169.6, δC 170.3. The HMBC correlations (Figure 2) of H-6 (δH 5.75, d, J = 10.0 Hz)/δC 169.6 and H-12 (δH 4.63, d, J = 5.0 Hz)/δC 170.3 suggested that two acetoxyl group were attached at C-6 and C-12. Compound 3 was the first example of a briarane-type diterpenoid that contains an ester group at C-6. The configuration of Compound 3 was determined by using NOESY correlations (Figure 4) and comparing the data with those of 1. The NOESY correlations of Me-15/Me-20, H-14; H-13/H-14 and H-7/H-17 indicated that all these atoms were β-oriented and the correlations of H-10/H-2, H-9, H-12; H-12/Me-19 suggested that all of these atoms were α-oriented. In addition, the NOESY correlations between OH-8 (δH 3.48, s) and H-6, H-10, Me-19 confirmed the α-orientation of H-6 and OH-8. On the basis of the above observations, the structure of briaviolide C (3) was assigned as the 2,6,9,12-tetraacetyl derivative of 1, having 1S, 2S, 6R, 9S, 10S, 11R, 12R, 13S and 14R configurations.
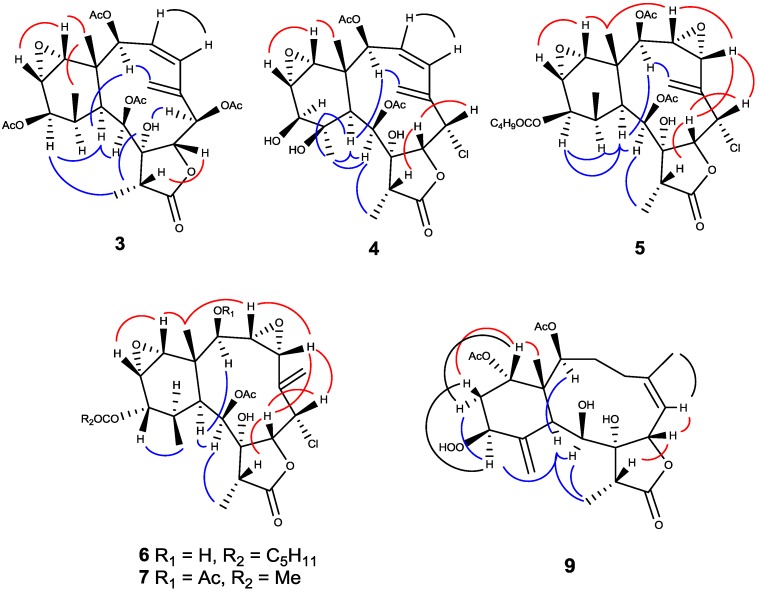
Figure 4.
Key NOESY correlations of Compounds 3–7 and 9.
A pseudo-molecular ion peak at m/z 537.1506 [M + Na]+ in the HRESIMS suggested that briaviolide D (4) had the molecular formula of C24H31O10Cl with nine degrees of unsaturation. The presence of a chlorine atom was supported by an isotope peak at m/z 539 in the LRESI spectrum, having one third of the intensity relative to m/z 537. The NMR spectroscopic data (Table 1 and Table 2) and IR spectrum revealed that Compound 4 possessed an 8-hydroxybriarane structure similar to that of Compound 1, except for one additional hydroxyl group at C-11 (δC 77.0). This finding was confirmed from 13C-NMR data and HMBC correlations between H-10/C-11, H-12/C-11 and Me-20/C-11. The NOESY correlations of H-10/Me-11, H-12 required that the methyl group at C-11 be α-oriented. The configurations of other chiral centers are similar to Compound 1 as ascertained by NOESY experiments (Figure 4). Thus, the structure of briaviolide D (4) was established as (1S,2S,6S,7R,8R,9S,10S,11R,12S,13S,14R,17R)-6-chloro-13,14-epoxy-2,9-diacetoxy-8,11,12-trihydroxybriaran-3(4),5(16)-dien-18,7-olide.
The HRESIMS data of 5 agreed with the molecular formula C29H39O11Cl containing a chlorine atom and 10 degrees of unsaturation. The IR bands at 3429, 1779 and 1738 cm−1 suggested the presence of hydroxyl group, ester groups and a γ-lactone moiety. A detailed inspection of 1H and 13C-NMR spectroscopic data (Table 1 and Table 2) of 5 indicated the presence of an 8-hydroxybriarane skeleton with two epoxides, two acetate esters and an isovalerate ester group. It was found that the spectroscopic data of Compound 5 were very similar to those of brianolide [23]. However, cross comparisons of 1H- and 13C-NMR spectra showed that the 12-acetoxyl signals in brianolide were replaced by an isovalerate ester group (δH 2.20, m; 2.10, m; 0.96 d, 6H, J = 6.3 Hz); (δC 171.9, 43.2, 25.7, 22.3 × 2) in Compound 5. The configurations of 5 was deduced by the NOESY analysis (Figure 4) and was based on the X-ray analysis of brianolide [24]. It was observed that the introduction of the 3,4-epoxide ring on the skeleton does not affect the boat-like conformation of the macro-ring, as revealed by the same NOESY interactions between H-16/H-2 and H-2/H-10 as those of Compound 1. Thus, the structure of briaviolide E (5) was established as (1R,2R,3R,4R,6S,7R,8R,9S,10S,11R,12R,13S,14R,17R)-6-chloro-3(4),13(14),-diepoxy-2,9-diacetoxy-12-isovaleryloxy-8-hydroxybriaran-5(16)-dien-18,7-olide.
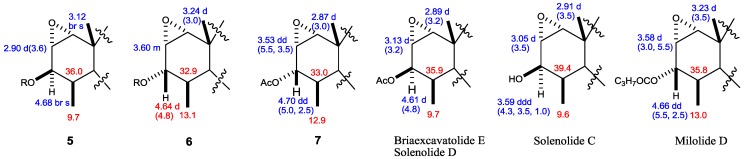
The HRESIMS of briaviolide F (6) showed a pseudo-molecular ion peak at m/z 593.2125 [M + Na]+, consistent with a molecular formula of C28H39O10Cl (Δ = 10). Cross comparison of 1H- and 13C-NMR spectroscopic data of 6 (Table 1 and Table 2) with those of 5 revealed that the differences were the substituents at C-2 and configurations at C-12. Comparison of the proton chemical shift of H-2 with that of milolide D [23] (Figure 5) could confirm the presence of a hydroxyl group at C-2 (δC 74.0) in 6. Signals of an acetyl group (δH 2.20; δC 169.8, 22.0) located at C-9 (δC 68.7) and one hexanoate group (δH 2.28, 1.61, 1.31, 1.31, 0.90; δC 173.1, 34.0, 24.8, 31.4, 22.4, 14.0) located at C-12 (δC 69.7) were determined by their HMBC correlations (Figure 2). The NOESY correlations (Figure 4) of H-10/H-2 and H-9, H-9/Me-19 suggested that H-2, H-9, H-10 and Me-19 were placed in α-orientation. Correlations of H-7/H-6 and H-17, H-12/H-13 and Me-20, H-14/H-13 and Me-15 agreed with β-orientations for all these groups. Thus, Compound 6 was established as a 2β-hydroxyl-12α-hexanoyl derivative of 5.
Figure 5.
Selected 1H- and 13C-NMR data of Compounds 5–7 and compared with known compounds.
Briaviolide G (7) had the molecular formula C26H33O11Cl, as determined by HRESIMS analysis. It was found that the 1H-, 13C-NMR (Table 1 and Table 2) and IR spectroscopic data were very similar to those of Compound 6. However, the hexanoate group at C-12 in 6 was replaced by an acetyl group at C-2 in 7. This finding was supported by the HMBC correlations between H-2 (δH 5.23, d, J = 9.5 Hz)/carbonyl carbon (δC 170.0) and H-12 (δH 4.70, dd, J = 5.0, 2.5 Hz)/carbonyl carbon (δC 169.4). Comparing the 1H- and 13C-NMR data of 7 with those of solenolide D [25] and briaexcavatolide E [22] indicated their resemblance, except for the chemical shifts around C-12 and C-13. Based on Kobayashi’s and Williams’s study [26], the configuration of 12-OH could be assigned (Figure 5). By comparison of the coupling constant of H-12/H-13 and the chemical shift of C-20, as well as the NOESY correlation between H-12 and Me-20 (δH 1.06), the acetyl group at C-12 was assigned to be α-face. The other NOSEY correlations (Figure 4) also indicated that Compound 7 had identical configurations as those of 6. Therefore, 7 was assigned a 2β-acetoxyl-12α-acetoxyl derivative of Compound 6.
Briaviolide H (8) was obtained as an amorphous gum. The molecular formula of 8 was determined as C24H34O9 by its HRESIMS. The IR spectrum showed absorption bands due to hydroxyl group (3440 cm−1), γ-lactone (1732 cm−1) and ester carbonyl (1712 cm−1). The 1H- and 13C-NMR spectra of 8 in CDCl3 gave broad signals, while those in acetone-d6 were well resolved. Characteristic resonances due to three methyl protons (δH 1.02, d, J = 7.2 Hz; 1.08, s; 1.85, s; each 3H) and two acetyl protons (δH 1.90, s, 6H) were observed in the 1H-NMR spectrum (Table 1). Signals of six oxygenated carbons (δC 74.8, 79.9, 83.6, 75.7, 69.4, 75.7), two acetyl carbons (δC 168.6 and 169.0) and a γ-lactone carbonyl carbon (δC 175.1) were observed in the 13C-NMR spectrum (Table 2). The HMBC correlations of H-2 (δH 5.34, d, J = 9.2 Hz)/carbonyl carbon (δC 168.6) and H-14 (δH 4.68, br s)/carbonyl carbon (δC 169.0) helped to locate two acetoxyl groups at C-2 and C-14. The above data suggested that Compound 8 is a highly oxygenated 8-hydroxybriarane-type diterpenoid. The 1H-NMR spectrum of 8 was similar to that of frajunolide A [19], except that the chemical shift of H-12 (δH 4.13, dd, J = 10.8, 5.2 Hz; in CDCl3) was located upfield as compared with frajunolide A (δH 5.35). The last remaining oxygen can only be accounted for by a hydroxyl group attached on C-9, because the chemical shift of H-9 (δH 4.68, br s) was also shifted upfield in comparison with frajunolide A (δH 5.37). The above data combined with HMBC correlations (Figure 2) revealed that 9- and 12-acetyl groups in frajunolide A were replaced by hydroxyl groups in 8. The NOESY correlations of H-2/H-10, H-10/H-9 and Me-19 in 8 suggested that the configurations of them were α-oriented. On the other hand, the correlations of H-6/H-7, H-7/H-17, H-14/Me-15 agreed with a β-configuration of H-7, H-14, Me-15 and H-17. The large coupling constant (J6,7 = 9.6 Hz) confirmed the anti-parallel arrangement of H-6 and H-7 and the β-orientation of H-7 [19]. It was concluded that briaviolide H (8) has the structure of (1S,2S,6Z,7S,8R,9S,10S,12S,14S,17R)-2,14-diacetoxy-12-hydroxy-8,9-dihydroxybriaran-5(6)-dien-18,7-olide.
The molecular formula C24H34O10 was assigned to Compound 9 from its HRESIMS and 13C NMR data (Table 2). The spectroscopic values of 9 suggested a briarane structure similar to that of 8 with one additional hydroperoxy group at C-12 (δC 83.8). The configuration of Compound 9 was further determined by the NOESY experiments (Figure 4), and the correlations revealed that 9 possessed the same relative configurations as those of 8. Thus, briaviolide I (9) was assigned as 12-hydroperoxyl derivative of Compound 8.
Briaviolide J (10) had the molecular formula C26H36O10, as determined by its HRESIMS and DEPT 13C NMR data. The IR absorptions of 10 were found at 3426, 1732 and 1675 cm−1, which indicated the presence of hydroxyl, a γ-lactone and ester groups. The 1H- and 13C-NMR data (Table 1 and Table 2) revealed that 10 was an 8-hydroxybriarane-type diterpenoid and was structurally similar to 8 and 9. Comparisons of their NMR and MS data showed that the only difference between 8 and 10 was the presence of an acetate group at C-12 in 10. Acetylation of 8 afforded a product identical to Compound 10. Thus, it was concluded that 10 is 12-acetoxyl derivative of Compound 8.
In addition, six known briaranes, solenolides A and D [25], excavatolide A [26], briaexcavatolide I [22], 4β-acetoxy-9-deacetystylatulide lactone and 9-deacetylstylatulide lactone [27], were identified. The new isolated briaranes 1–10 and derivative 11 were evaluated for anti-inflammatory activities on superoxide-anion generation and elastase release by human neutrophils in response to N-formyl-methionyl-leucyl-phenylalanine (fMLP)/Cytochalasin B (CB). The inhibition percentages of these compounds at the concentration of 10 μg/mL are summarized in Table 3. The bioassay data showed that Compounds 5 and 9 have moderate activities on both of superoxide-anion generation and elastase release, while Compound 11 has selective activity on the inhibition of elastase release.
Table 3.
Inhibitory effects of Compounds 1–11 on superoxide anion generation and elastase release by human neutrophils in response to fMLP/CB.
| Compound | Inhibition (%) a | |
|---|---|---|
| Superoxide Anion | Elastase Release | |
| 1 | 6.09 ±1.40 * | 11.04 ± 7.22 |
| 2 | 6.43 ± 2.17 * | 13.43 ± 2.66 ** |
| 3 | 16.87 ± 4.86 * | 6.40 ± 4.29 |
| 4 | 4.48 ± 1.47 * | 9.31 ± 6.64 |
| 5 | 34.17 ± 0.79 *** | 26.03 ± 9.51 |
| 6 | 17.35 ± 6.91 | 14.34 ± 5.28 |
| 7 | 3.25 ± 2.35 | 16.66 ± 3.12 ** |
| 8 | 6.01 ± 4.16 | 18.78 ± 2.29 ** |
| 9 | 28.66 ± 1.99 *** | 28.81 ± 6.37 * |
| 10 | 11.64 ± 3.92 * | 14.62 ± 4.41 ** |
| 11 | 6.09 ± 4.09 ** | 28.60 ± 7.54 * |
| genistein b | 65.05 ± 6.12 | 52.45 ± 6.34 |
a At a concentration of 10 μg/mL for each compound. Results are presented as the mean ± SEM (n = 3). * p < 0.05, ** p < 0.01, *** p < 0.001 compared with the control value; b Positive control.
3. Experimental Section
3.1. General Experimental Procedures
The melting point was measured on a BÜCHI Melting Point B-540 apparatus (Buchi, Flawil, Switzerland) and uncorrected. Optical rotations were recorded on a JASCO DIP-1020 polarimeter (Jasco, Tokyo, Japan). IR spectra were measured on a JASCO FT/IR-4100 spectrophotometer (Jasco, Tokyo, Japan). HR-ESI-MS were taken on a JEOL JMS-HX 110 mass spectrometer (Jeol, Tokyo, Japan). The NMR spectra were recorded either on a Bruker Avance 300, or a 400 NMR spectrometer, or on a Varian MR 400 NMR spectrometer (Varian, Santa Clara, CA, USA). The chemical shifts were given in δ (ppm) and coupling constants in Hz. Silica gel 60 (Merck, Darmstadt, Germany) was used for column chromatography (CC), and pre-coated silica gel plates (Merck, Darmstadt, Germany, Kieselgel 60 F-254, 1 mm) were used for preparative TLC. Sephadex LH-20 (Amersham Pharmacia Biotech AB, Sweden) was used for separation. LiChrospher® Si 60 (5 μm, 250–10, Merck, Darmstadt, Germany) and LiChrospher® 100 RP-18e (5 μm, 250–10, Merck, Darmstadt, Germany) were used for NP-HPLC and RP-HPLC (Merck, Darmstadt, Germany), respectively.
3.2. Animal Material
The gorgonian, Briareum violacea (Quoy and Gaimard), was collected in Pingtong County of southern Taiwan by scuba diving at a depth of 15 m, in May 2007. The fresh gorgonian was immediately frozen after collection and kept at −20 °C until being processed. A voucher specimen was deposited in the School of Pharmacy, College of Medicine, National Taiwan University.
3.3. Extraction and Isolation
The gorgonian, B. violacea (wet weight, 2.5 kg), was minced and extracted with acetone/MeOH (3 × 5 L) at room temperature, and the extracts were combined and concentrated under vacuum. The dark green crude residue was partitioned between EtOAc and H2O (1:1). The EtOAc-soluble portion was shaken with n-hexane/MeOH/H2O (4:3:1), and the MeOH layer (15 g) was evaporated and separated on Sephadex LH-20 to give eight fractions (L1 to L8). Fraction L3 (9.5 g) was subjected to flash column chromatography using silica gel and a gradient of n-hexane/EtOAc/MeOH to obtain 25 fractions (L3-1 to L3-25). Crystallization of L3-16 (n-hexane/EtOAc, 2:1; 640 mg) furnished 9-deacetylstylatulide lactone (123 mg). The MeOH-soluble portion of fraction L3-16 was separated on C18 reversed-phase (RP) HPLC using MeOH/H2O/CH3CN (50:45:10) to yield Compound 5 (9 mg), excavatolide A (6.5 mg) and stylatulide lactone (2 mg). Fraction L3-17 (n-hexane/EtOAc, 3:2; 1.6 g) was separated by RP-HPLC using MeOH/H2O/CH3CN (50:50:5) to afford Compound 6 (8.7 mg). Fraction L3-18 (n-hexane/EtOAc, 1:1; 2.0 g) was subjected to column chromatography using silica gel and a gradient of n-hexane/EtOAc/MeOH to obtain 10 fractions (L3-18-1 to L3-18-10). Fraction L3-18-2 (185 mg) was subjected to RP-HPLC using MeOH/H2O/CH3CN (65:30:5) to give Compounds 2 (3.5 mg), 3 (3.8 mg) and solenolide A (7.8 mg). Fraction L3-18-4 (231 mg) was separated by RP-HPLC using MeOH/H2O/CH3CN (65:30:5) to afford Compound 8 (11.0 mg) and solenolide D (9.4 mg). Fraction L3-18-7 was subjected on RP-HPLC using MeOH/H2O/CH3CN (55:45:5) to obtain Compounds 1 (12 mg), 7 (28 mg) and briaexcavatolide I (5.4 mg). Fraction L3-18-8 (64 mg) was purified by RP-HPLC using MeOH/H2O/CH3CN (65:30:5) to yield Compound 10 (10.5 mg). Fraction L3-20 (n-hexane/EtOAc, 1:2; 300 mg) was purified by RP-HPLC to afford Compounds 4 (3.5 mg) and 9 (4.3 mg).
Briaviolide A (1): Colorless amorphous prism; mp. 174–175 °C; −79 (c 0.5, CH2Cl2); IR νmax 3524, 3365, 2980, 2949, 1767, 1737, 1375, 1231 cm−1; 1H-NMR data (300 MHz, CDCl3), see Table 1; 13C-NMR data (75 MHz, CDCl3), see Table 2; ESIMS m/z 521 [M + Na]+, 523 [M + Na + 2]+; HRESIMS m/z 521.1554 [M + Na]+ (calcd. for C24H3135ClO9Na, 521.1557).
Briaviolide B (2): Colorless amorphous gum; −45 (c 0.1, CH2Cl2); IR νmax 3453, 2965, 2942, 1783, 1738, 1732, 1278 cm−1; 1H-NMR data (400 MHz, CDCl3), see Table 1; 13C-NMR data (100 MHz, CDCl3), see Table 2; ESIMS m/z 577 [M + Na]+, 579 [M + Na + 2]+; HRESIMS m/z 577.1812 [M + Na]+ (calcd. for C27H3535ClO10Na, 577.1816).
Briaviolide C (3): Colorless powder; −6 (c 0.1, CH2Cl2); IR νmax 3429, 2965, 2928, 1776, 1738, 1373, 1213 cm−1; 1H-NMR data (400 MHz, CDCl3), see Table 1; 13C-NMR data (100 MHz, CDCl3), see Table 2; ESIMS m/z 587 [M + Na]+; HRESIMS m/z 587.2101 [M + Na]+ (calcd. for C28H36O12Na, 587.2104).
Briaviolide D (4): Colorless amorphous gum; −11 (c 0.4, CH2Cl2); IR νmax 3433, 3015, 2989, 2938, 1776, 1734, 1373, 1220, 1019, 756 cm−1; 1H-NMR data (400 MHz, CDCl3), see Table 1; 13C-NMR data (100 MHz, CDCl3), see Table 2; ESIMS m/z 537 [M + Na]+, 539 [M + Na + 2]+; HRESIMS m/z 537.1506 [M + Na]+ (calcd. for C24H3135ClO10Na, 537.1503).
Briaviolide E (5): Colorless amorphous powder; −6 (c 0.1, CH2Cl2); IR νmax 3429, 2965, 2928, 1779, 1738, 1373, 1213 cm−1; 1H-NMR data (400 MHz, CDCl3), see Table 1; 13C-NMR data (100 MHz, CDCl3), see Table 2; ESIMS m/z 612 [M + Na]+, 614 [M + Na + 2]+; HRESIMS m/z 612.2075 [M + Na]+ (calcd. for C29H3935ClO11Na, 612.2079).
Briaviolide F (6): Colorless amorphous gum; +16 (c 0.4, CH2Cl2); IR νmax 3410, 2981, 2938, 1768, 1742, 1369, 1216, 1019, 756 cm−1; 1H-NMR data (300 MHz, CDCl3), see Table 1; 13C-NMR data (75 MHz, CDCl3), see Table 2; ESIMS m/z 593 [M + Na]+, 595 [M + Na + 2]+; HRESIMS m/z 593.2125 [M + Na]+ (calcd. for C28H3935ClO10Na, 593.2129).
Briaviolide G (7): Colorless amorphous gum; −18 (c 0.6, CH2Cl2); IR νmax 3544, 3447, 2981, 2937, 1782, 1739, 1375, 1223, 1021, 737 cm−1; 1H-NMR data (400 MHz, CDCl3), see Table 1; 13C-NMR data (100 MHz, CDCl3), see Table 2; ESIMS m/z 579 [M + Na]+, 581 [M + Na + 2]+; HRESIMS m/z 579.1605 [M + Na]+ (calcd. for C26H3335ClO11Na, 579.1609).
Briaviolide H (8): Colorless amorphous gum; +53 (c 0.2, CH2Cl2); IR νmax 3440, 2923, 1732, 1712, 1644, 1375, 1260, 1034, 953, 733 cm−1; 1H-NMR data (400 MHz, Acetone-d6), see Table 1; 13C-NMR data (100 MHz, Acetone-d6), see Table 2; ESIMS m/z 489 [M + Na]+; HRESIMS m/z 489.2106 [M + Na]+ (calcd. for C24H34O9Na, 489.2101).
Briaviolide I (9): Colorless amorphous powder; +20 (c 0.4, CH2Cl2); IR νmax 3425, 2928, 1738, 1715, 1373, 1259 cm−1; 1H-NMR data (400 MHz, CD3OD), see Table 1; 13C-NMR data (100 MHz, CD3OD), see Table 2; ESIMS m/z 505 [M + Na]+; HRESIMS m/z 505.2046 [M + Na]+ (calcd. for C24H34O10Na, 505.2050).
Briaviolide J (10): Colorless amorphous gum; +50 (c 0.1, CH2Cl2); IR νmax 3426, 2927, 2853, 1732, 1675, 1375, 1257, 1044, 737 cm−1; 1H-NMR data (400 MHz, pyridine- d5), see Table 1; 13C-NMR data (100 MHz, pyridine-d5), see Table 2; ESIMS m/z 531 [M + Na]+; HRESIMS m/z 531.2204 [M + Na]+ (calcd. for C26H36O10Na, 531.2206).
Benzyl briaviolide A (11): Colorless amorphous gum; −21 (c 0.2, CH2Cl2); IR νmax 3463, 2985, 2938, 1779, 1734, 1270, 1224 cm−1; 1H-NMR data (300 MHz, CDCl3), see Table 1; 13C-NMR data (75 MHz, CDCl3); see Table 2; ESIMS m/z 625 [M + Na]+, 627 [M + Na + 2]+; HRESIMS m/z 625.1820 [M + Na]+ (calcd. for C31H3535ClO10Na, 625.1816).
3.4. Benzoylation of Briaviolide (1)
Compound 1 (3.0 mg) was stirred with 0.1 mL of benzoyl chloride in pyridine (1.0 mL) for 20 h at room temperature. After evaporation, the residue was separated by a C18 reversed-phase HPLC column (MeOH/H2O, 4:1) to give pure Compound 11 (2.8 mg).
3.5. Single Crystal X-Ray Structure Determination of Briaviolide (1)
A suitable colorless crystal (0.20 × 0.15 × 0.10 mm3) of 1 for diffraction was obtained by simple evaporation from methanol solution. Crystal data: C24H31ClO9, orthorhombic, a = 10.4634(2) Å, b = 13.9191(3) Å, c =17.1825(3) Å, V = 2502.48(8) Å3, space group P212121, Z = 4, Dcalcd = 1.324 Mg/m3, λ= 1.54178 Å, μ(Mo Kα) 1.783 mm−1, F(000) = 1056, T = 295(2) K. A total of 17,948 reflections were collected, of which 4536 unique reflections (Rint = 0.0427) with I > 2σ(I) were used for the analysis. The data was solved using the direct method, and the structure was refined by a full-matrix least-squares procedure on F2 values. All non-hydrogen atoms were refined with anisotropic thermal parameters. The hydrogen atom positions were geometrically idealized and allowed to ride on their parent atoms. The final indices were R1 0.0499, wR2 0.1420 with goodness-of-fit = 1.149. The final X-ray molecular model is shown in Figure 3b.
3.6. Anti-Inflammatory Assays
3.6.1. Human Neutrophils Elastase Release
Degranulation of azurophilic granules was determined by elastase release, as described previously [28]. Experiments were performed using MeO-Suc-Ala-Ala-Pro-Val-p-nitroanilide as the elastase substrate. After supplementation with MeO-Suc-Ala-Ala-Pro-Val-p-nitroanilide (100 μM), neutrophils (6 × 105 cell/mL) were equilibrated at 37 °C for 2 min and incubated with each test compound for 5 min. Cells were activated by fMLP (100 nM)/CB (0.5 μg/mL), and changes in absorbance at 405 nm were monitored continuously for elastase release. The results are expressed as the percentage of the initial rate of elastase release in the fMLP/CB-activated, test compound-free (DMSO) control system.
3.6.2. Human Neutrophil Superoxide Generation
Human neutrophils were obtained by means of dextran sedimentation and Ficoll centrifugation. Superoxide anion production was assayed by monitoring the superoxide dismutase-inhibitable reduction of ferricytochrome c [29]. In brief, after supplementation with 0.5 mg/mL ferricytochrome c and 1.0 mM Ca2+, neutrophils were equilibrated at 37 °C for 2 min and incubated with drugs for 5 min. Cells were activated with 100 nM fMLP for 10 min. When fMLP was used as a stimulant, CB (1 μg/mL) was incubated for 3 min before activation by the peptide (fMLP/CB). Changes in absorbance with the reduction of ferricytochrome c at 550 nm were continuously monitored in a double-beam, six-cell positioner spectrophotometer with constant stirring (Hitachi U-3010, Tokyo, Japan). Calculations were based on differences in the reactions with and without SOD (100 U/mL) divided by the extinction coefficient for the reduction of ferricytochrome c.
4. Conclusions
Sixteen briarane diterpenoids, including ten new compounds briaviolides, A–J (1–10), were successfully isolated from the Taiwanese soft coral, Briareum violacea, and their structures determined. The inhibitory effects of the isolates and new derivative 11 on superoxide-anion generation and elastase release by human neutrophils in response to fMLP/CB were evaluated. Compounds 5 and 9 showed moderate anti-inflammatory activities at a concentration of 10 μg/mL. Compound 11, derived from Compound 1, showed better inhibition of elastase release than that of 1. Further comparison of the activities of those compounds may suggest that β-orientation and the chain length of ester groups at C-12 are important for the anti-inflammatory activities in briarane-type diterpenoids.
Acknowledgments
We thank Yi-Hung Liu, Instrumentation Center, National Taiwan University for providing X-ray crystallographic data. The financial support (NSC 98-2113-M-002-002-MY2) from the National Science Council, Taiwan, is also acknowledged.
Author Contributions
Ya-Ching Shen led the research team and supervised Ph.D. students, Chia-Ching Liaw, Yuan-Bin Cheng and Yun-Sheng Lin. Chia-Ching Liaw isolated the metabolites, measured various spectra and determined the structures. Yun-Sheng Lin and Chia-Ching Liaw collected the materials. Tsong-Long Hwang and Yao-Haur Kuo tested the biological activities. Ya-Ching Shen prepared the manuscript. Yuan-Bin Cheng edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bhakuni D.S., Rawat D.S. Bioactive Marine Natural Products. Springer; New York, NY, USA: 2005. [Google Scholar]
- 2.Shin J., Park M., Fenical W. The junceellolides, new anti-inflammatory diterpenoids of the briarane class from the Chinese gorgonian Junceella fragilis. Tetrahedron. 1989;45:1633–1638. doi: 10.1016/S0040-4020(01)80026-0. [DOI] [Google Scholar]
- 3.Rodriguez A.D. The natural products chemistry of west India gorgonian octocorals. Tetrahedron. 1995;51:4571–4618. doi: 10.1016/0040-4020(95)00216-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez J., Nieto R.M., Jimenez C. New briarane stecholide diterpenes from the Indonesian gorgonian Briareum sp. J. Nat. Prod. 1998;61:313–317. doi: 10.1021/np9703510. [DOI] [PubMed] [Google Scholar]
- 5.Sung P.J., Su J.H., Wang G.H., Lin S.F., Duh C.Y., Sheu J.H. Excavatolides F–M, new briarane diterpenes from the gorgonian Briareum excavatum. J. Nat. Prod. 1999;62:457–463. doi: 10.1021/np980446h. [DOI] [PubMed] [Google Scholar]
- 6.Wu S.L., Sung P.J., Chiang M.Y., Wu J.Y., Sheu J.H. New polyoxygenated briarane diterpenoids, briaexcavatolides O–R, from the gorgonian Briareum excavatum. J. Nat. Prod. 2001;64:1415–1420. doi: 10.1021/np010253l. [DOI] [PubMed] [Google Scholar]
- 7.Keifer P.A., Rinehart K.L., Hooper I.R. Renillafoulins, antifouling diterpenes from the sea pansy Renilla reniformis (Octocorallia) J. Org. Chem. 1986;51:4450–4454. doi: 10.1021/jo00373a020. [DOI] [Google Scholar]
- 8.Qi S.H., Zhang S., Qian P.Y., Xiao Z.H., Li M.Y. Ten new antifouling briarane diterpenoids from the south China sea gorgonian Junceella juncea. Tetrahedron. 2006;62:9123–9130. doi: 10.1016/j.tet.2006.07.049. [DOI] [Google Scholar]
- 9.Grode S.H., James T.R., Jr., Cardellina J.H., II Molecular structures of the briantheins, new insecticidal diterpenes from Briareum polyanthes. J. Org. Chem. 1983;48:5203–5207. doi: 10.1021/jo00174a010. [DOI] [Google Scholar]
- 10.Hamann M.T., Harrison K.N., Carroll A.R., Scheuer P.J. Briarane diterpenes from Micronesian gorgonians. Heterocycles. 1996;42:325–331. doi: 10.3987/COM-95-S39. [DOI] [Google Scholar]
- 11.Aoki S., Okano M., Matsui K., Itoh T., Satari R., Akiyama S., Kobayashi M. Brianthein A, a novel briarane-type diterpene reversing multidrug resistance in human carcinoma cell line, from the gorgonian Briareum excavatum. Tetrahedron. 2001;57:8951–8957. doi: 10.1016/S0040-4020(01)00894-8. [DOI] [Google Scholar]
- 12.Rodriguez A.D., Ramirez C., Cobar O.M. Briareins C–L, 10 new briarane diterpenoids from the common Caribbean gorgonian Briareum asbestinum. J. Nat. Prod. 1996;59:15–22. doi: 10.1021/np960001y. [DOI] [Google Scholar]
- 13.Burk J.E., Helm D.V.D., Chang C.Y., Ciereszko L.S. The crystal and molecular structure of briarein A, a diterpenoid from the gorgonian Briareum asbestinum. Acta Cryst. 1977;B33:704–709. doi: 10.1107/S0567740877004518. [DOI] [Google Scholar]
- 14.Sung P.J., Sheu J.H., Xu J.P. Survey of briarane-type diterpenoid of marine origin. Heterocycle. 2002;56:535–579. doi: 10.3987/REV-01-546. [DOI] [Google Scholar]
- 15.Sung P.J., Chang P.C., Fang L.S., Sheu J.H., Chen W.C., Chen Y.P., Lin M.R. Suvery of briarane-related diterpenoids-Part II. Heterocycles. 2005;65:195–204. doi: 10.3987/REV-04-589. [DOI] [Google Scholar]
- 16.Sung P.J., Gwo H.H., Fan T.Y., Li J.J., Dong J., Han C.C., Wu L.S., Fang L.S. Natural product chemistry of gorgonian corals of the genus Junceella. Biochem. Syst. Ecol. 2004;32:185–196. [Google Scholar]
- 17.Shen Y.C., Lin Y.C., Chiang M.Y. Juncenolide A, a new briarane from the Taiwanese gorgonian Junceella juncea. J. Nat. Prod. 2002;65:54–56. doi: 10.1021/np010408p. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y.C., Lin Y.C., Huang Y.L. Juncenolide E, a new briarane from Taiwanese gorgonian Junceella juncea. J. Chin. Chem. Soc. (Taipei) 2003;50:1267–1270. [Google Scholar]
- 19.Shen Y.C., Chen Y.H., Hwang T.L., Guh J.H., Khalil A.T. Four new briarane diterpenoids from the gorgonian coral Junceella fragilis. Helv. Chim. Acta. 2007;90:1391–1398. doi: 10.1002/hlca.200790141. [DOI] [Google Scholar]
- 20.Liaw C.C., Shen Y.C., Lin Y.S., Hwang T.L., Kuo Y.H., Khalil A.T. Frajunolides E–K, briarane diterpenes from Junceella fragilis. J. Nat. Prod. 2008;71:1551–1556. doi: 10.1021/np800126f. [DOI] [PubMed] [Google Scholar]
- 21.Chang Y.-C., Huang I.-C., Chiang M.Y.-N., Hwang T.-L., Kung T.-H., Lin C.-S., Sheu J.-H., Sung P.-J. Briaviodiol A, a new cembranoid from a soft coral Briareum violacea. Chem. Pharm. Bull. 2010;58:1666–1668. doi: 10.1248/cpb.58.1666. [DOI] [PubMed] [Google Scholar]
- 22.Sheu J.H., Sung P.J., Su J.H., Liu H.Y., Duh C.Y., Chiang M.Y. Briaexcavatolides A–J, new diterpenes from the gorgonian Briareum excavatum. Tetrahedron. 1999;55:14555–14564. doi: 10.1016/S0040-4020(99)00931-X. [DOI] [Google Scholar]
- 23.Kwak J.H., Schmitz F.J., Williams G.C. Milolides, new briarane diterpenoids from the western pacific octocoral Briareum stechei. J. Nat. Prod. 2001;64:754–760. doi: 10.1021/np010009u. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi J., Cheng J.F., Nakamura H., Ohizumi Y., Tomotake Y., Matsuzaki T., Grace K.J.S., Jacobs R.S., Kato Y., Brinen L.S., et al. Structure and stereochemistry of brianolide, a new antiinflammatory diterpenoid from the Okinawan gorgonian Briareum sp. Cell. Mol. Life Sci. 1991;47:501–502. doi: 10.1007/BF01959955. [DOI] [PubMed] [Google Scholar]
- 25.Groweiss A., Look S.A., Fenical W. Solenolides, new antiinflammatory and antiviral diterpenoids from a marine octocoral of the genus Solenopodium. J. Org. Chem. 1988;53:2401–2406. doi: 10.1021/jo00246a001. [DOI] [Google Scholar]
- 26.Sheu J.H., Sung P.J., Cheng M.C., Liu H.Y., Fang L.S., Duh C.Y., Chiang M.Y. Novel cytotoxic diterpenes, excavatolides A–E, isolated from the Formosan gorgonian Briareum excavatum. J. Nat. Prod. 1998;61:602–608. doi: 10.1021/np970553w. [DOI] [PubMed] [Google Scholar]
- 27.Sheu J.H., Sung P.J., Huang L.H., Lee S.F., Wu T., Chang B.Y., Duh C.Y., Fang L.S., Soong K., Lee T.J. New cytotoxic briarane diterpene from the formosan gorgonian Briareum sp. J. Nat. Prod. 1996;59:935–938. doi: 10.1021/np960218s. [DOI] [PubMed] [Google Scholar]
- 28.Sklar L.A., McNeil V.M., Jesaitis A.J., Painter R.G., Cochrane C.G. A continuous, spectroscopic analysis of the kinetics of elastase secretion by neutrophils. The dependence of secretion upon receptor occupancy. J. Biol. Chem. 1982;257:5471–5475. [PubMed] [Google Scholar]
- 29.Hwang T.L., Leu Y.L., Kao S.H., Tang M.C., Chang H.L. Viscolin, a new chalcone from Viscum coloratum, inhibits human neutrophil superoxide anion and elastase release via a cAMP-dependent pathway. Free Radic. Biol. Med. 2006;41:1433–1441. doi: 10.1016/j.freeradbiomed.2006.08.001. [DOI] [PubMed] [Google Scholar]