Abstract
Complex I (NDH-1) translocates protons across the membrane using electron transfer energy. Two different coupling mechanisms are currently being discussed for complex I: direct (redox-driven) and indirect (conformation-driven). Semiquinone (SQ) intermediates are suggested to be key for the coupling mechanism. Recently, using progressive power saturation and simulation techniques, three distinct SQ species were resolved by EPR analysis of E. coli complex I reconstituted into proteoliposomes. The fast-relaxing SQ (SQNf) signals completely disappeared in the presence of the uncoupler gramicidin D or the potent E. coli complex I inhibitor squamotacin. The slow-relaxing SQ (SQNs) signals were insensitive to gramicidin D, but they were sensitive to squamotacin. The very slow-relaxing SQ (SQNvs) signals were insensitive to both gramicidin D and squamotacin. Interestingly, no SQNs signal was observed in the ΔNuoL mutant, which lacks transporter module subunits NuoL and NuoM. Furthermore, we sought out the effect of using menaquinone (which has a lower redox potential compared to that of ubiquinone) as an electron acceptor on the proton pumping stoichiometry by in vitro reconstitution experiments with ubiquinone-rich or menaquinone-rich double knock-out membrane vesicles, which contain neither complex I nor NDH-2 (non-proton translocating NADH dehydrogenase). No difference in the proton pumping stoichiometry between menaquinone and ubiquinone was observed in the ΔNuoL and D178N mutants, which are considered to lack the indirect proton pumping mechanism. However, the proton pumping stoichiometry with menaquinone decreased by half in the wild-type. The roles and relationships of SQ intermediates in the coupling mechanism of complex I are discussed.
Keywords: NADH dehydrogenase, complex I, EPR, semiquinone, iron-sulfur clusters, Flavin, energy coupling, proton pumping, reactive oxygen species
1. Introduction
Complex I (NADH:quinone oxidoreductase: EC 1.6.5.3) is an entry point for electrons into the respiratory chains of mitochondria and many other aerobic organisms. Complex I transfers two electrons from NADH to ubiquione, translocates protons across the membrane, and generates a transmembrane electric potential and proton gradient which is essential for cellular maintenance and ATP production (Brandt, 2006; Hirst, 2013; Sazanov, 2007; Yagi and Matsuno-Yagi, 2003). The electron acceptor ubiquinone is a highly lipophilic molecule and is located mainly in the inner mitochondrial membrane. It is composed of a redox active 2,3-dimethoxy benzoquinone ring conjugated to an isoprenoid chain. The length of the isoprenoid side chain ranges from 6 to 10 units depending on the species. Ubiquinone also functions as a proton transferring agent as it takes up two protons when ubiquinone is reduced and releases them when it is oxidized. Thus, the energy conversion role of ubiquinone has been postulated for decades (Mitchell, 1961, 1979). However, the mechanism of how electron transfer is linked to vectorial proton translocation in complex I remains unknown.
1.1 Semiquinone species
Two-electron reduction of quinones to quinols (hydroquinones) usually occurs in two sequential one-electron steps although some enzymes such as NAD(P)H:quinone oxidoreductase 1 (NQO1) reduce quinones (Q) to hydroquinones (QH2) in a single two-electron step. The partially-reduced free-radical form (Q•−) is called semiquinone (SQ). The g = 2.00 EPR signals of SQ (or ubisemiquinone) in isolated bovine heart complex I upon reduction by NADH was first reported by Suzuki and King in 1983 (Suzuki and King, 1983). There were at least two populations of stable SQ radicals where 80% of which were sensitive to rotenone and the remaining 20% was insensitive to rotenone. Then, Vinogradov’s group discovered a rotenone-sensitive, low-temperature EPR signal in tightly coupled bovine heart submitochondrial particles (SMP) upon steady-state oxidation of NADH or succinate in 1990 (Kotlyar et al., 1990). This SQ signal is seen only in the presence of oligomycin that induces the respiratory control, and which disappears in the presence of an uncoupler (CCCP or gramicidin D). Subsequently, in 1994, Albracht’s group also confirmed SQ as obligatory intermediates in the electron transfer from NADH to ubiquinone, although it reported that SQ formation in isolated complex I or in tightly coupled SMP is not related to the degree of coupling in the preparation (De Jong and Albracht, 1994). After 1995, Vinogradov-Ohnishi’s (Magnitsky et al., 2002) and Albracht-Dunham’s groups (van Belzen et al., 1997), further characterized the SQ signals using bovine heart SMP and isolated complex I.
Physicochemical properties of SQ species differ considerably in their spin relaxation behavior. At least two types of the complex I-associated SQ species were detected by cryogenic EPR (Magnitsky et al., 2002): the fast-relaxing ubisemiquinone (SQNf) and the slowly-relaxing ubisemiquinone (SQNs). The SQNf signals were seen better in the presence of oligomycin, which was added to increase the respiratory control ratio to 7–9. The SQNf signals were sensitive to uncouplers, while the SQNs signals were insensitive to uncouplers. Both SQ species are equally sensitive to piericidin A, while SQNf is 10 times more sensitive to rotenone than SQNs (Magnitsky et al., 2002). It suggests some difference in their protein microenvironment, favoring the idea that SQNf and SQNs are different entities, and accommodated in different specific quinone binding sites. Furthermore, a direct spin-spin interaction between cluster N2 (the terminal iron-sulfur cluster in the main electron pathway from NADH) and SQNf was found below 25 K by both groups mentioned above (Chevallet et al., 1997; Ohnishi et al., 1998; Vinogradov et al., 1995). This led to the estimation of center-to center distance of 12 Å which allows efficient electron transfer between cluster N2 and quinone (Moser et al., 2006).
Based on their experiments, it has been believed that SQ intermediates appearing during complex I catalysis are key for the coupling mechanism in complex I (Magnitsky et al., 2002; Vinogradov et al., 1995). Therefore, the understanding of molecular properties and functions of the individual semiquinone species is a prerequisite to elucidate the energy-coupling mechanism of complex I.
1.2 Proposed energy coupling mechanisms of complex I
Investigating the proton pumping mechanism of complex I is one of the most challenging bioenergetic problems, not only because of the structural/functional complexity of complex I, but also because of its exceptionally high proton pumping stoichiomentry of 4H+/2e− (proton/electron) compared with other mitochondrial/bacterial energy-converting enzymes (Brandt, 2006; Sazanov, 2007; Yagi and Matsuno-Yagi, 2003). Several hypothetical energy coupling mechanisms have been proposed for complex I over the years, but none have been proven experimentally. The complex I coupling mechanisms proposed are:
“Mixed mode.” Based on the sequence similarity between complex I and hydrogenases, Friedrich proposed a “mixed model” in which complex I contains two coupling sites, one being a redox-driven “direct” and the other a conformation-driven “indirect” proton pump (Friedrich, 2001).
“Long range conformational energy transfer mechanism.” Brandt’s group suggested a “long range conformational energy transfer mechanism” in which conformational energy released by redox reactions in the peripheral arm of complex I is transmitted to proton pumping devices in the membrane arm (Brandt et al., 2003).
“Conformation-driven Semiquinone- gated proton pump.” Ohnishi’s group proposed a “conformation-driven SQ-gated proton pump mechanism” based on EPR analyses of protein-bound SQ radicals which are directly spin-coupled with cluster N2 (the electron donor to Q) and are observable only in the presence of membrane potential (Ohnishi and Salerno, 2005).
After X-ray crystal structures of complex I was determined by Sazanov’s group (Baradaran et al., 2013), “long-range conformational energy transfer mechanism” became most likely.
In principle, two different coupling mechanisms have been discussed for complex I: direct (redox-driven) and indirect (conformation-driven) (Fig. 1).
In the direct (redox-driven) coupling model, it is postulated that Q is directly involved in proton translocation. Indeed, numerous pieces of data support the idea that proton translocation is directly coupled to electron transfer from cluster N2 (in the NuoB subunit) to Q (Fig. 1), bound in a pocket surrounded by the NuoB, NuoD, and NuoH subunits. The data include an X-ray crystallographic structure (Sazanov and Hinchliffe, 2006), photoaffinity labeling/cross-linking studies (Berrisford et al., 2008; Murai et al., 2009), extensive mutagenesis studies of subunits NuoD and NuoB homologs in Y. lipolytica complex I (Fendel et al., 2008; Tocilescu et al., 2007), and the NuoH subunit in E. coli complex I (Sinha et al., 2009).
In the indirect (conformation-driven) coupling model, energy transduction is proposed to involve long-range conformational changes connecting the electron transfer module to distant proton pumping modules. Even long before X-ray crystal structures of complex I were determined, experiments involving detergent disruption of purified bovine complex I into subcomplexes (Iα, Iβ, Iγ, and Iλ) (Sazanov et al., 2000) and electron microscopy (EM) analyses (Baranova et al., 2007a; Baranova et al., 2007b) have supported distal locations for subunits NuoL and NuoM (Fig. 1) which are believed to drive proton translocation based on their high sequence similarity to multi-subunit K+ or Na+/H+ antiporters (Mathiesen and Hagerhall, 2002). As the high proton stoichiometry of 4H+/2e- is confirmed for complex I (Wikstrom, 1984), it seems reasonable to suggest that complex I utilizes an indirect mechanism, to achieve this high proton pumping stoichiometry.
Figure 1.
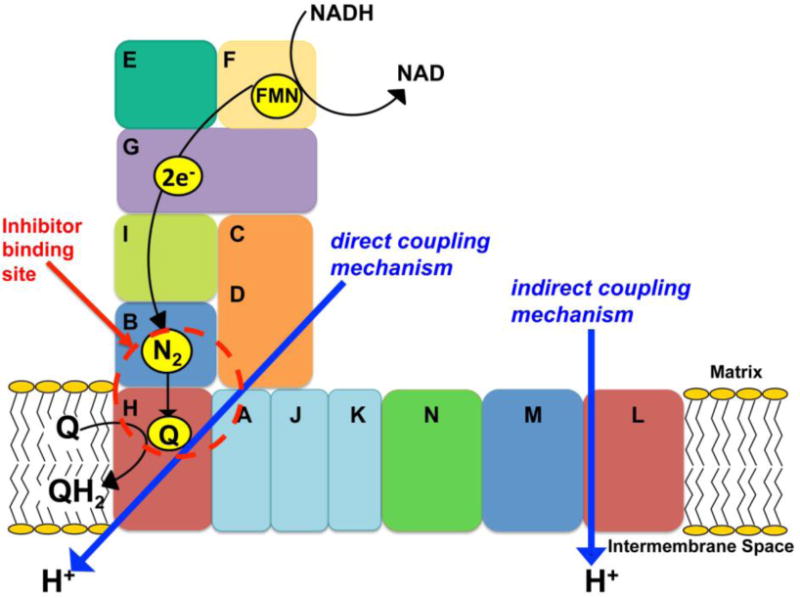
A schematic representation of E. coli complex I reaction mechanism.
2. Three SQ intermediates in isolated E. coli complex I reconstituted into proteoliposomes
Until recently, SQ signals have been characterized only in the bovine heart complex I (Magnitsky et al., 2002; Vinogradov et al., 1995) but not yet directly in complex I isolated from bacteria or fungi by EPR. Bacterial complex I catalyzes the same reaction and harbors the same set of cofactors as in mitochondrial complex I and consists of only 13–17 subunits (Sazanov, 2007; Yagi et al., 1998; Yip et al.) but, at least 13–14 of which have homologs in the mitochondrial enzyme (Yagi et al., 1998). Taking the advantage of its simplicity and ease of genetic manipulation, we have chosen E. coli complex I as a model system to study the structure and function of complex I. We also generated a knock-out NuoL (ΔNuoL) mutant which is the E. coli homolog for mitochondrial ND5, regarded as a transporter module at the distal end of the membrane domain, to compare biochemical/biophysical profiles of SQ signals between the wild type and ΔNuoL mutant.
In contrast to intact SMP where there is significant EPR spectral overlapping from SQ signals arising from other respiratory enzyme complexes, in isolated complex I, only SQ signals associated with complex I could be detected. However, their SQ characteristics might not be the same as those observed in an SMP system in situ because there is no membrane potential or proton motive force, and the protein microenvironment surrounding Q binding sites might be different. To minimize these problems, we reconstituted purified complex I into proteoliposomes, which mimics membrane environment.
Recently, we have established our purification method and obtained highly pure and active complex I from E. coli (Narayanan et al., 2013). Using our preparations, we were able to resolve three distinct SQ species in E. coli complex I for the first time by EPR analyses using progressive power saturation and simulation techniques, and we investigated their biochemical/biophysical properties (Leung, submitted). The three SQ species were distinguished by their relaxation rates. They are: fast-relaxing SQ (SQNf) with P1/2 (half-saturation power level) > 50 mW and a wider linewidth (12.8 G); slow-relaxing SQ (SQNs) with P1/2 = ~ 3 mW and a 10 G linewidth; and very slow-relaxing SQ (SQNvs) with P1/2 = ~ 0.1 mW and a 7.5 G linewidth (Leung, submitted). Our data are consistent with the characteristics of bovine counterparts for which SQNf has a wider line width (8.4 G) (Yano et al., 2005) than SQNs (7.0 G) (Ohnishi et al., 2005). The spectral line shape was also very different among those SQ species. The Gaussian and Lorentzian broadening ratios for SQNf, SQNs, and SQNvs were 1 to 0, 0.75 to 0.25, and 0 to 1, respectively, indicating that SQNf is confined in a dense environment surrounded by amino acid side chains while SQNvs is localized in a free environment, probably close to the outside of the protein (Bales et al., 1998).”
2.1. SQNf
The fast-relaxing SQNf signals completely disappeared in the presence of the uncoupler gramicidin D or the potent E. coli complex I inhibitor squamotacin. The pH dependency of the SQNf signals correlated with the proton-pumping activities and with NADH:decylubiquinone (DQ) activities (Leung, submitted). These features are the same as those reported for SQNf in bovine heart SMP (Yano et al., 2005). We further characterized the SQNf signals. When NADPH, a non-physiological substrate, was used to reduce iron-sulfur clusters, almost no SQNf signal (less than 1% of the control signal amplitude with NADH) were observed. This indicates that NADPH failed to trigger a conformational change required for the formation of SQNf. The SQNf signal was greatly diminished in the ΔNuoL mutant in which the initial proton pumping rate was reduced to only 10% of the control (Leung, submitted). The ΔNuoL mutant complex I does not contain transporter module subunits NuoL and NuoM. These results mentioned above all support the idea that the SQNf species is the key redox intermediate for the coupling reaction.
Regarding the spin interaction with cluster N2, unlike the SQNf signals in tightly coupled bovine SMP, the SQNf signals in E. coli complex I showed a very weak magnetic interaction only at much lower temperatures (< 10 K) and no splitting signals resulting from the magnetic interactions between SQNf and cluster N2 were detected.
The largest difference between the SQNf profiles we found in E. coli complex I and those in bovine complex I was that SQNf is protonated at pH 7. The protonation of SQNf was expected based on its wider linewidth (12.8 G) much larger than those of the corresponding SQ anion radicals (7-9 G). Because, it is known that protonation increases spectral linewidth because of an asymmetric perturbation of the spin density on the quinone ring (Bowyer, 1985; Hales and Case, 1981). The protonation of SQNf in E. coli was confirmed by a deuterium exchange experiment in which the linewidth of SQNf decreased to 10.2 G (Leung, submitted).
We further investigated that the effects of deuteration of complex I on electron transfer and proton pumping activities. NADH:DQ activities decreased to 54% of the control in the wild type complex I while they remained the same ~100% of the control in the ΔNuoL mutant. NADH:ferricyanide activities decreased to 70% in both the wild type and the ΔNuoL mutant. It has been suggested that when an electron acceptor undergoes protonation, the rate of electron transfer is accelerated by the protonation (Yuasa et al., 2008). Our data suggest that the protonation event is critically involved in the redox reaction in the wild type, but not in the ΔNuoL mutant in which the indirect mechanism does not occur. However, the initial proton pumping rate decreased to 63% in the deuterated ΔNuoL mutant complex I (Fig. 2, right). This implies an isotope effect on the proton pumping rate by the direct mechanism, resulting in a lower Proton/electron stoichiometry. On the other hand, the initial proton pumping rate decreased only to 43% in the deuterated wild type complex I (Fig. 2, left), despite of the significant decrease (54%) in the electron transfer rate. This data can be interpreted to mean that the initial proton pumping rate more or less slows down proportionately with electron transfer rate. This seems reasonable to support the idea that conformational change also takes place in the deuterated complex I.
Figure 2.
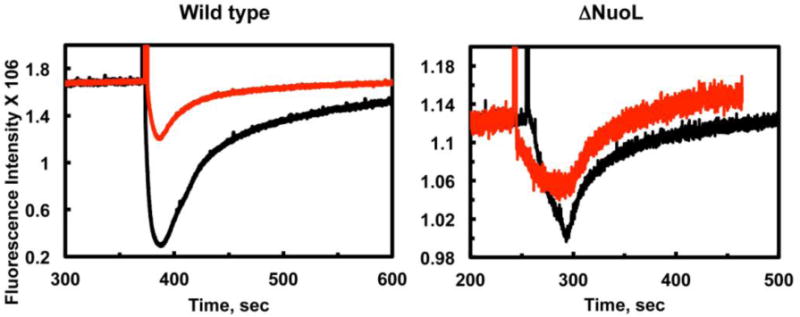
The deuterium effect on the proton pumping activities in the wild type and the ΔNuoL variant. Generation of a proton gradient was monitored by the quench of the ACMA fluorescence. Proteoliposomes (PL) were reconstituted with purified complex 1 in a H2O buffer (5mM MOPS pH 7.0+ 50mM KCl) and a D2O buffer (5mM MOPS pD 6.6+ 50mM KCl). The data were normalized based on the complex I concentrations and orientation factor in the proteoliposomes.
The highly conserved Tyr84 in the NuoD subunit (or Tyr273 in the NuoCD, as NuoC and NuoD subunits are fused in E. coli complex I) subunit of complex I is only ~ 7 Å away from cluster N2, and it faces the quinone binding site based on the crystal structure of the hydrophilic domain of Thermus thermophiles complex I (Baradaran et al., 2013; Sazanov and Hinchliffe, 2006). Mutational analyses of the corresponding Tyr144 in Y. lipolytica revealed that this residue is essential for complex I activities in both electron and proton transfer (Tocilescu et al., 2010). Therefore, it is highly likely that this Tyr84 residue and/or possibly the neighboring residue Glu83 could be a proton donor for the SQNf. Raising the pH of the solution tends to deprotonate SQ. Since the pKa for SQH•/ SQ•− is estimated to be ~ pH 7.2 (Wraight, 2004), it is not clear why SQNf at pH 7 is protonated in E. coli complex I but not in bovine heart complex I at the same pH. However, the unique pH profile of SQNf that disappears at higher pH was shared in both E. coli and bovine SMP. This profile is completely opposite to the common profile of known SQ radicals such as SQH in cytochrome bo3 (Ingledew et al., 1995) or SQi (Robertson et al., 1984) in the cytochrome bc1 complex. The disappearance of the SQNf signals at higher pH strongly suggests that SQNf is bound to a negatively charged residue or region, thus, the pH increase introduces a negative charge and decreases the stability constant of SQ anion radicals.
Redox-coupled proton translocation in the membrane domain requires long-range energy transfer through the protein complex. It is probable that the protonation of SQNf triggers and facilitates conformational changes to operate proton pump machinery far away from the catalytic site.
2.2. SQNs
The SQNs signals were insensitive to gramicidin D, but they were sensitive to squamotacin and decreased to ~30 % of the control intensity. These responses were previously observed in SQNs from intact bovine SMP with the uncoupler FCCP and the complex I inhibitor rotenone (Magnitsky et al., 2002). We recently discovered that there was no SQNs signal in the ΔNuoL mutant, while the SQNf signals are still detectable, even though they drastically decreased. Furthermore, we found that this ΔNuoL variant contains only one quinone per one mole of complex I, in contrast to the wild-type which contains two quinones. Since we confirmed that NuoL and NuoM are lost in the ΔNuoL mutant (Leung, submitted), the secondary quinone binding site is likely to be located in subunit NuoL, NuoM, or in the interface between NuoN and NuoM. Based on our findings that there are no SQNs signals and no secondary bound Q in ΔNuoL, it is tempting to speculate that the SQNs signal arises from this remote secondary quinone binding site away from the catalytic core. Indeed, SQNs was previously suggested to be remotely located from cluster N2 (estimated > 30 Å) (Magnitsky et al., 2002). However, this raises a serious question. How is it possible that a quinone molecule at the secondary Q binding site far from the catalytic site can still be reduced with an electron from NADH and become SQNs? Our data oppose the recent notion based on the recent X-ray crystal structures that there is only one ubiquinone-reactive site in complex I (Baradaran et al., 2013).
In terms of the proton to electron stoichiometry, it was found that this ΔNuoL mutant has a very low ratio of Proton/electron as it has only ~10% and 40% of proton pumping and the electron transfer activity (NADH:DQ) activities of the wild type. Considering the fact that this ΔNuoL mutant is missing NuoL and NuoM, no indirect conformation-driven proton pumping mechanism exists in the ΔNuoL mutant. It can be concluded that SQNs is also very critical of the direct coupling mechanism of complex I as well.
2.3. SQNvs
The SQNvs signals were insensitive to both gramicidin D and squamotacin (Fig. 3, left). SQNvs could possibly be equivalent to SQNx that was originally reported in SMP (Magnitsky et al., 2002), which was later dismissed as a non- intrinsic complex I component (Ohnishi et al., 2005). Therefore, to avoid any possible confusion, we designated this very slow-relaxing SQ species as SQNvs signals (Leung, submitted). The SQNvs signal appeared after the addition of NADH, reached its maximum level at 5 sec before the SQNf signal peaked at 10 sec. It disappeared faster than SQNs as NADH is consumed. Its insensitivity to the complex I inhibitor and the 100% Lorentzian broadening features suggest that SQNvs is in a free environment, likely very close to the Q pool. However, interestingly, when NADPH was used to reduce iron-sulfur clusters, the signal amplitude of SQNvs increased by 16%, and even more increased in the presence of squamotacin by 40%, compared to the SQNvs signal intensity with NADH. In addition, the SQNvs signals in ΔNuoL were found to be sensitive to gramicidin and squamotacin (Fig. 3, right). All these data above suggest that SQNvs is a legitimate complex I-associated SQ species.
Figure 3.
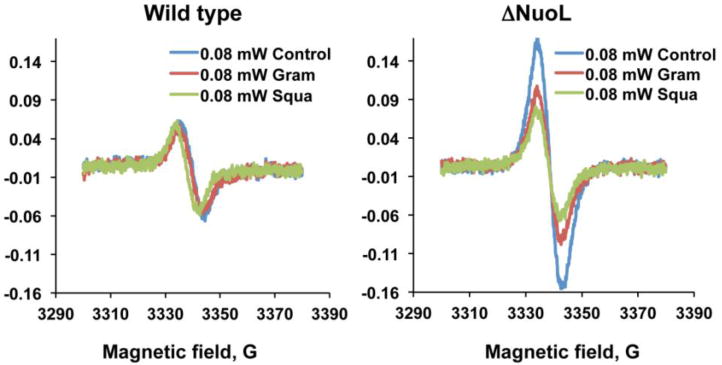
The effects of gramicidin D and squamotacin on the SQNvs spectra in the wild type and the ΔNuoL variant. The EPR data were taken at 150 K and 0.08 mW, and accumulated 10 times. The EPR conditions were: microwave frequency, 9.45 GHz; modulation frequency, 100 kHz; modulation amplitude, 6 G; time constant, 82 ms.
The role of SQNvs in the complex I catalytic mechanism is unknown at this moment. However, it has been reported in a study of steady state kinetics that the rotenone-insensitive reaction in bovine complex I is also physiologically relevant (Nakashima et al., 2002). The SQNvs species might be involved in this complex I inhibitor insensitive reaction. It can be speculated that this SQNvs binding site is in a dynamic equilibrium with the Q pool in the membrane and provides a route to release some electrons under certain conditions. This electron release through the SQNvs site might play a larger part in electron transfer activities in bacterial complex I, than those in mitochondrial complex I. If it is the case, then, this can explain why almost no respiratory control (no increase in electron transfer activities in the presence of uncoupler) is observed in bacterial complex I. Indeed, only up to a RCR (respiratory control ratio) = 1.2 was observed in our E. coli complex I proteoliposomes.
3. Roles of SQ species in ROS production from complex I
Complex I is inhibited by more than 60 different families of compounds including synthetic herbicides and insecticides (Degli Esposti, 1998). It is commonly accepted that complex I is a main site of reactive oxygen species (ROS) production in mitochondria. The sites of electron leaks in this process have been suggested to be cluster N2, SQNf, FMN, and cluster N1a, but the direct oxygen reducing site has still not been identified. Based on ROS production patterns by complex I, these inhibitors were grouped into two classes (Fato et al., 2008). Class A inhibitors include rotenone and piericidin A which increase ROS production in the presence of NADH. The effect is potentiated in the presence of the hydrophilic ubiquinone Q1. Class B inhibitors include stigmatellin and capsaicin, which decrease ROS production below the baseline regardless of the presence of Q1. In addition, it is reported that during catalytic electron transfer from NADH to DQ, the superoxide generation site was mostly shifted to the SQ (Ohnishi et al., 2010). However, when the catalytic formation of SQ species was suppressed in the presence of rotenone or piericidin A, complex I enhances the formation of flavin semiquinone and increases the overall superoxide generation (Ohnishi et al., 2010). Combined with our data on three distinct SQ species, it is temping to speculate that: i) Under physiological conditions, SQNs is a main source of ROS production in complex I because it is expected that SQNs is located in a more open environment where oxygen can be accessible from the outside, compared to the SQNf site (Fig. 4A). ii) Class A inhibitors such as rotenone, bind to the SQNf binding site and kills SQNf formation. This results in pushing electrons back all the way to the FMN (flavin mononucleotide) binding site where NADH is oxidized, and it increases ROS production via flavin semiquinone (Fig. 4B). A small portion of electrons goes to SQNs via SQNvs, and slightly contributes ROS production (Fig. 4B) iii) Class B inhibitors such as capsaicin, bind to the SQNs binding site, kill SQNs formation, and stop the electron flow from cluster N2 to SQNf. However, in contrast to Class A inhibitors, Class B inhibitors do not push electrons back to the iron-sulfur clusters. Instead, electrons slowly escape through the SQNvs site, where dismutation between SQ species takes place before they react with oxygen to produce ROS (Fig. 4C).
Figure 4.
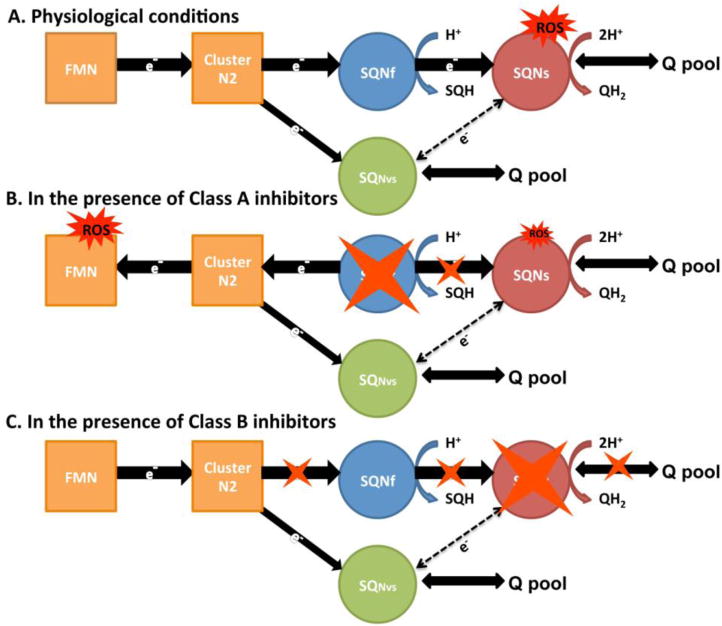
A proposed schematic representation of an intramolecular electron transfer from cluster N2 to the Q pool and suggested ROS production sites in complex I. SQNvs is formed by direct electron transfer from cluster N2, or it can be formed with electrons from SQNs under certain conditions. SQNs and SQNvs are accessible to the Q pool. (A) under physiological conditions, SQNs is a major ROS production site. SQNf and SQNvs sites are less likely to be ROS production sites. Because SQNf is bound to the enzyme deep inside the protein where oxygen is not accessible, while SQNvs is in a relatively free environment and it eventually dismutates with each other. (B) Class A inhibitors presumably bind to the SQNf binding site. The FMN binding site becomes a major ROS production site. Small amounts of ROS could still be produced at SQNs that is formed with electrons coming from SQNvs. (C) Class B inhibitors presumably bind to the SQNs binding site. No ROS is produced in complex I.
3. Proton/electron stoichiometry with ubiquinone or menaquinone
It was shown that bovine complex I translocates two protons per electron using intact mitochondria (Wikstrom, 1984). Later, the proton pumping stoichiometry of complex I was measured using bovine heart SMP (Galkin et al., 1999), purified complex I from Y. lipolytica reconstituted into proteoliposomes (Galkin et al., 2006), and more recently, in living cells using multiwavelength cell spectroscopy techniques (Ripple et al., 2013). These works all support the conclusion that the Proton/electron stoichiometry of mitochondrial complex I has the value of 2. However, the Proton/electron stoichiometry for bacterial complex I is still under debate. So far, there is only one report on proton pumping stoichiometry in E. coli complex I, showing that complex I pumps 1.5 protons per electron (Bogachev et al., 1996). This was measured using E. coli cells grown anaerobically. Low stoichiometry was explained by the fact that in anaerobic conditions menaquinone works as an electron acceptor in the E. coli respiratory chain. Since menaquinone (E0′ = −74 mV) has a more negative redox potential than ubiquinone (E0′ = 100 mV), the energy released by complex I upon its reduction with NADH may not be enough for the translocation of four protons across the membrane (Bongaerts et al., 1995). Therefore, the obtained 1.5 protons per electron stoichiometry for E. coli complex I could be underestimated when ubiquinone is used as an electron acceptor in aerobically grown E. coli cells.
As an attempt to see whether menaquinone change the proton/electron stoichiometry, we generated a double knockout (DKO) E. coli strain which contains neither NADH dehydrogenases complex I (NDH-1) nor NDH-2 (non-proton translocating type 2 NADH dehydrogenase). We also prepared ubiquinone-rich and menaquinone-rich membrane vesicles from E. coli DKO cells grown aerobically and anaerobically, respectively. Purified E. coli complex I was then reconstituted into those DKO membranes in vitro. Proton pumping activities and electron transfer activities (NADH:DQ activities) were measured. We found that the initial proton pumping rate was almost double when the wild type complex I was reconstituted into ubiquinone-rich membranes compared to that with menaquinone-rich membranes (Fig. 5Aa and 5Ab) while the electron transfer activities were the same (Fig. 5Ac). In a striking contrast, no difference in initial proton pumping and electron transfer activities between complex I reconstituted with ubiquinone-rich and menaquinone-rich membranes were found when the ΔNuoL mutant, or the D178N mutant that lacks the indirect proton pumping mechanism (Nakamaru-Ogiso et al., 2010), were used (Fig. 5B and 5C). Our data strongly suggest that menaquinone is insufficient to trigger the conformation-driven indirect proton pumping mechanism, and it leads only to a low proton/electron stoichiometry decreased by ~50%. The relationships between initial proton pumping rates and the intensity of each SQ species using menaquinone are currently under investigation.
Figure 5.
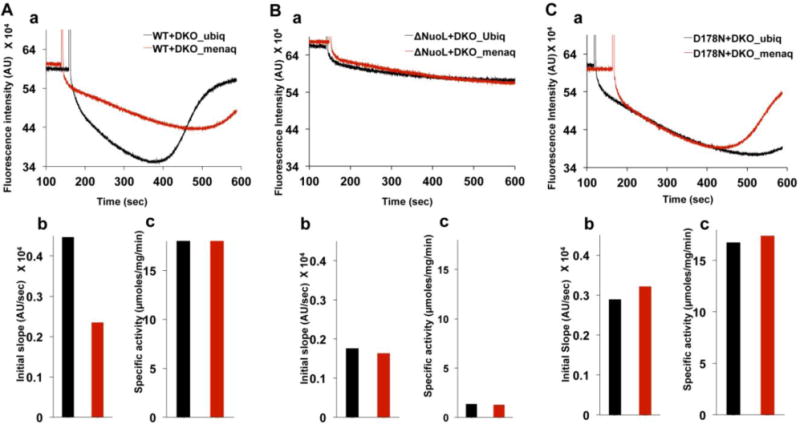
Proton pumping and NADH-DBQ activities in ubiquinone-rich and menaquinone-rich DKO membranes reconstituted with purified E. coli complex I. DKO membranes are devoid of complex I (NDH-1) and NDH-2. (A) 5 μL of reconstituted complex I (1 mg/mL) in 1 mL of reconstitution buffer (50 mM MOPS, pH 7.0, 50 mM KCl, 20 mM MgCl2) containing 0.2 μM ACMA dye was used for the H+ pumping assay. (B) Initial acidification rate was calculated from the initial slope of fluorescence quenching of ACMA.
Acknowledgments
This work was supported by NIH grant RO1GM097409 to E.N.-O. and AHA grant 11SDG5560001 to E.N.-O.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bales BL, Peric M, Lamy-Freund MT. Contributions to the Gaussian line broadening of the proxyl spin probe EPR spectrum due to magnetic-field modulation and unresolved proton hyperfine structure. Journal of magnetic resonance. 1998;132:279–286. doi: 10.1006/jmre.1998.1414. [DOI] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova EA, Holt PJ, Sazanov LA. Projection structure of the membrane domain of Escherichia coli respiratory complex I at 8 A resolution. J Mol Biol. 2007a;366:140–154. doi: 10.1016/j.jmb.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Baranova EA, Morgan DJ, Sazanov LA. Single particle analysis confirms distal location of subunits NuoL and NuoM in Escherichia coli complex I. J Struct Biol. 2007b;159:238–242. doi: 10.1016/j.jsb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Berrisford JM, Thompson CJ, Sazanov LA. Chemical and NADH-induced, ROS-dependent, cross-linking between subunits of complex I from Escherichia coli and Thermus thermophilus. Biochemistry. 2008;47:10262–10270. doi: 10.1021/bi801160u. [DOI] [PubMed] [Google Scholar]
- Bogachev AV, Murtazina RA, Skulachev VP. H+/e− stoichiometry for NADH dehydrogenase I and dimethyl sulfoxide reductase in anaerobically grown Escherichia coli cells. Journal of bacteriology. 1996;178:6233–6237. doi: 10.1128/jb.178.21.6233-6237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Molecular microbiology. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- Bowyer JRaOT. EPR spedtrostudy in the study of ubisemiquinones in redox chains. In: Lenaz G, editor. Coenxyme Q. New York: John Wiley & Sons; 1985. pp. 409–432. [Google Scholar]
- Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annual review of biochemistry. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- Brandt U, Kerscher S, Drose S, Zwicker K, Zickermann V. Proton pumping by NADH:ubiquinone oxidoreductase. A redox driven conformational change mechanism? FEBS letters. 2003;545:9–17. doi: 10.1016/s0014-5793(03)00387-9. [DOI] [PubMed] [Google Scholar]
- Chevallet M, Dupuis A, Lunardi J, van Belzen R, Albracht SP, Issartel JP. The NuoI subunit of the Rhodobacter capsulatus respiratory Complex I (equivalent to the bovine TYKY subunit) is required for proper assembly of the membraneous and peripheral domains of the enzyme. European journal of biochemistry / FEBS. 1997;250:451–458. doi: 10.1111/j.1432-1033.1997.0451a.x. [DOI] [PubMed] [Google Scholar]
- De Jong AM, Albracht SP. Ubisemiquinones as obligatory intermediates in the electron transfer from NADH to ubiquinone. European journal of biochemistry / FEBS. 1994;222:975–982. doi: 10.1111/j.1432-1033.1994.tb18948.x. [DOI] [PubMed] [Google Scholar]
- Degli Esposti M. Inhibitors of NADH-ubiquinone reductase: an overview. Biochimica et biophysica acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- Fato R, Bergamini C, Bortolus M, Maniero AL, Leoni S, Ohnishi T, Lenaz G. Differential effects of mitochondrial Complex I inhibitors on production of reactive oxygen species. Biochimica et biophysica acta. 2008 doi: 10.1016/j.bbabio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendel U, Tocilescu MA, Kerscher S, Brandt U. Exploring the inhibitor binding pocket of respiratory complex I. Biochimica et biophysica acta. 2008;1777:660–665. doi: 10.1016/j.bbabio.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Friedrich T. Complex I: a chimaera of a redox and conformation-driven proton pump? Journal of bioenergetics and biomembranes. 2001;33:169–177. doi: 10.1023/a:1010722717257. [DOI] [PubMed] [Google Scholar]
- Galkin A, Drose S, Brandt U. The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochimica et biophysica acta. 2006;1757:1575–1581. doi: 10.1016/j.bbabio.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Galkin AS, Grivennikova VG, Vinogradov AD. –>H+/2e− stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS letters. 1999;451:157–161. doi: 10.1016/s0014-5793(99)00575-x. [DOI] [PubMed] [Google Scholar]
- Hales BJ, Case EE. Immobilized radicals IV. Biological semiquinone anions and neutral semiquinones. Biochimica et biophysica acta. 1981;637:291–302. [Google Scholar]
- Hirst J. Mitochondrial complex I. Annual review of biochemistry. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- Ingledew WJ, Ohnishi T, Salerno JC. Studies on a stabilisation of ubisemiquinone by Escherichia coli quinol oxidase, cytochrome bo. European journal of biochemistry / FEBS. 1995;227:903–908. doi: 10.1111/j.1432-1033.1995.tb20217.x. [DOI] [PubMed] [Google Scholar]
- Kotlyar AB, Sled VD, Burbaev DS, Moroz IA, Vinogradov AD. Coupling site I and the rotenone-sensitive ubisemiquinone in tightly coupled submitochondrial particles. FEBS letters. 1990;264:17–20. doi: 10.1016/0014-5793(90)80753-6. [DOI] [PubMed] [Google Scholar]
- Leung SA, Narayanan M, Inaba Y, Elguindy MM, Nakamaru-Ogiso E. Semiquinone Intermediates are involved in the energy coupling mechanism of E. coli. complex I. doi: 10.1016/j.bbabio.2015.04.004. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnitsky S, Toulokhonova L, Yano T, Sled VD, Hagerhall C, Grivennikova VG, Burbaev DS, Vinogradov AD, Ohnishi T. EPR characterization of ubisemiquinones and iron-sulfur cluster N2, central components of the energy coupling in the NADH-ubiquinone oxidoreductase (complex I) in situ. Journal of bioenergetics and biomembranes. 2002;34:193–208. doi: 10.1023/a:1016083419979. [DOI] [PubMed] [Google Scholar]
- Mathiesen C, Hagerhall C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochimica et biophysica acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Keilin’s respiratory chain concept and its chemiosmotic consequences. Science. 1979;206:1148–1159. doi: 10.1126/science.388618. [DOI] [PubMed] [Google Scholar]
- Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochimica et biophysica acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Murai M, Sekiguchi K, Nishioka T, Miyoshi H. Characterization of the inhibitor binding site in mitochondrial NADH-ubiquinone oxidoreductase by photoaffinity labeling using a quinazoline-type inhibitor. Biochemistry. 2009;48:688–698. doi: 10.1021/bi8019977. [DOI] [PubMed] [Google Scholar]
- Nakamaru-Ogiso E, Kao MC, Chen H, Sinha SC, Yagi T, Ohnishi T. The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. The Journal of biological chemistry. 2010;285:39070–39078. doi: 10.1074/jbc.M110.157826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima Y, Shinzawa-Itoh K, Watanabe K, Naoki K, Hano N, Yoshikawa S. The second coenzyme Q1 binding site of bovine heart NADH: coenzyme Q oxidoreductase. Journal of bioenergetics and biomembranes. 2002;34:89–94. doi: 10.1023/a:1015119808009. [DOI] [PubMed] [Google Scholar]
- Narayanan M, Gabrieli DJ, Leung SA, Elguindy MM, Glaser CA, Saju N, Sinha SC, Nakamaru-Ogiso E. Semiquinone and cluster N6 signals in His-tagged proton-translocating NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. The Journal of biological chemistry. 2013;288:14310–14319. doi: 10.1074/jbc.M113.467803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi ST, Shinzawa-Itoh K, Ohta K, Yoshikawa S, Ohnishi T. New insights into the superoxide generation sites in bovine heart NADH-ubiquinone oxidoreductase (Complex I): the significance of protein-associated ubiquinone and the dynamic shifting of generation sites between semiflavin and semiquinone radicals. Biochimica et biophysica acta. 2010;1797:1901–1909. doi: 10.1016/j.bbabio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Johnson JE, Jr, Yano T, Lobrutto R, Widger WR. Thermodynamic and EPR studies of slowly relaxing ubisemiquinone species in the isolated bovine heart complex I. FEBS letters. 2005;579:500–506. doi: 10.1016/j.febslet.2004.11.107. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Salerno JC. Conformation-driven and semiquinone-gated proton-pump mechanism in the NADH-ubiquinone oxidoreductase (complex I) FEBS letters. 2005;579:4555–4561. doi: 10.1016/j.febslet.2005.06.086. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Sled VD, Yano T, Yagi T, Burbaev DS, Vinogradov AD. Structure-function studies of iron-sulfur clusters and semiquinones in the NADH-Q oxidoreductase segment of the respiratory chain. Biochimica et biophysica acta. 1998;1365:301–308. doi: 10.1016/s0005-2728(98)00082-6. [DOI] [PubMed] [Google Scholar]
- Ripple MO, Kim N, Springett R. Mammalian complex I pumps 4 protons per 2 electrons at high and physiological proton motive force in living cells. The Journal of biological chemistry. 2013;288:5374–5380. doi: 10.1074/jbc.M112.438945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DE, Prince RC, Bowyer JR, Matsuura K, Dutton PL, Ohnishi T. Thermodynamic properties of the semiquinone and its binding site in the ubiquinol-cytochrome c (c2) oxidoreductase of respiratory and photosynthetic systems. The Journal of biological chemistry. 1984;259:1758–1763. [PubMed] [Google Scholar]
- Sazanov LA. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE. Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry. 2000;39:7229–7235. doi: 10.1021/bi000335t. [DOI] [PubMed] [Google Scholar]
- Sinha PK, Torres-Bacete J, Nakamaru-Ogiso E, Castro-Guerrero N, Matsuno-Yagi A, Yagi T. Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J Biol Chem. 2009 doi: 10.1074/jbc.M809468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, King TE. Evidence of an ubisemiquinone radical(s) from the NADH-ubiquinone reductase of the mitochondrial respiratory chain. The Journal of biological chemistry. 1983;258:352–358. [PubMed] [Google Scholar]
- Tocilescu MA, Fendel U, Zwicker K, Drose S, Kerscher S, Brandt U. The role of a conserved tyrosine in the 49-kDa subunit of complex I for ubiquinone binding and reduction. Biochimica et biophysica acta. 2010;1797:625–632. doi: 10.1016/j.bbabio.2010.01.029. [DOI] [PubMed] [Google Scholar]
- Tocilescu MA, Fendel U, Zwicker K, Kerscher S, Brandt U. Exploring the ubiquinone binding cavity of respiratory complex I. The Journal of biological chemistry. 2007;282:29514–29520. doi: 10.1074/jbc.M704519200. [DOI] [PubMed] [Google Scholar]
- van Belzen R, Kotlyar AB, Moon N, Dunham WR, Albracht SP. The iron-sulfur clusters 2 and ubisemiquinone radicals of NADH:ubiquinone oxidoreductase are involved in energy coupling in submitochondrial particles. Biochemistry. 1997;36:886–893. doi: 10.1021/bi9612982. [DOI] [PubMed] [Google Scholar]
- Vinogradov AD, Sled VD, Burbaev DS, Grivennikova VG, Moroz IA, Ohnishi T. Energy-dependent Complex I-associated ubisemiquinones in submitochondrial particles. FEBS letters. 1995;370:83–87. doi: 10.1016/0014-5793(95)00803-h. [DOI] [PubMed] [Google Scholar]
- Wikstrom M. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS letters. 1984;169:300–304. doi: 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- Wraight CA. Proton and electron transfer in the acceptor quinone complex of photosynthetic reaction centers from Rhodobacter sphaeroides. Frontiers in bioscience : a journal and virtual library. 2004;9:309–337. doi: 10.2741/1236. [DOI] [PubMed] [Google Scholar]
- Yagi T, Matsuno-Yagi A. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry. 2003;42:2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Procaryotic complex I (NDH-1), an overview. Biochimica et biophysica acta. 1998;1364:125–133. doi: 10.1016/s0005-2728(98)00023-1. [DOI] [PubMed] [Google Scholar]
- Yano T, Dunham WR, Ohnishi T. Characterization of the delta muH+-sensitive ubisemiquinone species (SQ(Nf)) and the interaction with cluster N2: new insight into the energy-coupled electron transfer in complex I. Biochemistry. 2005;44:1744–1754. doi: 10.1021/bi048132i. [DOI] [PubMed] [Google Scholar]
- Yip CY, Harbour ME, Jayawardena K, Fearnley IM, Sazanov LA. Evolution of respiratory complex I: “supernumerary” subunits are present in the alpha-proteobacterial enzyme. The Journal of biological chemistry. 286:5023–5033. doi: 10.1074/jbc.M110.194993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa J, Yamada S, Fukuzumi S. One-step versus stepwise mechanism in protonated amino acid-promoted electron-transfer reduction of a quinone by electron donors and two-electron reduction by a dihydronicotinamide adenine dinucleotide analogue. Interplay between electron transfer and hydrogen bonding. Journal of the American Chemical Society. 2008;130:5808–5820. doi: 10.1021/ja8001452. [DOI] [PubMed] [Google Scholar]


