Abstract
The symbiotic association between Medicago truncatula and Sinorhizobium meliloti is a well-established model system in the legume–Rhizobium community. Despite its wide use, the symbiotic efficiency of this model has been recently questioned and an alternative microsymbiont, S. medicae, has been proposed. However, little is known about the physiological mechanisms behind the higher symbiotic efficiency of S. medicae WSM419. In the present study, we inoculated M. truncatula Jemalong A17 with either S. medicae WSM419 or S. meliloti 2011 and compared plant growth, photosynthesis, N2-fixation rates, and plant nodule carbon and nitrogen metabolic activities in the two systems. M. truncatula plants in symbiosis with S. medicae showed increased biomass and photosynthesis rates per plant. Plants grown in symbiosis with S. medicae WSM419 also showed higher N2-fixation rates, which were correlated with a larger nodule biomass, while nodule number was similar in both systems. In terms of plant nodule metabolism, M. truncatula–S. medicae WSM419 nodules showed increased sucrose-catabolic activity, mostly associated with sucrose synthase, accompanied by a reduced starch content, whereas nitrogen-assimilation activities were comparable to those measured in nodules infected with S. meliloti 2011. Taken together, these results suggest that S. medicae WSM419 is able to enhance plant carbon catabolism in M. truncatula nodules, which allows for the maintaining of high symbiotic N2-fixation rates, better growth and improved general plant performance.
Keywords: Medicago truncatula, Sinorhizobium medicae, Sinorhizobium meliloti, symbiosis, efficiency, nitrogen fixation, carbon metabolism
INTRODUCTION
One of the most studied plant–microbe symbiosis is the one established between members of the Leguminosae family and soil bacteria from diverse genera collectively termed rhizobia. When compatible symbiotic partners interact, the microsymbiont is able to invade the host root hair cells, typically (but not exclusively) through infection threads, reaching the root cortex, where they are released and differentiate into nitrogen-fixing forms; the bacteroids. In such differentiated forms, bacteria express an enzyme complex, the nitrogenase, which catalyzes the reduction of atmospheric dinitrogen (N2) to ammonium during the highly energy-demanding process known as symbiotic N2-fixation. During this complex symbiotic interaction the plant provides a carbon source, mainly in the form of malate (Udvardi et al., 1988), to be used as a respiratory substrate to fuel the N2-fixation process (Lodwig and Poole, 2003). Symbiotic N2-fixation is estimated to contribute to nearly half of the global biological N2-fixation reactions worldwide, representing a key process for sustainable natural and agricultural systems (Gruber and Galloway, 2008).
In recent years Medicago truncatula (barrel medic) has been one of the model legume species most widely studied by the symbiotic community (Barker et al., 1990; Cook, 1999). The development of mutant collections (Tadege et al., 2008; Calderini et al., 2011), optimization of transformation techniques (Boisson-Dernier et al., 2001) and availability of its genome sequence (Young et al., 2011) have greatly contributed to progress in the field.
So far at least two Sinorhizobium [renamed Ensifer (Young, 2003)] species have been described to nodulate Medicago spp: Sinorhizobium meliloti and S. medicae (Rome et al., 1996a). Although M. truncatula is able to establish N2-fixing symbiosis with both symbionts, most plant molecular biology studies have been carried out using the sequenced S. meliloti 1021 strain (Galibert et al., 2001). In recent years, however, the suitability of the M. truncatula–S. meliloti model has been questioned based on evidences that suggest that N2-fixation in this model is only partially effective (Moreau et al., 2008; Terpolilli et al., 2008). Instead, S. medicae WSM419, for which genomic sequence is also available (Reeve et al., 2010), has been suggested as a more efficient symbiont for M. truncatula (Terpolilli et al., 2008).
Phylogenetic analysis has shown that S. meliloti and S. medicae form a tight cluster within the Sinorhizobium group (Gaunt et al., 2001). Furthermore, application of several molecular markers to genetically analyze this relationship suggests that S. medicae was originated from an ancestral S. meliloti population (Biondi et al., 2003). Nowadays, these rhizobial species can be differentiated both at the phenotypic and genotypic level: S. meliloti is more specific for the tetraploid M. sativa and is preferentially found in alkaline or neutral soils, while S. medicae prefers diploid Medicago species such as M. truncatula and is predominantly found in moderately acid environments (Biondi et al., 2003; Garau et al., 2005). These host and environment preferences may have been a consequence of the various interspecific horizontal gene transfers that occurred during species diversification (Bailly et al., 2007; Epstein et al., 2014).
Nevertheless, to date, the physiological mechanisms underlying the higher symbiotic efficiency in the M. truncatula A17–S. medicae WSM419 association remain largely unknown. Comparative genomic studies of multiple S. meliloti and S. medicae strains have shed some light, suggesting that differences in gene content between the two species, particularly in genes involved in sulfur assimilation, conjugation and secretion, can be related to the differential symbiotic interaction and N2-fixation efficiency (Sugawara et al., 2013). Understanding which are the factors that underpin N2-fixation efficiency in legumes has potentially profound implications for sustainable agricultural systems and the environment.
In the current work, we analyzed the differences at the physiological and metabolic levels between the currently established model M. truncatula–S. meliloti and the more efficient M. truncatula–S. medicae symbiosis. We hypothesized that plant nodule metabolism may be enhanced in the S. medicae symbiosis compared to less efficient strains. To test this hypothesis, two sets of M. truncatula Jemalong A17 plants were grown under symbiotic conditions either with S. meliloti 2011 or S. medicae WSM419. Plant growth parameters, photosynthesis, N2-fixation, and plant nodule carbon and nitrogen metabolic activities were determined. Results presented here show that S. medicae WSM419-derived nodules generate a stronger sink in the plant, through the activation of sucrose-hydrolyzing enzymes. This allows the maintenance of high N2-fixation rates, increased nodule growth, and, therefore, a generally improved plant performance.
MATERIALS AND METHODS
GROWTH CONDITIONS
Medicago truncatula Gaertn cv. Jemalong A17 plants were grown in 1-L pots with a mixture of perlite:vermiculite (2:5, v/v) as substrate under controlled environmental conditions (14 h day/10 h night; 450 μmol m-2 s-1 light intensity; 22°C/16°C day/night temperature; 60–70% relative humidity). After germination, plantlets were separated into two sets: one was inoculated with S. meliloti strain 2011 (Meade and Signer, 1977) and the other was inoculated with S. medicae strain WSM419 (Rome et al., 1996b). Bacterial cultures were grown on a rotary shaker (175 rpm) at 28°C for 48 h in yeast extract mannitol broth containing (g L-1) K2HPO4 (0.5), 0.2 MgSO4⋅7H2O, NaCl (0.1), mannitol (10), and yeast extract (0.4), pH adjusted to 6.8, to an OD600 of 0.7–0.8, which corresponds to ∼3 × 108 cells (Vincent, 1970). 1 ml of the cultures was inoculated onto each seedling at sowing.
Plants were watered with a nutrient solution containing (values in mg L-1): MgSO4⋅7H2O (493), K2SO4 (279), K2HPO4 (145), CaCl2 (56), KH2PO4 (23), EDTA-Fe (17), H3BO3 (1.43), CaSO4⋅2H2O (1.03), MnSO4⋅7H2O (0.77), ZnSO4⋅7H2O (0.22), CoCl2⋅6H2O (0.12), CuSO4⋅5H2O (0.08), NaMoO4⋅2H2O (0.05). For the first 3 weeks, 0.25 mM ammonium nitrate was added to the nutrient solution. Eight weeks after planting, symbiotic N2-fixation was measured, nodules collected, divided into aliquots, frozen in liquid N2 and stored at -80°C for analytical determinations. Two nodule aliquots per plant were used for nodule number estimation based on total nodule weight. Shoots and roots were weighed for fresh weight (FW) determinations and, subsequently, oven-dried at 80°C for 48 h before dry weight (DW) was measured.
NITROGEN FIXATION AND CHLOROPHYLL CONTENT DETERMINATIONS
Symbiotic N2-fixation was measured in intact plants as apparent nitrogenase activity (ANA). H2 evolution from sealed roots systems was measured in an open flow-through system under N2:O2 (79%:21%, v/v) according to Witty and Minchin (1998) using an electrochemical H2-sensor (Qubit System, Canada).
Photosynthesis was determined in the apical leaves with an open system mode (model LC pro+; ADC BioScientific Ltd., Great Amwell, UK) using an ADC PLC-7504 leaf chamber. To estimate leaf chlorophyll content a Minolta SPAD-502 system was employed (Konica Minolta Sensing Europe BV, UK).
NODULE PROTEIN EXTRACTION AND ENZYMES ASSAY
Nodules (100 mg FW) were homogenized in a mortar and pestle with 500–600 μL of extraction buffer (50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), 5 mM MgCl2, 20 mM KCl, 1 mM EDTA, 20% polyvinylpolypyrrolidone, pH 7) where 1.5 mg mL-1 of DTT, 0.7 μL mL-1 of β-mercaptoethanol and 20 μL mL-1 plant protease inhibitor cocktail (Sigma-Aldrich) were freshly added. Homogenates were centrifuged at 12,000 g and 4°C for 15 min and supernatants were collected as nodule plant fractions. The nodule plant fraction was desalted using Bio Gel P6DG columns (Bio-Rad) equilibrated with 250 mM MOPS (pH 7), 100 mM KCl and 25 mM MgCl2. The desalted extract was used to measure the following enzyme activities according to Gonzalez et al. (1998): sucrose synthase (EC 2.4.1.13), alkaline invertase (EC 3.2.1.26), NADH-dependent glutamate synthase (GOGAT; EC 1.4.1.14), and aspartate aminotransferase (AAT; EC 2.6.1.1). The protein content in crude and desalted extracts was quantified using a Bradford-based dye-binding assay (Bio-Rad) employing bovine serum albumin as standard.
CARBOHYDRATE AND STARCH DETERMINATION
100 mg-FW nodule aliquots were extracted in 80% (v/v) ethanol and ultrasonicated in a water bath system. After sonication, samples were centrifuged at 7,500 g and 4°C for 5 min and supernatants were collected. These steps were repeated three times. Afterward the supernatants were dried in a Turbovap LV evaporator (Zymark Corp, Hopkinton, MA, USA) and soluble compounds were redissolved in 1 mL distilled water, homogenized and stored at -20°C. The ethanol-insoluble residue was extracted for starch determination as in Macrae (1971). Carbohydrates were analyzed by high-performance capillary electrophoresis (Warren and Adams, 2000) using 10 mM benzoate (pH 12) containing 0.5 mM myristyltrimethylammonium bromide as a buffer under the following conditions: -15 kV potential, 50 μm-internal diameter and 30/40.2 cm-long capillary tube, indirect UV detection at 225 nm.
STATISTICAL ANALYSIS
All data are reported as mean ± standard deviation of n = 5 independent measurements. Statistical analysis was conducted using Student’s t-test and p≤ 0.05 was considered as statistically significant. The homogeneity of variances was tested using Levene’s test.
RESULTS
In general terms, M. truncatula plants inoculated with S. medicae WSM419 outperformed those inoculated with S. meliloti 2011. Total plant biomass in the M. truncatula–S. medicae system was more than two-fold higher than when using the S. meliloti strain and the difference was most notable for shoots (Figure 1; Table 1). Plants inoculated with the S. medicae strain maintained a 1:1 shoot-to-root ratio, while this declined to ∼3:4 in plants inoculated with S. meliloti 2011 (Table 1).
FIGURE 1.
Medicago truncatula cv. Jemalong A17 plants 8 weeks after inoculation with either Sinorhizobium meliloti 2011 (left) or Sinorhizobium medicae WSM419 (right). Scale bar = 2 cm.
Table 1.
Plant biomass.
| Biomass (g FW) | M. truncatula–S. meliloti 2011 | M. truncatula–S. medicae WSM419 |
|---|---|---|
| Shoot | 9.10 ± 1.81 | 26.48 ± 1.00* |
| Root | 12.85 ± 2.55 | 25.70 ± 2.73* |
| Total plant | 22.40 ± 4.43 | 52.95 ± 2.66* |
| Shoot:root ratio | 0.71 ± 0.02 | 1.08 ± 0.13* |
M. truncatula plant biomass values when grown in symbiosis with S. meliloti 2011 or S. medicae WSM419. Values are mean ± standard deviation of five biological replicates. An asterisk (*) denotes significant differences (Student’s t-test at p ≤ 0.05). FW = fresh weight.
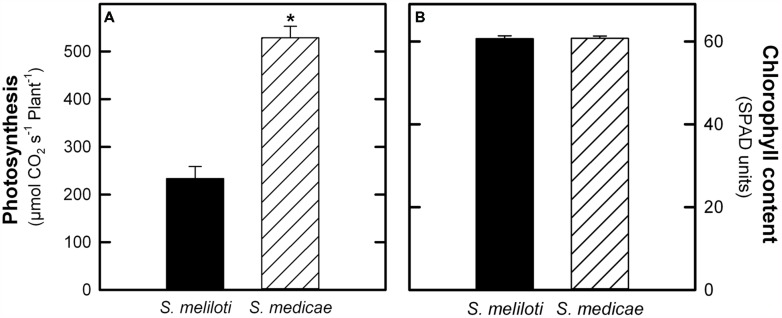
Regarding photosynthetic CO2 assimilation, M. truncatula–S. medicae plants showed a 55.8% increase in photosynthesis when expressed on a plant basis (Figure 2A). However, when expressed on a leaf area basis, M. truncatula–S. meliloti showed higher photosynthetic rates (86.37 ± 2.09 μmol CO2 s-1 cm-2) compared to S. medicae-inoculated plants (67.19 ± 2.43 μmol CO2 s-1 cm-2). These higher photosynthetic rates were, however, not correlated with increased leaf chlorophyll content values, with both plant systems presenting similar values (Figure 2B).
FIGURE 2.
Photosynthesis rates (A) and chlorophyll content (B) in M. truncatula inoculated with either S. meliloti 2011 or S. medicae WSM419. Values represent mean ± standard deviation (n = 5). An asterisk (*) denotes significant differences (Student’s t-test at p ≤ 0.05).
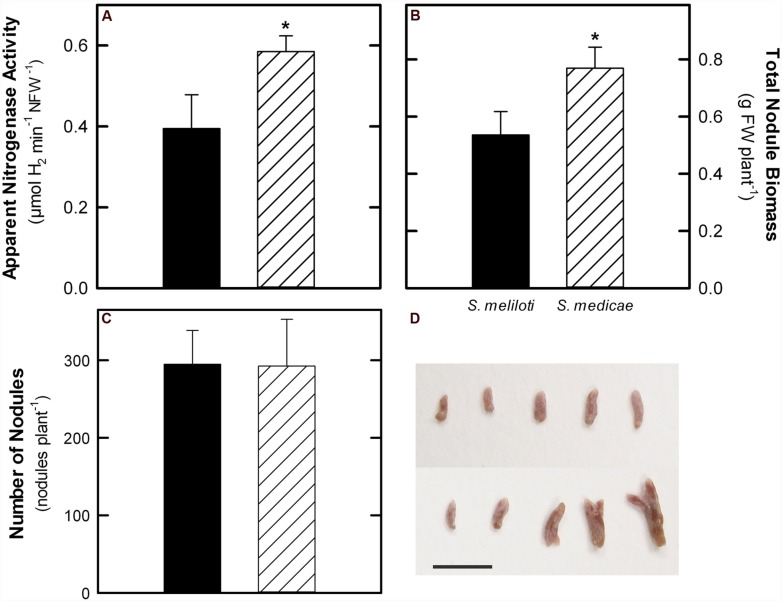
To accurately estimate the rates of N2-fixation, ANA was measured as H2 evolution in intact plants (Witty and Minchin, 1998). The M. truncatula–S. medicae symbiosis showed increased N2-fixation values both when expressed on a plant (+57%) and nodule FW basis (Figure 3A). Plants inoculated with S. medicae showed higher nodule biomass (Figure 3B), although the number of root nodules was similar in both cases (Figure 3C). The increase in nodule biomass was, therefore, correlated with higher biomass per nodule. Plants inoculated with the S. medicae strain presented larger and more frequently bifurcated nodules compared to plants inoculated with the S. meliloti strain (Figure 3D). Furthermore, the plant fraction of M. truncatula–S. medicae nodules showed a significantly higher protein content than that of nodules infected with S. meliloti (25.18 ± 3.32 vs. 20.53 ± 3.58 mg protein g FW-1, mean ± standard deviation, respectively).
FIGURE 3.
N2-fixation rates measured as apparent nitrogenase activity (ANA, A), total nodule biomass (B), nodule number (C) in M. truncatula plants inoculated with either S. meliloti 2011 or S. medicae WSM419. D, representative image of nodules sampled from plants inoculated with S. meliloti 2011 (top) or S. medicae WSM419 (bottom). Scale bar = 500 μm. Values represent mean ± standard deviation (n = 5). An asterisk (*) denotes significant differences (p ≤ 0.05).
To better understand the metabolic differences in nodules following inoculation with the two microsymbionts, we measured the activity of the two main sucrose-degrading enzymes in nodules, sucrose synthase and alkaline invertase, as well as the activity of two key enzymes involved in ammonium assimilation, GOGAT and AAT. In both systems the specific activity of sucrose synthase was on average more than 25-fold higher than that of alkaline invertase (data not shown). Comparing the activity levels across systems, only sucrose synthase showed a significant increase in S. medicae-infected nodules (Figure 4A). In terms of nodule nitrogen metabolism, neither GOGAT nor AAT activities showed significantly different rates when comparing the two inoculants (data not shown).
FIGURE 4.
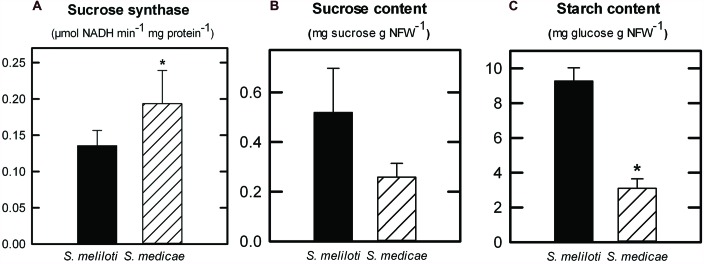
Sucrose synthase (A) enzymatic activity in M. truncatula nodules inoculated with either S. meliloti 2011 or S. medicae WSM419. Values are given in μmol NADH min-1 mg plant protein-1 and represent mean ± standard deviation (n = 5). Sucrose (B) and starch content (C) in M. truncatula nodules inoculated with either S. meliloti 2011 or S. medicae WSM419. Values (in mg g NFW-1) represent mean ± standard deviation of five biological replicates. An asterisk (*) denotes significant differences (p ≤ 0.05).
Given that nodule sucrose catabolism was found to be more active in the M. truncatula–S. medicae symbiosis, the main carbon metabolites in nodules were quantified; sucrose and starch (Figures 4B,C). As a general trend, S. medicae-infected nodules presented lower levels of carbohydrates compared to those infected by the S. meliloti strain, with significant differences found in terms of starch content (Figure 4C).
DISCUSSION
The efficiency of a legume–Rhizobium symbiosis is usually evaluated by comparing plant growth parameters (e.g., biomass, N content) of inoculated versus N-fed plants. These types of study, mostly analyzed from the bacterial perspective, have demonstrated that symbiotic efficiency varies depending upon the specific bacterial strain used (Miller and Sirois, 1982; Mhadhbi et al., 2005; Parra-Colmenares and Kahn, 2005; Heath and Tiffin, 2007; Rangin et al., 2008; Terpolilli et al., 2008; Oono and Denison, 2010). However, the plant contribution to these variable efficiencies has received much less attention.
In this work, we analyzed the effectiveness of the symbiosis of M. truncatula A17 with two Sinorhizobium strains, S. meliloti 2011 and S. medicae WSM419, with special emphasis on understanding the main differences at the nodule metabolic level. Under our experimental growth conditions, M. truncatula plants grown almost exclusively on fixed N upon inoculation with S. meliloti 2011 did not show symptoms of N deficiency, presenting leaf chlorophyll contents comparable to those of plants inoculated with the S. medicae strain (Figure 2B). We did, however, observe a general outperformance of plants inoculated with the S. medicae strain in terms of plant biomass (Figure 1; Table 1), photosynthesis per plant (Figure 2A) and N2-fixation rates (Figure 3A). Interestingly, this improved fixation performance was correlated with a larger biomass per nodule, leading to a higher total nodule biomass per plant, but not to increased nodule number (Figure 3).
Nodules are strong sink tissues due to the high-energy demand that symbiotic N2-fixation represents for the plant (Silsbury, 1977; Schuize et al., 1999). These high-energy requirements are met by allocating photoassimilates from the aerial part to nodules, mostly in the form of sucrose, where they are hydrolyzed by either sucrose synthase or alkaline invertase (Morell and Copeland, 1984; Flemetakis et al., 2006). Sucrose synthase is considered to be primarily responsible for sucrose metabolism in mature nodules and its role has been shown to be essential for symbiotic N2-fixation in legumes (Gordon et al., 1999; Baier et al., 2007; Horst et al., 2007), while alkaline invertase appears to have a secondary role (Welham et al., 2009). In this study, the predominant role of sucrose synthase as the main sucrose-degrading enzyme in nodules was corroborated, showing a significantly higher specific activity than that of alkaline invertase in both symbiotic systems (>20-fold higher in average). Nodules from plants inoculated with the more efficient S. medicae strain showed higher sucrose synthase activity compared to S. meliloti 2011 nodules (Figure 4A). Furthermore, S. medicae WSM419-inoculated plants maintained nodule starch at significantly lower levels compared to those inoculated with the S. meliloti strain (Figure 4C), despite the higher photosynthetic rates of the former (Figure 2A). This inverse correlation between symbiotic efficiency and starch accumulation has been similarly observed in alfalfa plants when inoculated with a fix - strain (Aleman et al., 2010). Indeed, in non-fixing alfalfa nodules, the products from sucrose breakdown are re-directed to starch biosynthesis due to the lower energy demand. Taken together, these results suggest that S. medicae WSM419 activates plant carbon catabolic reactions in nodules to keep up with the high nitrogenase demand for ATP and, as a consequence, they become stronger metabolic sinks in the plant (Sung et al., 1989). This positive feedback keeps N2-fixation rates high, promoting plant growth and, therefore, increasing the plant photosynthetic capacity. A similar mechanism has been described when bacteroid respiration is enhanced in nodules by the overexpression of a cytochrome oxidase (Soberon et al., 1999; Silvente et al., 2002; Talbi et al., 2012).
Despite the differences in N2-fixation rates, plants inoculated with the S. meliloti strain did not show significant differences in terms of nodule number (Figure 3C). Differences were, however, found in the plant protein fraction of nodules, most likely related to the metabolic activation discussed above. It is interesting, though, that these differences are mostly observed at the level of carbon metabolism, while the specific activity of enzymes involved in N assimilation did not differ significantly when the two symbiotic systems were compared (data not shown).
In conclusion, results presented here suggest that at least one of the factors contributing to the higher effectiveness of the M. truncatula–S. medicae WSM419 symbiosis is the activation of plant carbon catabolism in nodules, which allows the maintenance of high N2-fixation rates and, ultimately, leads to an improved plant performance. In agreement with previous studies (Moreau et al., 2008; Terpolilli et al., 2008), the use of S. medicae WSM419 as the partner of choice for M. truncatula symbiotic studies is highly recommended.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Euan K. James (The James Hutton Institute, Dundee, UK) and Dr. Jason J. Terpolilli (Murdoch University, Western Australia, Australia) for sharing the S. medicae WSM419 strain, and Dr. Gustavo Garijo for technical assistance. This work has been partially funded by the Spanish National Research and Development Programmes (AGL2011-23738 and AGL2011-30386-C02-01). Estíbaliz Larrainzar and Erena Gil-Quintana are funded by the European FP7-PEOPLE program (253141). Amaia Seminario is funded by a predoctoral fellowship from the Public University of Navarre.
REFERENCES
- Aleman L., Ortega J. L., Martinez-Grimes M., Seger M., Holguin F. O., Uribe D. J., et al. (2010). Nodule-enhanced expression of a sucrose phosphate synthase gene member (MsSPSA) has a role in carbon and nitrogen metabolism in the nodules of alfalfa (Medicago sativa L.). Planta 231 233–244 10.1007/s00425-009-1043-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M. C., Barsch A., Kuster H., Hohnjec N. (2007). Antisense repression of the Medicago truncatula nodule-enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol. 145 1600–1618 10.1104/pp.107.106955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly X., Olivieri I., Brunel B., Cleyet-Marel J. C., Béna G. (2007). Horizontal gene transfer and homologous recombination drive the evolution of the nitrogen-fixing symbionts of Medicago species. J. Bacteriol. 189 5223–5236 10.1128/JB.00105-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. G., Bianchi S., Blondon F., Dattee Y., Duc G., Essad S., et al. (1990). Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 8 40–49 10.1007/BF02668879 [DOI] [Google Scholar]
- Biondi E. G., Pilli E., Giuntini E., Roumiantseva M. L., Andronov E. E., Onichtchouk O. P., et al. (2003). Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol. Lett. 220 207–213 10.1016/S0378-1097(03)00098-3 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Becard G., Rosenberg C., Barker D. G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700 10.1094/MPMI.2001.14.6.695 [DOI] [PubMed] [Google Scholar]
- Calderini O., Carelli M., Panara F., Biazzi E., Scotti C., Tava A., et al. (2011). Collection of mutants for functional genomics in the legume Medicago truncatula. Plant Genet. Resour. 9 174–176 10.1017/S1479262111000165 [DOI] [Google Scholar]
- Cook D. R. (1999). Medicago truncatula- a model in the making! Curr. Opin. Plant Biol. 2 301–304 10.1016/S1369-5266(99)80053-3 [DOI] [PubMed] [Google Scholar]
- Epstein B., Sadowsky M. J., Tiffin P. (2014). Selection on horizontally transferred and duplicated genes in Sinorhizobium (Ensifer), the root-nodule symbionts of Medicago. Genome Biol. Evol. 6 1199–1209 10.1093/gbe/evu090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemetakis E., Efrose R. C., Ott T., Stedel C., Aivalakis G., Udvardi M. K., et al. (2006). Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline. Plant Mol. Biol. 62 53–69 10.1007/s11103-006-9003-4 [DOI] [PubMed] [Google Scholar]
- Galibert F., Finan T. M., Long S. R., Puhler A., Abola P., Ampe F., et al. (2001). The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293 668–672 10.1126/science.1060966 [DOI] [PubMed] [Google Scholar]
- Garau G., Reeve W. G., Brau L., Deiana P., Yates R. J., James D., et al. (2005). The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 276 263–277 10.1007/s11104-005-0374-0 [DOI] [Google Scholar]
- Gaunt M. W., Turner S. L., Rigottier-Gois L., Lloyd-Macgilp S. A., Young J. P. (2001). Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 51 2037–2048 10.1099/00207713-51-6-2037 [DOI] [PubMed] [Google Scholar]
- Gonzalez E. M., Aparicio-Tejo P. M., Gordon A. J., Minchin F. R., Royuela M., Arrese-Igor C. (1998). Water-deficit effects on carbon and nitrogen metabolism of pea nodules. J. Exp. Bot. 49 1705–1714 10.1093/jxb/49.327.1705 [DOI] [Google Scholar]
- Gordon A. J., Minchin F. R., James C. L., Komina O. (1999). Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol. 120 867–877 10.1104/pp.120.3.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber N., Galloway J. N. (2008). An Earth-system perspective of the global nitrogen cycle. Nature 451 293–296 10.1038/nature06592 [DOI] [PubMed] [Google Scholar]
- Heath K. D., Tiffin P. (2007). Context dependence in the coevolution of plant and rhizobial mutualists. Proc. Biol. Sci. 274 1905–1912 10.1098/rspb.2007.0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst I., Welham T., Kelly S., Kaneko T., Sato S., Tabata S., et al. (2007). TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 144 806–820 10.1104/pp.107.097063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig E., Poole P. (2003). Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 22 37–78 10.1080/713610850 [DOI] [Google Scholar]
- Macrae J. C. (1971). Quantitative measurement of starch in very small amounts of leaf tissue. Planta 96 101–108 10.1007/BF00386360 [DOI] [PubMed] [Google Scholar]
- Meade H. M., Signer E. R. (1977). Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 74 2076–2078 10.1073/pnas.74.5.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhadhbi H., Jebara M., Limam F., Huguet T., Aouani M. E. (2005). Interaction between Medicago truncatula lines and Sinorhizobium meliloti strains for symbiotic efficiency and nodule antioxidant activities. Physiol. Plant. 124 4–11 10.1111/j.1399-3054.2005.00489.x [DOI] [Google Scholar]
- Miller R. W., Sirois J. C. (1982). Relative efficacy of different alfalfa cultivar-Rhizobium meliloti strain combinations for symbiotic nitrogen fixation. Appl. Environ. Microbiol. 43 764–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau D., Voisin A. S., Salon C., Munier-Jolain N. (2008). The model symbiotic association between Medicago truncatula cv. Jemalong and Rhizobium meliloti strain 2011 leads to N-stressed plants when symbiotic N2 fixation is the main N source for plant growth. J. Exp. Bot. 59 3509–3522 10.1093/jxb/ern203 [DOI] [PubMed] [Google Scholar]
- Morell M., Copeland L. (1984). Enzymes of sucrose breakdown in soybean nodules: alkaline invertase. Plant Physiol. 74 1030–1034 10.1104/pp.74.4.1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono R., Denison R. F. (2010). Comparing symbiotic efficiency between swollen versus nonswollen rhizobial bacteroids. Plant. Physiol. 154 1541–1548 10.1104/pp.110.163436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Colmenares A., Kahn M. L. (2005). Determination of nitrogen fixation effectiveness in selected Medicago truncatula isolates by measuring nitrogen isotope incorporation into pheophytin. Plant Soil 270 159–168 10.1007/s11104-004-1308-y [DOI] [Google Scholar]
- Rangin C., Brunel B., Cleyet-Marel J. C., Perrineau M. M., Béna G. (2008). Effects of Medicago truncatula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural Sinorhizobium species community. Appl. Environ. Microbiol. 74 5653–5661 10.1128/AEM.01107-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve W., Chain P., O’Hara G., Ardley J., Nandesena K., Bräu L., et al. (2010). Complete genome sequence of the Medicago microsymbiont Ensifer (Sinorhizobium) medicae strain WSM419. Stand. Genomic Sci. 2 77–86 10.4056/sigs.43526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S., Brunel B., Normand P., Fernandez M., Cleyet-Marel J. C. (1996a). Evidence that two genomic species of Rhizobium are associated with Medicago truncatula. Arch. Microbiol. 165 285–288 10.1007/s002030050328 [DOI] [PubMed] [Google Scholar]
- Rome S., Fernandez M. P., Brunel B., Normand P., Cleyet-Marel J. C. (1996b). Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int. J. Syst. Bacteriol. 46 972–980 10.1099/00207713-46-4-972 [DOI] [PubMed] [Google Scholar]
- Schuize J., Adgo E., Merbach W. (1999). Carbon costs associated with N2 fixation in Vicia faba L and Pisum sativum L. over a 14-day period. Plant Biol. 1 625–631 10.1111/j.1438-8677.1999.tb00273.x [DOI] [Google Scholar]
- Silsbury J. H. (1977). Energy requirement for symbiotic nitrogen fixation. Nature 267 149–150 10.1038/267149a0 [DOI] [PubMed] [Google Scholar]
- Silvente S., Blanco L., Camas A., Ortega J. L., Ramírez M., Lara-Flores M. (2002). Rhizobium etli mutant modulates carbon and nitrogen metabolism in Phaseolus vulgaris nodules. Mol. Plant Microbe Interact. 15 728–733 10.1094/MPMI.2002.15.7.728 [DOI] [PubMed] [Google Scholar]
- Soberon M., Lopez O., Morera C., Girard M. L., Tabche M. L., Miranda J. (1999). Enhanced nitrogen fixation in a Rhizobium etli ntrC mutant that overproduces the Bradyrhizobium japonicum symbiotic terminal oxidase cbb3. Appl. Environ. Microbiol. 65 2015–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S. J., Xu D. P., Black C. C. (1989). Identification of actively filling sucrose sinks. Plant Physiol. 89 1117–1121 10.1104/pp.89.4.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara M., Epstein B., Badgley B. D., Unno T., Xu L., Reese J., et al. (2013). Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 14:R17 10.1186/gb-2013-14-2-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54 335–347 10.1111/j.1365-313X.2008.03418.x [DOI] [PubMed] [Google Scholar]
- Talbi C., Sánchez C., Hidalgo-Garcia A., González E. M., Arrese-Igor C., Girard L., et al. (2012). Enhanced expression of Rhizobium etli cbb3 oxidase improves drought tolerance of common bean symbiotic nitrogen fixation. J. Exp. Bot. 63 5035–5043 10.1093/jxb/ers101 [DOI] [PubMed] [Google Scholar]
- Terpolilli J. J., O’Hara G. W., Tiwari R. P., Dilworth M. J., Howieson J. G. (2008). The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytol. 179 62–66 10.1111/j.1469-8137.2008.02464.x [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Price G. D., Gresshoff P. M., Day D. A. (1988). A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett. 231 36–40 10.1016/0014-5793(88)80697-5 [DOI] [Google Scholar]
- Vincent J. M. (1970). A Manual for the Practical Study of Root-Nodule Bacteria. IBP Handbook Number 15. International Biological Programme. Oxford: Blackwell Scientific [Google Scholar]
- Warren C. R., Adams M. A. (2000). Capillary electrophoresis for the determination of major amino acids and sugars in foliage: application to the nitrogen nutrition of Sclerophyllous species. J. Exp. Bot. 51 1147–1157 10.1093/jexbot/51.347.1147 [DOI] [PubMed] [Google Scholar]
- Welham T., Pike J., Horst I., Flemetakis E., Katinakis P., Kaneko T., et al. (2009). A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J. Exp. Bot. 60 3353–3365 10.1093/jxb/erp169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witty J. F., Minchin F. R. (1998). Methods for the continuous measurement of O2 consumption and H2 production by nodulated legume root systems. J. Exp. Bot. 49 1041–1047 10.1093/jxb/49.323.1041 [DOI] [Google Scholar]
- Young J. M. (2003). The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination “Sinorhizobium adhaerens” (Casida 1982) Willems et al. 2003 legitimate? Request for an opinion. Int. J. Syst. Evol. Microbiol. 53 2107–2110 10.1099/ijs.0.02665-0 [DOI] [PubMed] [Google Scholar]
- Young N. D., Debellé F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480 520–524 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]