Abstract
CXCL12-CXCR4-CXCR7 signaling promotes tumor growth and metastasis in breast cancer. Alternative splicing of CXCL12 produces isoforms with distinct structural and biochemical properties, but little is known about isoform-specific differences in breast cancer subtypes and patient outcomes. We investigated global expression profiles of the six CXCL12 isoforms, CXCR4, and CXCR7 in The Cancer Genome Atlas breast cancer cohort using next-generation RNA sequencing in 948 breast cancer and benign samples and seven breast cancer cell lines. We compared expression levels with several clinical parameters, as well as metastasis, recurrence, and overall survival (OS). CXCL12-α, -β, and -γ are highly co-expressed, with low expression correlating with more aggressive subtypes, higher stage disease, and worse clinical outcomes. CXCL12-δ did not correlate with other isoforms but was prognostic for OS and showed the same trend for metastasis and recurrence-free survival. Effects of CXCL12-δ remained independently prognostic when taking into account expression of CXCL12,CXCR4, and CXCR7. These results were also reflected when comparing CXCL12-α, -β, and -γ in breast cancer cell lines. We summarized expression of all CXCL12 isoforms in an important chemokine signaling pathway in breast cancer in a large clinical cohort and common breast cancer cell lines, establishing differences among isoforms in multiple clinical, pathologic, and molecular subgroups. We identified for the first time the clinical importance of a previously unstudied isoform, CXCL12-δ.
Introduction
Nearly all human genes undergo alternative splicing, substantially increasing diversity in protein structure and function [1]. Genome-wide analyses of several different cancers demonstrate extensive perturbations in splicing during tumor initiation and metastasis [2], [3]. Alternatively spliced proteins regulate fundamental processes in cancer, including apoptosis, metabolism, and metastasis, suggesting that dysregulated splicing is critical to malignancy [4], [5], [6]. As prominent examples of alternative splicing in cancer, a switch from pyruvate kinase M1 to the M2 isoform drives anabolic metabolism in malignant cells, and a novel splice variant of the transmembrane protein CD44 promotes metastasis [5], [7], [8], [9]. Isoforms of these and other genes preferentially expressed in malignant versus normal tissues provide potential biomarkers for detection of cancer and may contribute to drug resistance of cancer cells. Identifying changes in protein isoform expression in cancer will improve understanding of key signaling pathways in tumorigenesis and point to novel therapeutic targets to improve cancer therapy [10], [11].
Chemokine CXCL12 and its chemokine receptors CXCR4 and CXCR7 (recently renamed as ACKR3) comprise a signaling axis strongly linked to tumor growth and metastasis in breast cancer and more than 20 other malignancies [12], [13]. CXCL12 binding to CXCR4 activates pathways including phosphatidylinositol-3 kinase and mitogen-activated protein kinases to promote growth, survival, and chemotaxis of breast cancer cells. High levels of CXCL12 are expressed in common sites of breast cancer metastasis such as lung, liver, bone, and brain [14]. CXCR4 commonly is upregulated on breast cancer cells, and numerous studies have demonstrated both gene and protein overexpression of CXCR4 on cancer cells in primary breast tumors [15], [16], [17], [18]. The anatomic distribution of CXCL12 and studies in mouse models of cancer suggest that gradients of this chemokine drive local invasion and subsequent homing of CXCR4 + breast cancer cells to secondary sites [18], [19]. CXCR7 also is expressed by breast cancer cells and stromal cells, such as endothelium on tumor vasculature, in primary breast cancers [20]. CXCR7 functions as a scavenger receptor for CXCL12, functioning in part to decrease amounts of this chemokine in the extracellular space and establish chemotactic gradients [21], [22]. CXCR7 also promotes survival and invasion of malignant cells [23].
Although six different isoforms of human CXCL12 (α, β, γ, δ, ε, and φ) have been described, most studies of CXCL12 focus only on the α isoform or do not distinguish among isoforms [24]. CXCL12 may be secreted by malignant cells in primary breast cancers in addition to carcinoma-associated fibroblasts and/or mesenchymal stem cells in the tumor microenvironment [17], [25], [26]. Fibroblasts isolated from primary breast tumors secrete CXCL12 at higher levels than fibroblasts from normal mammary tissue despite no genetic mutations in stroma [27], [28]. These findings suggest that cancer cells stimulate adjacent fibroblasts to produce higher levels of total CXCL12 in breast tumors than normal mammary tissue [28]. However, while these data demonstrate that carcinoma-associated fibroblasts are characterized by increased CXCL12, it remains unclear to what extent total CXCL12 in breast cancers differs from normal tissue and affects prognosis. On the basis of expression of CXCL12-α and -β in two different breast cancer microarray data sets and immunohistochemistry (IHC) of primary breast tumors, Mirisola and colleagues reported that higher expression levels of CXCL12-α and -β correlate with better disease-free survival [29]. However, a separate high throughput analysis of CXCL12 expression concluded that higher CXCL12 levels correlate with increased metastasis and local recurrence in breast cancer [17]. Determining effects of high versus low CXCL12 on prognosis and disease progression in breast cancer is essential to direct optimal use of therapeutic antibodies and other agents being developed for CXCL12-targeted cancer therapy [30].
Prior genetic analyses of mRNA for CXCL12 isoforms have used microarrays, which frequently lack probes to detect specific isoforms of these genes. However, next-generation sequencing overcomes this limitation. Using bioinformatics analysis of publicly available data sets from The Cancer Genome Atlas (TCGA), we investigated expression of CXCL12 isoforms, as well as CXCR4 and CXCR7 in breast cancer. We then correlated patterns of expression with important molecular phenotypes, clinical parameters, and outcomes in these patients. These analyses revealed distinct differences in expression for various isoforms of these genes. We show that low levels of expression of CXCL12 correlate with worse prognosis in breast cancer with isoform-specific differences among α, β, γ, and δ isoforms. These data demonstrate the impact of CXCL12 isoforms in breast cancer and underscore the need to better understand functional differences among these molecules in disease progression and therapy.
Methods
Study Design and RNA Sequencing
Publicly available RNA next-generation sequencing and clinical data (844 breast cancer and 104 benign breast samples) were retrieved from TCGA for breast cancer [31]. Additional clinical data such as PAM50 clustering and clinical follow-up for the TCGA were obtained from the UCSC Cancer Genomics Browser [32]. RNA sequencing data for seven breast cancer cell lines (two samples each) were obtained from the Illumina iDEA database (www.illumina.com). Three of these cell lines have been shown to have metastatic potential (BT20, MDA-MB-231, and MDA-MB-468), and four cell lines have been shown to have no metastatic potential (BT474, MCF7, T47D, and ZR-75-1) [33], [34], [35]. RNA sequencing reads were aligned to the genome with Tophat [36] using Genome Reference Consortium Human Build 37 (GRCh37 or hg19) (www.ncbi.nlm.nih.gov) as the reference genome.
Seven hundred eighty-five of the cancer samples had clinical data from TCGA, and 832 had data from UCSC Cancer Genome Browser. Her2 status was not included as a column, so we calculated it based on the IHC data column. For cases with equivocal IHC, we then used the in situ hybridization data column.
Normalization was performed using Fragments per Kilobase per Million, and isoform expression values were generated using Cufflinks with Ensembl version 69 as the reference transcriptome [37]. Cufflinks calculates isoform expression levels using a statistical model in which the probability of observing a given fragment is a linear function of the transcript abundance. Gene level expression is the sum of transcript level expression, as each read is assigned to a single transcript. Tophat was chosen because it is the standard sequence aligner used by Cufflinks [38].
Statistical and Bioinformatics Analysis
Correlation coefficients were generated using Spearman's correlation. Hierarchical clustering was performed on the covariance matrices to generate heat maps. Expression levels of the isoforms and at the gene level were compared across clinical and pathologic groups such as cancer versus normal, tumor stage, histology, hormone receptor status, and PAM50 cluster [39]. Means between groups were compared using analysis of variance. Expression was divided into high versus low expression using the median expression value. Kaplan-Meier curves were generated for the high and low expression groups and compared using the log-rank test for metastasis-free survival (MFS), recurrence-free survival (RFS), and overall survival (OS). Hazard ratios (HRs) were generated using univariate Cox regression. Multi-gene analysis was performed using Cox regression with expression of each gene/isoform as a covariate. Comparison of expression between metastatic versus non-metastatic cell lines was performed using Student's t-test. Statistics and plots were generated using the R statistical computing software and GraphPad Prism.
Results
Differential Expression of CXCL12 Isoforms, CXCR4, and CXCR7 in Breast Cancer versus Normal Breast Tissue
Studies of isoforms of CXCL12 in cancer and other diseases have been limited by the lack of isoform-specific probes on microarrays and antibodies for IHC. As a result, studies have focused predominantly on only the α and β isoforms of CXCL12. To overcome limitations of microarrays and antibodies, we investigated expression levels of all isoforms of CXCL12 and receptors CXCR4 and CXCR7 in breast cancer using the TCGA RNA sequencing data set. The clinical and pathologic characteristics of the tumor samples and patients in this data set are shown in Table 1.
Table 1.
Cancer Patient Characteristics Compiled from TCGA.
| Mean Age (Years) |
58.0 ± 13.3 |
Median Follow-Up (Months) |
23.7 |
|---|---|---|---|
| Variable (n) | Variable (n) | ||
| ER | Node stage | ||
| Positive | 579 | 0 | 361 |
| Negative | 170 | 1 + | 409 |
| PR | Death | ||
| Positive | 505 | Positive | 104 |
| Negative | 241 | Negative | 675 |
| Her2 receptor | Recurrence | ||
| Positive | 141 | Positive | 53 |
| Negative | 508 | Negative | 315 |
| PAM50 status | Metastasis | ||
| Basal | 138 | Positive | 48 |
| Her2 | 66 | Negative | 324 |
| Luminal A | 414 | Gender | |
| Luminal B | 190 | Female | 776 |
| Normal | 24 | Male | 9 |
| Overall stage | Menopause | ||
| 1 | 127 | Pre-menopause | 181 |
| 2 | 127 | Post-menopause | 493 |
| 3 | 171 | Race | |
| 4 | 15 | White | 591 |
| Tumor stage | Black or African American | 55 | |
| 1 | 207 | Asian | 51 |
| 2 | 467 | American Indian or Alaskan Native | 1 |
| 3 | 76 | ||
| 4 | 32 |
The Cufflinks analysis program assigns each read to individual isoforms such that the sum of expression levels for a specific isoform is equal to the gene level of expression. On the basis of this analysis, we determined that the most common isoform of CXCL12 in breast cancer is α (65%), followed by β (27%) > γ (5%) > δ (2%). We detected only very low levels of expression for CXCL12-ε (0.1%) and -φ (0.2%) and therefore refrained from statistical inference using these isoforms.
To establish for the first time correlations among CXCL12 isoforms, CXCR4, and CXCR7, we examined the Spearman's covariance matrix with hierarchal clustering for breast cancer and normal tissues, respectively (Figure 1, A and B). In breast cancer, CXCL12-α and -β were highly correlated (correlation coefficient of 0.91), and these two isoforms also correlated highly with gene-level expression of CXCL12 (correlation coefficients of 0.99 and 0.95 for CXCL12-α and -β, respectively; Figure 1A). By comparison, the γ, ε, and φ isoforms of CXCL12 correlated moderately with gene-level expression of α and β (correlation coefficients of 0.44 to 0.59). Interestingly, the δ isoform, which is not well characterized in the literature, correlated very poorly with the other CXCL12 isoforms (correlation coefficients of − 0.11 to 0.27) and in cancer samples, clustered with CXCR4 and CXCR7 rather than with the other CXCL12 isoforms. CXCR4 and CXCR7 displayed a weak positive correlation with gene-level expression of CXCL12 and its α and β isoforms but did not correlate with each other. These same general correlations were present in normal samples (Figure 1B). However, in normal samples, CXCR7 tended to correlate inversely with CXCR4, and CXCR7 also exhibited modest to strong correlations with CXCL12-α and -β and overall gene-level expression of this chemokine.
Figure 1.
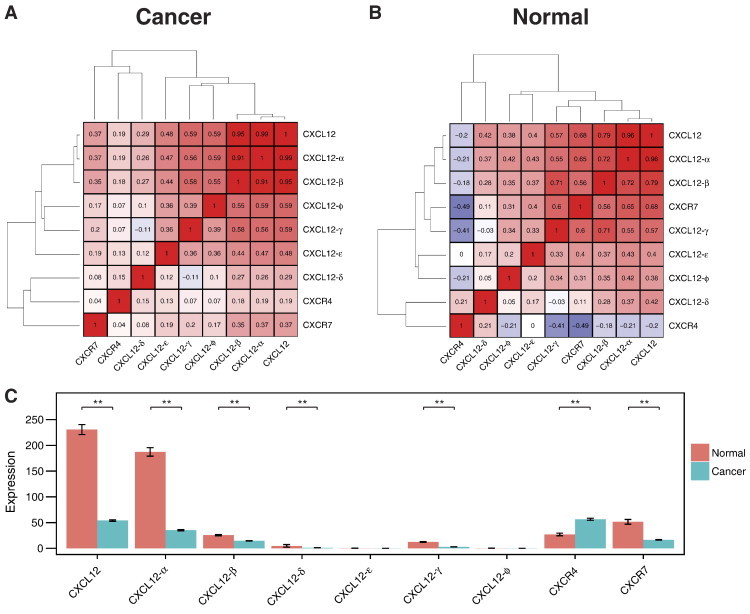
Spearman's covariance matrix with hierarchal clustering of CXCL12 isoforms, CXCR4, and CXCR7 for (A) breast cancer and (B) normal tissues. (C) Expression levels of CXCL12 isoforms, CXCR4, and CXCR7 in breast cancer and normal tissues. Expression levels are means ± SEM. *P < .05 and **P < .001.
We next investigated levels of expression for various chemokine and receptor isoforms in cancer and normal tissues. While previous publications report discordant results for CXCL12 in breast cancer versus normal breast, our analysis showed significant down-regulation of CXCL12-α, -β, and -γ in cancer (Figure 1C). Expression of CXCL12-δ also decreased in cancer as compared with normal, although differences were not significant. Similarly, CXCR7 was downregulated in cancer. CXCR4 demonstrated the opposite pattern with up-regulation in cancer, consistent with prior literature [15], [17], [18].
Variations among CXCL12 Isoforms, CXCR4, and CXCR7 with Clinical and Molecular Staging Parameters
Within cancer samples, CXCL12-α, -β, and -γ varied significantly with tumor stage (Figure 2A). For these isoforms of CXCL12, lower stage tumors had higher levels of expression with the highest amounts of each isoform present in stage I primary breast tumors. We observed a similar trend for gene-level expression of CXCL12. We also compared differences in expression of various isoforms with histologic classifications of breast cancer. Invasive ductal and invasive lobular carcinomas comprise the majority of the TCGA data set, and most of the mixed histology samples contain features of both invasive ductal and lobular cancer. Gene-level expression of CXCL12, as well as α, β, and γ isoforms, showed significant variations across different histologic groups (Figure 2B). Amounts of total CXCL12 and these three isoforms were highest in invasive lobular cancer with a rank order of invasive lobular > mixed > invasive ductal carcinoma. We note that lowest levels of expression for CXCL12 and the α, β, and γ isoforms occurred in less common histologic types of breast cancer, medullary and mucinous. Other isoforms of CXCL12 did not vary significantly with tumor stage or histologic type, and we also did not identify significant correlations with gene-level CXCR4 or CXCR7.
Figure 2.
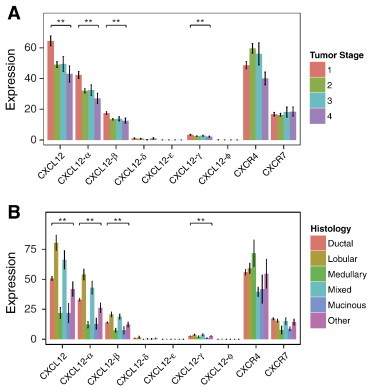
Expression levels of CXCL12-α, -β and -γ vary significantly by (A) tumor and (B) histology. Expression levels are means ± SEM. *P < .05 and **P < .001.
In clinical oncology, breast cancers are categorized on the basis of hormone receptors [estrogen (ER) and progesterone (PR)] and amplification of the oncogene Her2. These categories determine prognosis and treatment options [40]. We analyzed expression of CXCL12, CXCR4, and CXCR7 and individual isoforms in tumors positive for both ER and PR, Her2 only, and all three receptors (triple positive), as well as primary cancers lacking expression of these three receptors (triple negative). Gene-level expression of CXCL12 and the α and β isoforms each varied significantly across these subtypes with highest amounts in ER/PR positive and triple positive cancers (Figure 3A). By comparison, levels of overall CXCL12, CXCL12-α, and CXCL12-β decreased in triple negative cancer and to an even greater extent in Her2 positive tumors. Other isoforms of CXCL12 did not vary significantly with receptor status. CXCR7 varied with receptor status in a pattern comparable to CXCL12 (Figure 3A). Levels of CXCR7 were highest in ER/PR positive and triple positive tumors with lower expression in triple negative and Her2 positive cancers. Interestingly, we identified a distinct pattern of expression for CXCR4, which was elevated in triple negative breast cancer relative to the other groups [41].
Figure 3.
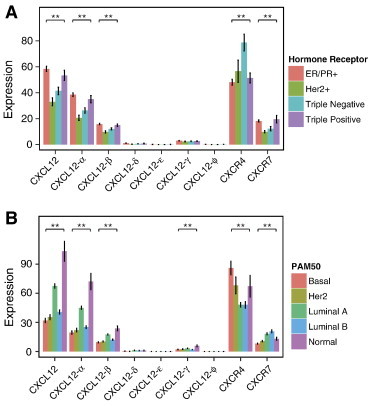
Expression levels of CXCL12 isoforms, CXCR4, and CXCR7 vary with (A) hormone receptor status and (B) molecular subtype. Expression levels are means ± SEM. *P < .05 and **P < .001.
More recently, breast cancers have been classified into intrinsic molecular subtypes (Normal-like, Luminal A, Luminal B, Her2-enriched, and Basal-like) defined by a 50-gene panel referred to as PAM50. Intrinsic subtypes add prognostic and predictive information to standard metrics used to categorize breast cancer. When analyzed across intrinsic subtypes, CXCL12 and its α, β, and γ isoforms varied significantly (Figure 3B). Expression was highest in the Normal-like cluster, which is consistent with our data in Figure 1A showing up-regulation of these isoforms in normal samples. Luminal A had the next highest expression with Luminal B, Her2-enriched, and Basal clusters exhibiting lower expression. We also identified significant variations of receptors with intrinsic subtypes of breast cancer. CXCR4 showed differential expression among clusters with lowest levels in Luminal A and Luminal B subtypes and highest expression in Basal cancers. By comparison, levels of CXCR7 were highest in Luminal A and Luminal B subtypes.
CXCL12 and its α, β, and γ isoforms vary significantly with race. We identified higher expression in whites than Asians or African-Americans (Figure W1A). Gene-level CXCL12 and the α isoform also changed significantly by age group with levels peaking in the 50 to 60 year age group relative to younger or older patients (Figure W1B). CXCL12-β and -γ showed a similar pattern across age groups, although differences were not significant. We did not identify significant correlations for race or age groups for CXCR4 or CXCR7.
CXCL12 isoforms correlate with patient outcomes
We next examined the correlation of gene and isoform expression with important clinical outcomes in breast cancer: metastasis, recurrence, and OS. Kaplan-Meier curves for high versus low expression of gene-level CXCL12 demonstrated that low-expression corresponded with a significantly worse MFS (P < .008, HR = 2.2) but not RFS or OS (Figure 4, A–C). Similarly, low expression of CXCL12-α corresponded with significantly worse MFS (P < .033, HR = 1.9) but not RFS or OS (Figure 4, D–F). Unlike CXCL12-α, low levels of both CXCL12-β and -γ correlated with significantly worse MFS (β isoform P < .0015, HR = 2.6; γ isoform P < .011, HR = 2.2) and RFS (β isoform P < .028, HR = 2.1; γ isoform P < .024, HR = 2.1) but not OS (Figure 4, G–L).
Figure 4.
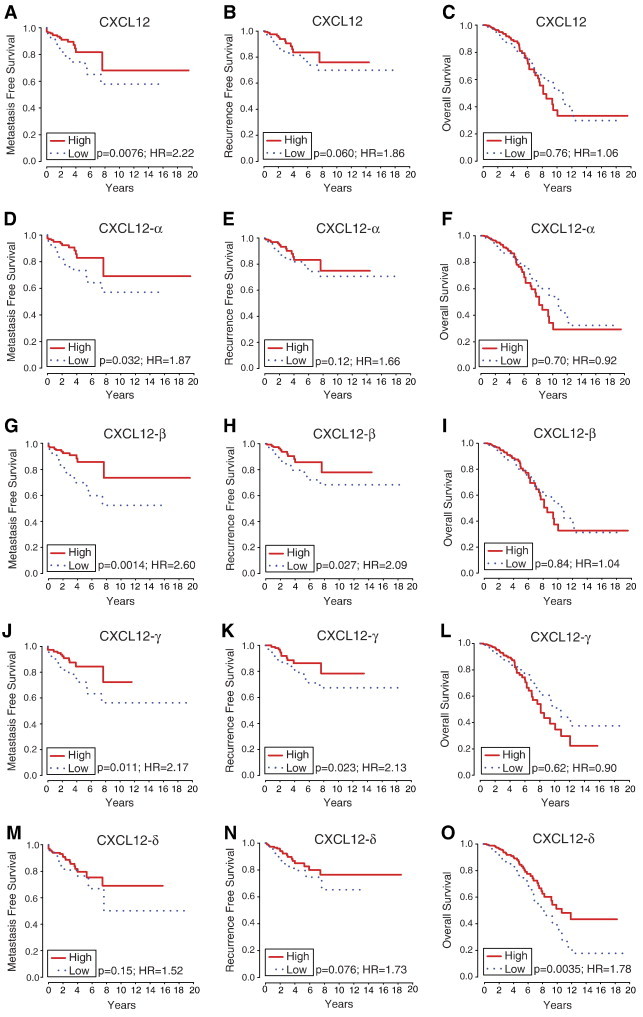
MFS, RFS, and OS curves for CXCL12 isoforms. Higher levels of gene-level CXCL12 (A–C), CXCL12-α (D–F), -β (G–I), and -γ (J–L) generally correlate with improved MFS and RFS. Higher expression of CXCL12-δ (M–O) correlates with better OS.
CXCL12-δ, the isoform that does not correlate with expression patterns of other isoforms in breast cancer or normal breast tissue, had a different association with outcomes. Low expression of the δ isoform also showed trends for reduced MFS and RFS (Figure 4, M and N), although not statistically significant (MFS, P < .16, HR = 1.5; RFS, P < .077, HR = 1.7). Notably, low CXCL12-δ was the only CXCL12 isoform correlated with worse OS (P < .0035, HR = 1.8; Figure 4O).
CXCL12, CXCR4, and CXCR7 do not operate independently but as important components in a complex network. We examined the expression levels of CXCL12-δ, the least understood isoform in the context of the expression of the other genes in the pathway. Low CXCL12-δ is independently prognostic for OS even after taking into account CXCL12, CXCR4, and CXCR7 expression (P < .004, HR = 0.56) and shows the same trend in MFS and RFS (Figure 5, A–C) multi-gene analyses.
Figure 5.
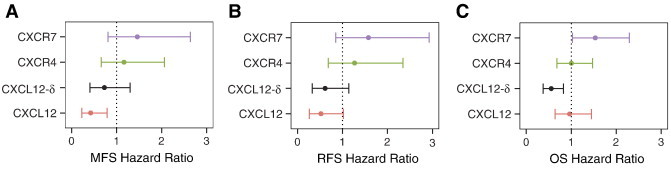
Multi-gene analysis reveals that expression of CXCL12-δ is independently prognostic for (A) MFS, (B) RFS, and (C) OS when accounting for gene-level CXCL12, CXCR4, and CXCR7. Error bars represent 95% confidence intervals.
CXCL12 Isoforms Correlate with Metastatic Potential in Breast Cancer Cell Lines
By nature, clinical samples such as the TCGA contain a mix of cell types, including tumor cells, normal breast tissue, and vasculature, making it difficult to identify the cell type(s) producing each transcript. To overcome this limitation, we examined RNAseq data in seven breast cancer cell lines for CXCL12 isoforms. Surprisingly, we found that isoform expression shows a different trend than those in the TCGA samples, with γ showing the highest expression proportion (42%), followed by α (33%) > β (24%). We detected only very low levels of expression for CXCL12-δ (0.5%), -ε (0.1%) and -φ (0.2%).
We compared CXCL12 isoform expression levels between cell lines with metastatic potential and those without metastatic potential (Figure 6) and found that CXCL12 and its α and β isoforms were expressed significantly lower in samples with metastatic potential, which is in agreement with the trends of isoform expression in clinical samples. The same trend was seen with CXCL12-γ, though not statistically significant.
Figure 6.
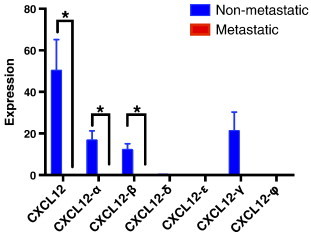
Expression of gene-level CXCL12 and those of CXCL12-α and -β isoforms are significantly higher in cell lines without metastatic potential compared to cell lines with metastatic potential. Expression levels are means ± SEM. *P < .05.
Discussion
While alternative splicing formerly appeared to be limited to a small number of genes, studies now demonstrate that almost all human genes undergo alternative splicing to create protein diversity [42]. Variants generated by loss of splicing fidelity or regulated transitions between isoforms drive cancer and many other human diseases, generating a large number of novel transcripts and proteins expressed predominantly or uniquely in cancer. For example, up to 36 different isoforms of the Wilms tumor gene 1 have been identified with specific variants specifically upregulated in acute and chronic myeloid leukemias, suggesting key functions in cancer initiation and/or progression [43], [44]. Similarly, isoforms of vascular endothelial growth factor exhibit distinct functional activities in tumor angiogenesis that vary on the basis of anatomic site, emphasizing the importance of tumor environments on isoforms [45], [46], [47]. In addition to conferring unique functions to cancer cells and tumor environments, alternative splicing offers a rich source of potential prognostic and predictive biomarkers. Biomarkers and targeted therapies based on alternative splicing may have a higher likelihood for success than conventional approaches centered on a whole gene or protein. Collectively, these studies highlight the clinical relevance of identifying disease-associated changes in alternative splicing.
Prior research has established central functions of CXCL12 in cancer growth and metastasis, but very few studies have investigated isoforms of CXCL12 in cancer. In renal cell carcinoma, an analysis limited to CXCL12-α and -β revealed that only the β isoform correlated with tumor grade and infiltration of CD8 T cells [48]. CXCL12-β also was upregulated in bladder cancer, a disease in which expression of this isoform predicted metastasis and disease-specific mortality [49]. This study of bladder cancer also showed that amounts of CXCL12-α did not change between normal and malignant tissues, while CXCL12-γ was undetectable. Neither these studies nor any others have investigated the other three CXCL12 isoforms (δ, ε, or φ) in cancer due to the lack of antibodies against these isoforms and limitations in high throughput technology.
Next-generation sequencing allows our study to fill notable gaps in knowledge about the CXCL12/CXCR4/CXCR7 pathway by providing the first characterization of expression levels of all known alternative splicing variants of CXCL12 in breast cancer or any other malignancy. We found that primary human breast cancers express four different isoforms of CXCL12 in rank order of α > β > γ > δ, while ε and φ essentially were undetectable in the TCGA breast cancer samples. Expression of CXCL12 isoforms varied significantly across many different clinical and molecular categories of breast cancer, including stage, histologic type, intrinsic molecular subtype, and hormone receptor status. Changes in abundance of transcripts typically occurred in parallel for each CXCL12 isoform as would be expected for an mRNA regulated by the same common promoter elements. We also discovered lower levels of CXCL12 transcripts in subtypes of breast cancer regarded as more aggressive, such as triple negative and Her2 amplified, and with progression to higher stage. These findings corresponded with Kaplan-Meier analyses, where low expression of CXCL12 and specific isoforms was associated with worse outcomes. We identified isoform-specific effects on MFS, RFS, and OS, with low levels of CXCL12-α, -β and -γ significantly correlated with worse MFS and RFS. Most notably, we note that low levels of CXCL12-δ associated with worse OS and showed the same trend for RFS and MFS, despite the fact that CXCL12-δ expression does not correlate with expression of the other isoforms. This relationship is robust and persists even after taking into account CXCL12, CXCR4, and CXCR7 expression in multi-gene analysis, indicating the independent prognostic significance of CXCL12-δ. These data provide the first evidence that CXCL12-δ is expressed in human cancer and correlate with a patient outcome.
Expression levels of CXCL12 in breast cancer cell lines generally mirror conclusions from the clinical samples that lower levels of CXCL12 correlate with worse prognosis. We found that breast cancer cell lines without metastatic potential (in mouse models) had higher levels of CXCL12 expression than cell lines that metastasize more widely. Studies of CXCL12 in breast cancer focus on secretion of this chemokine by stromal cells in primary and metastatic sites, frequently overlooking effects of CXCL12 produced by cancer cells. However, epigenetic silencing of the CXCL12 promoter has been reported in breast cancer cells with greater metastatic potential, and re-expressing CXCL12 limits metastatic disease in mouse xenograft models [25]. Our analysis of cell lines may inform likely sources of various CXCL12 isoforms in tumor microenvironments. Breast cancer cells express CXCL12-α, -β, and -γ with very minimal expression of δ, which could indicate that stromal cells are the predominant source of the δ isoform in primary breast cancers. We also note that CXCL12-γ is higher than α and β in our panel of breast cancer cell lines, which is opposite the pattern in primary tumors. Differences between data from cell lines versus tumors may reflect dynamic regulation of CXCL12 isoforms in vivo, greater contributions of stromal cells to overall expression of CXCL12-α and -β in breast tumors, or simply genomic changes as the original cancer samples were transformed into immortalized cell lines. In addition, CXCL12 levels within the tumor microenvironment may be affected by posttranslational modification, such as cleavage by CD26 or matrix-metalloproteinase-2 [50], [51].
Isoform-specific differences in expression and breast cancer outcomes suggest distinct functions of individual splice variants of CXCL12 on disease progression. Recent studies have begun to identify unique biochemical properties of CXCL12 isoforms, particularly α, β, and γ. While all isoforms share the same core structure, CXCL12-β, -γ, -δ, -ε, and -φ differ by inclusion of exons that add 4, 40, 51, 1, or 11 additional amino acids, respectively, to the carboxy terminus of the molecule [24]. Particularly for CXCL12-γ, the added carboxy-terminal amino acids are enriched with basic residues that enhance binding to heparan sulfates and other negatively charged extracellular matrix molecules [52]. By comparison, CXCL12-β and, to a greater extent, -γ have reduced binding affinities for receptors CXCR4 and CXCR7. Biochemical differences in binding to receptors and extracellular matrix molecules translate to different functional outcomes. In mouse models, CXCL12-γ promotes chemotaxis of immune cells and endothelial progenitors to a significantly greater extent than other isoforms [53], [54]. Greater binding to heparan sulfates and extracellular matrix molecules also limits proteolytic degradation of CXCL12 [55]. These studies highlight functional differences among CXCL12 isoforms in receptor binding, chemotaxis, and stability that could alter outcomes in breast cancer. Our data also support further studies analyzing functional differences among CXCL12 isoforms, especially for CXCL12-δ.
Correlation between gene transcript data and protein expression is dependent on the gene and tissue type. However, mRNA expression is generally a good proxy for protein expression and is frequently used as biomarkers.[56], [57], [58] Gene expression also forms the basis of the PAM50 molecular subtyping of breast cancer as well as Oncotype Dx, a widely used predictive model for chemotherapy response in breast cancer.[59], [60], [61], [62] Specifically for CXCL12-α, -β, and -γ, mRNA levels as measured by quantitative reverse transcription–polymerase chain reaction correlate with protein levels as measured by ELISA.[63] We also recognize that this study has limitations based on the data publicly available through the TCGA. While the data set contains transcript data for a large number of patients, the median follow-up time is relatively short, and therefore, the number of metastasis and recurrence events is small, thus limiting our statistical power. This likely accounts for why the P values for CXCL12-δ MFS and RFS do not reach significance. We also do not know the full treatment history for all patients, such as exact chemotherapy and radiation regimens, and there is likely significant heterogeneity in treatments given the multi-institutional nature of the data. Even with these limitations, we were able to identify significant differences in outcomes for isoforms of CXCL12.
In summary, our data reveal new associations of CXCL12, CXCR4, and CXCR7 gene expression with molecular, histologic, and clinical categories of human breast cancer. In addition, we have identified isoform-specific differences in CXCL12 for outcomes in breast cancer, suggesting distinct biochemical functions of isoforms in disease progression. These compelling results establish the foundation for mechanistic preclinical studies of these isoforms in breast cancer. Additional studies are also warranted to elucidate the biologic and functional differences between the CXCL12 isoforms and validate them as potential biomarkers.
The following are the supplementary data related to this article.
(A) CXCL12 and its α, β, and γ isoforms vary significantly with race. (B) Overall CXCL12 and CXCL12-α vary significantly with age. Expression levels are means ± SEM. *P < .05, **P < .001.
Footnotes
This research was supported by United States National Institutes of Health grants R01CA136553, R01CA142750, R01CA170198, P50CA093990, R01GM096040, and R01EB012579.
This article refers to supplementary material, which is designated by Figure W1 and is available online at www.transonc.com.
References
- 1.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oltean S., Bates D.O. Hallmarks of alternative splicing in cancer. Oncogene. 2013 doi: 10.1038/onc.2013.533. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., Manley J.L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babic I., Anderson E.S., Tanaka K., Guo D., Masui K., Li B., Zhu S., Gu Y., Villa G.R., Akhavan D. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–1008. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin S., Jansen F., Bokelmann J., Kolb H. Soluble CD44 splice variants in metastasizing human breast cancer. Int J Cancer. 1997;74:443–445. doi: 10.1002/(sici)1097-0215(19970822)74:4<443::aid-ijc14>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 6.Schwerk C., Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., Cantley L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 8.Christofk H.R., Vander Heiden M.G., Wu N., Asara J.M., Cantley L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann M., Rudy W., Zöller M., Tölg C., Ponta H., Herrlich P., Günthert U. CD44 splice variants confer metastatic behavior in rats: homologous sequences are expressed in human tumor cell lines. Cancer Res. 1991;51:5292–5297. [PubMed] [Google Scholar]
- 10.Brinkman B.M. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Pajares M.J., Ezponda T., Catena R., Calvo A., Pio R., Montuenga L.M. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- 12.Duda D.G., Kozin S.V., Kirkpatrick N.D., Xu L., Fukumura D., Jain R.K. CXCL12 (SDF1α)-CXCR4/CXCR7 pathway inhibition: an emerging sensitizer for anticancer therapies? Clin Cancer Res. 2011;17:2074–2080. doi: 10.1158/1078-0432.CCR-10-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X., Cheng G., Hao M., Zheng J., Zhou X., Zhang J., Taichman R.S., Pienta K.J., Wang J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29:709–722. doi: 10.1007/s10555-010-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teicher B.A., Fricker S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 15.Cabioglu N., Sahin A., Doucet M., Yavuz E., Igci A., Yildirim E.O., Aktas E., Bilgic S., Kiran B., Deniz G. Chemokine receptor CXCR4 expression in breast cancer as a potential predictive marker of isolated tumor cells in bone marrow. Clin Exp Metastasis. 2005;22:39–46. doi: 10.1007/s10585-005-3222-y. [DOI] [PubMed] [Google Scholar]
- 16.Holm N.T., Byrnes K., Li B.D., Turnage R.H., Abreo F., Mathis J.M., Chu Q.D. Elevated levels of chemokine receptor CXCR4 in HER-2 negative breast cancer specimens predict recurrence. J Surg Res. 2007;141:53–59. doi: 10.1016/j.jss.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Kang H., Watkins G., Parr C., Douglas-Jones A., Mansel R.E., Jiang W.G. Stromal cell derived factor-1: its influence on invasiveness and migration of breast cancer cells in vitro, and its association with prognosis and survival in human breast cancer. Breast Cancer Res. 2005;7:R402–R410. doi: 10.1186/bcr1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 19.Smith M.C., Luker K.E., Garbow J.R., Prior J.L., Jackson E., Piwnica-Worms D., Luker G.D. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 20.Miao Z., Luker K.E., Summers B.C., Berahovich R., Bhojani M.S., Rehemtulla A., Kleer C.G., Essner J.J., Nasevicius A., Luker G.D. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naumann U., Cameroni E., Pruenster M., Mahabaleshwar H., Raz E., Zerwes H.G., Rot A., Thelen M. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajagopal S., Kim J., Ahn S., Craig S., Lam C.M., Gerard N.P., Gerard C., Lefkowitz R.J. β-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci U S A. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns J.M., Summers B.C., Wang Y., Melikian A., Berahovich R., Miao Z., Penfold M.E., Sunshine M.J., Littman D.R., Kuo C.J. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu L., Cecil J., Peng S.B., Schrementi J., Kovacevic S., Paul D., Su E.W., Wang J. Identification and expression of novel isoforms of human stromal cell-derived factor 1. Gene. 2006;374:174–179. doi: 10.1016/j.gene.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Wendt M.K., Cooper A.N., Dwinell M.B. Epigenetic silencing of CXCL12 increases the metastatic potential of mammary carcinoma cells. Oncogene. 2008;27:1461–1471. doi: 10.1038/sj.onc.1210751. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X.H., Jin X., Malladi S., Zou Y., Wen Y.H., Brogi E., Smid M., Foekens J.A., Massagué J. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allinen M., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Orimo A., Gupta P.B., Sgroi D.C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V.J., Richardson A.L., Weinberg R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 29.Mirisola V., Zuccarino A., Bachmeier B.E., Sormani M.P., Falter J., Nerlich A., Pfeffer U. CXCL12/SDF1 expression by breast cancers is an independent prognostic marker of disease-free and overall survival. Eur J Cancer. 2009;45:2579–2587. doi: 10.1016/j.ejca.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Zhong C., Wang J., Li B., Xiang H., Ultsch M., Coons M., Wong T., Chiang N.Y., Clark S., Clark R. Development and preclinical characterization of a humanized antibody targeting CXCL12. Clin Cancer Res. 2013;19:4433–4445. doi: 10.1158/1078-0432.CCR-13-0943. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldman M., Craft B., Swatloski T., Ellrott K., Cline M., Diekhans M., Ma S., Wilks C., Stuart J., Haussler D. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 2013;41:D949–D954. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan A.L., George R., Vantyghem S.A., Lee M.W., Hodgson N.C., Engel C.J., Holliday R.L., Girvan D.P., Scott L.A., Postenka C.O. Role of the integrin-binding protein osteopontin in lymphatic metastasis of breast cancer. Am J Pathol. 2006;169:233–246. doi: 10.2353/ajpath.2006.051152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chekhun S., Bezdenezhnykh N., Shvets J., Lukianova N. Expression of biomarkers related to cell adhesion, metastasis and invasion of breast cancer cell lines of different molecular subtype. Exp Oncol. 2013;35:174–179. [PubMed] [Google Scholar]
- 35.Matrone M.A., Whipple R.A., Thompson K., Cho E.H., Vitolo M.I., Balzer E.M., Yoon J.R., Ioffe O.B., Tuttle K.C., Tan M. Metastatic breast tumors express increased tau, which promotes microtentacle formation and the reattachment of detached breast tumor cells. Oncogene. 2010;29:3217–3227. doi: 10.1038/onc.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 40.Onitilo A.A., Engel J.M., Greenlee R.T., Mukesh B.N. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu Q.D., Panu L., Holm N.T., Li B.D., Johnson L.W., Zhang S. High chemokine receptor CXCR4 level in triple negative breast cancer specimens predicts poor clinical outcome. J Surg Res. 2010;159:689–695. doi: 10.1016/j.jss.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 42.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 43.Haber D.A., Sohn R.L., Buckler A.J., Pelletier J., Call K.M., Housman D.E. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci U S A. 1991;88:9618–9622. doi: 10.1073/pnas.88.21.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopotová T., Polák J., Schwarz J., Klamová H., Moravcová J. Expression of four major WT1 splicing variants in acute and chronic myeloid leukemia patients analyzed by newly developed four real-time RT PCRs. Blood Cells Mol Dis. 2012;49:41–47. doi: 10.1016/j.bcmd.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Guo P., Xu L., Pan S., Brekken R.A., Yang S.T., Whitaker G.B., Nagane M., Thorpe P.E., Rosenbaum J.S., Su Huang H.J. Vascular endothelial growth factor isoforms display distinct activities in promoting tumor angiogenesis at different anatomic sites. Cancer Res. 2001;61:8569–8577. [PubMed] [Google Scholar]
- 46.Ladomery M.R., Harper S.J., Bates D.O. Alternative splicing in angiogenesis: the vascular endothelial growth factor paradigm. Cancer Lett. 2007;249:133–142. doi: 10.1016/j.canlet.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Nowak D.G., Amin E.M., Rennel E.S., Hoareau-Aveilla C., Gammons M., Damodoran G., Hagiwara M., Harper S.J., Woolard J., Ladomery M.R. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J Biol Chem. 2010;285:5532–5540. doi: 10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wehler T.C., Graf C., Altherr K., Zimmermann T., Brenner W., Thuroff J.W., Biesterfeld S., Gockel I., Theobald M., Galle P.R. SDF1β expression in renal cell carcinoma correlates with grading and infiltration by CD8+ T-cells. Anticancer Res. 2011;31:2797–2803. [PubMed] [Google Scholar]
- 49.Gosalbez M., Hupe M.C., Lokeshwar S.D., Yates T.J., Shields J., Veerapen M.K., Merseburger A.S., Rosser C.J., Soloway M.S., Lokeshwar V.B. Differential Expression of SDF-1 Isoforms in Bladder Cancer. J Urol. 2013 doi: 10.1016/j.juro.2013.11.053. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De La Luz Sierra M., Yang F., Narazaki M., Salvucci O., Davis D., Yarchoan R., Zhang H.H., Fales H., Tosato G. Differential processing of stromal-derived factor-1α and stromal-derived factor-1β explains functional diversity. Blood. 2004;103:2452–2459. doi: 10.1182/blood-2003-08-2857. [DOI] [PubMed] [Google Scholar]
- 51.Peng H., Wu Y., Duan Z., Ciborowski P., Zheng J.C. Proteolytic processing of SDF-1α by matrix metalloproteinase-2 impairs CXCR4 signaling and reduces neural progenitor cell migration. Protein Cell. 2012;3:875–882. doi: 10.1007/s13238-012-2092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altenburg J.D., Broxmeyer H.E., Jin Q., Cooper S., Basu S., Alkhatib G. A naturally occurring splice variant of CXCL12/stromal cell-derived factor 1 is a potent human immunodeficiency virus type 1 inhibitor with weak chemotaxis and cell survival activities. J Virol. 2007;81:8140–8148. doi: 10.1128/JVI.00268-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rueda P., Balabanian K., Lagane B., Staropoli I., Chow K., Levoye A., Laguri C., Sadir R., Delaunay T., Izquierdo E. The CXCL12γ chemokine displays unprecedented structural and functional properties that make it a paradigm of chemoattractant proteins. PLoS One. 2008;3:e2543. doi: 10.1371/journal.pone.0002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rueda P., Richart A., Récalde A., Gasse P., Vilar J., Guérin C., Lortat-Jacob H., Vieira P., Baleux F., Chretien F. Homeostatic and tissue reparation defaults in mice carrying selective genetic invalidation of CXCL12/proteoglycan interactions. Circulation. 2012;126:1882–1895. doi: 10.1161/CIRCULATIONAHA.112.113290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadir R., Imberty A., Baleux F., Lortat-Jacob H. Heparan sulfate/heparin oligosaccharides protect stromal cell-derived factor-1 (SDF-1)/CXCL12 against proteolysis induced by CD26/dipeptidyl peptidase IV. J Biol Chem. 2004;279:43854–43860. doi: 10.1074/jbc.M405392200. [DOI] [PubMed] [Google Scholar]
- 56.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghazalpour A., Bennett B., Petyuk V.A., Orozco L., Hagopian R., Mungrue I.N., Farber C.R., Sinsheimer J., Kang H.M., Furlotte N. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 59.Albain K.S., Barlow W.E., Shak S., Hortobagyi G.N., Livingston R.B., Yeh I.T., Ravdin P., Bugarini R., Baehner F.L., Davidson N.E. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin M., Prat A., Rodriguez-Lescure A., Caballero R., Ebbert M.T., Munarriz B., Ruiz-Borrego M., Bastien R.R., Crespo C., Davis C. PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Res Treat. 2013;138:457–466. doi: 10.1007/s10549-013-2416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paik S., Tang G., Shak S., Kim C., Baker J., Kim W., Cronin M., Baehner F.L., Watson D., Bryant J. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 62.Prat A., Parker J.S., Fan C., Perou C.M. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135:301–306. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cavnar S.P., Ray P., Moudgil P., Chang S.L., Luker K.E., Linderman J.J., Takayama S., Luker G.D. Microfluidic source-sink model reveals effects of biophysically distinct CXCL12 isoforms in breast cancer chemotaxis. Integr Biol (Camb) 2014;6:564–576. doi: 10.1039/c4ib00015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) CXCL12 and its α, β, and γ isoforms vary significantly with race. (B) Overall CXCL12 and CXCL12-α vary significantly with age. Expression levels are means ± SEM. *P < .05, **P < .001.


