Abstract
Melanoma is the leading cause of death from skin cancer in industrialized countries. Several melanoma-related biomarkers and signaling pathways have been identified; however, their relevance to melanoma development/progression or to clinical outcome remains to be established. Aberrant activation of Wnt/β-catenin pathway is implicated in various cancers including melanoma. We have previously demonstrated Rad6, an ubiquitin-conjugating enzyme, as an important mediator of β-catenin stability in breast cancer cells. Similar to breast cancer, β-catenin-activating mutations are rare in melanomas, and since β-catenin signaling is implicated in melanoma, we examined the relationship between β-catenin levels/activity and expression of β-catenin transcriptional targets Rad6 and microphthalmia-associated transcription factor-M (Mitf-M) in melanoma cell models, and expression of Rad6, β-catenin, and Melan-A in nevi and cutaneous melanoma tissue specimens. Our data show that Rad6 is only weakly expressed in normal human melanocytes but is overexpressed in melanoma lines. Unlike Mitf-M, Rad6 overexpression in melanoma lines is positively associated with high molecular weight β-catenin protein levels and β-catenin transcriptional activity. Double-immunofluorescence staining of Rad6 and Melan-A in melanoma tissue microarray showed that histological diagnosis of melanoma is significantly associated with Rad6/Melan-A dual positivity in the melanoma group compared to the nevi group (P = .0029). In contrast to strong β-catenin expression in normal and tumor areas of superficial spreading malignant melanoma (SSMM), Rad6 expression is undetectable in normal areas and Rad6 expression increases coincide with increased Melan-A in the transformed regions of SSMM. These data suggest a role for Rad6 in melanoma pathogenesis and that Rad6 expression status may serve as an early marker for melanoma development.
Introduction
Melanoma is the leading cause of death from skin cancer in industrialized countries. Numerous potential biomarkers have been identified by high throughput technologies; however, their relevance to melanoma development, progression, or clinical outcome remains to be established [1]. Currently used histological criteria such as primary tumor invasion and lymph node status fail to identify early-stage disease and cases that will eventually progress. Thus, there is a clear clinical need for markers that can aid in the early diagnosis of melanoma, predict melanoma progression, or identify patients with subclinical metastatic disease. While several biomarkers identified previously (e.g., p53, c-myc, CD44, and bcl-2) have shown limited clinical utility, others have potential prognostic significance and include microphthalmia-associated transcription factor (Mitf), p16, β-catenin, tissue plasminogen activator, and ephrin A1 among others [1, and references within].
β-Catenin functions as a structural protein to regulate cell adhesion via interactions with E-cadherin, and is also involved in activation of the canonical Wnt signaling pathway. Aberrant activation of Wnt/β-catenin pathway results from β-catenin accumulation and is implicated in development and progression of various cancers including colon cancer, breast cancer, prostate cancer, esophageal cancer, and melanoma [2]. Levels of β–catenin are kept low through a multiprotein APC/Axin/β-TrCP-regulated 26S proteasomal degradation system [3], [4], [5], [6]. However, overexpression of certain Wnt ligands, loss of Wnt inhibitory factors, or mutations in key components of the multiprotein β-catenin degradation complex contribute to accumulation of β-catenin and activation of the canonical Wnt signaling pathway [2]. Aberrant accumulation of β-catenin in the cytoplasm/nucleus is correlated with poor prognosis for several cancer types [2], [7].
Nearly one-third of human primary melanoma specimens and melanoma cell lines have been reported to display β-catenin accumulation [8], [9], implying a significant functional role for the Wnt/β-catenin pathway in human melanoma. Much of the tumor promoting effects of β-catenin arise from its function as a transcription factor in complex with T-cell factor or LEF-1 (lymphocyte enhancer factor 1) proteins to activate its target genes involved in tumorigenesis such as c-myc [10], Mitf [11], and cyclin D1 [12]. Mitf, a basic/helix-loop–helix/leucine-zipper transcription factor [13] was first identified in mouse, mutation of which results in loss of pigmentation [14]. Mitf exists in multiple isoforms (Mitf-A, Mitf-B, Mitf-C, Mitf-D, Mitf-H and Mitf-M) that are expressed from distinct promoters [15] and yield different expression profiles. The Mitf-M isoform is melanocyte-specific and functions in melanocyte differentiation and survival [16]. Functional studies place Mitf as an essential lineage-specific target of Wnt/β-catenin signaling both in melanocyte development, and melanoma tumorigenesis [11], [17].
Previous studies from our laboratory demonstrated Rad6B, an ubiquitin conjugating enzyme and a key component of the postreplication DNA repair pathway [18], [19], [20], [21], [22], [23], and as an important mediator of β-catenin stability in breast cancer cells [24]. Rad6B enhances β-catenin stability and transcriptional activity by inducing lysine 63-linked polyubiquitin modifications in β-catenin that render β-catenin insensitive to 26S proteasomal degradation [24]. Rad6B is also a transcriptional target of β-catenin [25], thus activating a positive feedback loop between β-catenin-induced Rad6B gene expression and Rad6-induced β-catenin stabilization [24], [25], [26]. Rad6 expression is low in normal breast tissues, and increases in Rad6 expression become detectable in early breast cancer with continued overexpression in invasive breast carcinomas and metastatic breast cancer [27], [28]. As in breast cancer [29], β-catenin activating mutations are rare in melanomas [9], and since β-catenin signaling is implicated in melanoma development and progression, we examined the relationship between Rad6, Mitf, and β-catenin levels/activity in melanoma cell models, and expression of Rad6 in nevi and cutaneous melanomas. Our data show that Rad6 is only weakly expressed in normal human epidermal melanocytes, but is overexpressed in melanoma lines, and unlike Mitf-M, Rad6 expression correlates with elevated levels of high molecular weight β-catenin and β-catenin transcriptional activity. Immunofluorescence analysis of Rad6 and Melan-A in melanoma tissue microarray showed weak or low Rad6 expression in nevi compared to malignant melanomas. Furthermore, while Rad6 expression is negligible in normal areas of skin, increases in Rad6 expression coinciding with increases in Melan-A positive cells are observed in superficial spreading malignant melanoma (SSMM), suggesting that Rad6 expression status could serve as an early marker of neoplastic conversion to melanoma.
Materials and Methods
Cell Culture
Normal human primary epidermal melanocytes (HeMa-LP; (Life Technologies, Carlsbad, California) were cultured in Dermal Cell Basal Medium supplemented with melanocyte growth supplements insulin (5 μg/ml), ascorbic acid (50 μg/ml), L-glutamine (6 mmol/L), epinephrine (1.0 μmol/L), calcium chloride (0.2 mmol/L) and M8 supplement (ATCC, Manassas, VA). Cultures were used within 5 to 10 passages. Human melanoma cell lines A2058 (ATCC), A375 (ATCC), MelJuso (DSMZ, Braunschweig, Germany), M14 (National Cancer Institute, Frederick, Maryland), Malme-3 M and G361 (ATCC) were cultured in RPMI 1640 medium with 10% fetal bovine serum. The human breast cancer cell line MDA-MB-231 cells (ATCC) were maintained in DMEM/F12 medium supplemented with 5% fetal bovine serum [30].
Migration and Invasion Assays
Migration/invasion assays were performed in Boyden chambers (Neuroprobe, Cabin John, MD) containing 8 μm pore size polycarbonate membrane coated with Matrigel basement membrane matrix (BD Biocoat, BD Biosciences, Bedford, MA) as described previously [30].
100 × 103 Cells in serum-free media were seeded in transwell chambers and following incubation overnight at 37°C and 5% CO2, the migrated/invaded cells were fixed and counted after staining with Protocol Hema 3 stain set (Fisher Scientific, Pittsburgh, PA). Stained membranes were scanned and density of spots quantitated with NIH Imaging J Version 1.62. Assays were performed in sextuplets.
Western blot Analysis
Whole cell lysates were prepared as previously described [24]. Nuclear and cytoplasmic subfractions were prepared using a nuclear/cytosol fractionation kit (MBL International, Woburn, MA). Aliquots of whole cell lysates, nuclear, or cytoplasmic fractions containing 25 μg protein were subjected to SDS-PAGE and western blot analysis with antibodies to Rad6, β-catenin (SantaCruz Biotechnology, Inc., Dallas, TX), β-actin (Sigma-Aldrich, St. Louis, MO), MITF (clone C5, Abcam, Cambridge, MA), lamin A/C (SantaCruz), or GAPDH (SantaCruz). In humans, the yeast homologous Rad6 gene is duplicated and the proteins are encoded by two genes HHR6A (or Rad6A) and HHR6B (Rad6B) from chromosomes Xq24-q25 and 5q23-q31, respectively. Rad6A and Rad6B share 95% identical amino acid residues [31], and the Rad6 antibody is unable to distinguish between Rad6A and Rad6B proteins [27]. Therefore, rather than referring to the protein detected by the antibody as Rad6A or Rad6B, we refer to it as Rad6.
Luciferase Reporter Assay
Melanoma and normal melanocytes (300 × 103 cells) were seeded in 35 mm dishes and grown overnight. Cells were transfected with TOP/Flash or FOP/Flash vectors (1.0 μg) and pSV40-Renilla (50 ng) as previously described [24]. 48 h after transfection, cells were lysed with Passive lysis buffer (Promega, Madison, WI), and firefly and Renilla luciferase activities measured using the Dual Luciferase reporter assay kit (Promega). RLU firefly values from FOP/Flash and TOP/Flash transfections were normalized against Renilla luciferase and protein content.
Melanoma Tissue Array and Skin Specimen Analysis
The melanoma tissue microarray ME1004 (contains 24 and 56 cases of nevi and malignant melanoma, respectively; US Biomax Inc., Rockville, MD) was used to assess potential differences in Rad6 expression between benign and malignant melanocytic tumors. We analyzed the first nine sequential primary melanomas and the first nine sequential nevi in the tissue microarray that closely corresponded to the anatomical sites of the melanoma tumors. We selected common cutaneous melanomas, and excluded rare forms of melanoma in the mucosa, genitalia or on volar surfaces. In addition, six cases of superficial spreading cutaneous melanoma were retrieved from the files of the Pinkus Dermatopathology Laboratory, a private dermatopathology laboratory located in Monroe, Michigan. Preserved paraffin-embedded tissue specimens collected for each case were assigned an accession code that excluded patient identifier information. These non-identifiable archived tumor samples were acquired after review and approval by the Wayne State University Human Investigation Committee.
Immunofluorescence Analysis
HeMa-LP and melanoma cells were fixed with buffered formalin, and permeabilized with methanol/acetone prior to incubation with Rad6 and β-catenin antibodies [24]. Expression of Rad6, Melan-A, or β-catenin was analyzed in melanoma tissue microarray ME1004, and in clinical superficial spreading melanoma. Tissues were deparaffinized, rehydrated, and boiled in 1 mmol/L sodium citrate buffer (pH 6.0) by microwave for 10 minutes and blocked with Super Block (Skytek Laboratories, Logan, UT) for 1 hour at room temperature. Following incubation with the primary antibodies, slides were incubated with FITC- or Texas Red-labeled secondary antibodies (Molecular Probes), and nuclei counterstained with 4’,6-diamidino-2-phenylindole (DAPI). Slides were also stained in the absence of primary antibody or with isotype matched nonimmune IgG to assess nonspecific reactions. Images were collected on an Olympus BX60 microscope equipped with a Sony DXC-979 MD 3CCD video camera. Results were evaluated by two investigators in blinded manner. Melanocytic and melanoma cells were identified with Melan-A (MART-1) marker, and Rad6 staining in the tissues was evaluated as a percentage of Melan-A stained cells in each section. The histologic morphology of the tissue cores were confirmed by counterstaining the slides with hematoxylin.
Statistical Analysis
Statistical analyses were performed with Student's t test and P values < 0.05 were considered statistically significant. To compare the number of immunostained cells in nevi and melanoma, a two sample-2-sided t test was utilized. Poisson regression model was employed with SAS version 9 to analyze the association between histological diagnosis (melanoma versus nevi) and occurrence of dual (Rad6 and Melan-A) positive cells among Melan-A positive cell populations. The number of Melan-A positive cells was used as an off-set variable, while the number of Rad6 positive cells among them was used as a response variable, and histological diagnosis as an explanatory variable.
Results
Rad6 and β-Catenin are Co-Upregulated in Melanoma lines
Whereas several reports have linked increased expression of β-catenin and activity with melanoma development and progression [9], [32], [33], [34], [35], [36], others have found correlation between elevated β-catenin levels and improved survival of patients [37], [38]. We have previously reported that Rad6 overexpression induces polyubiquitin modifications of β-catenin that render it insensitive to 26S proteasomal degradation and confer increased transcriptional activity [24].
Western blot analysis of whole cell lysates prepared from normal human primary epidermal melanocytes (HeMa-LP cells) and a panel of primary (MelJuso, A375, G361) and metastatic (A2058, M14) melanoma lines for Rad6 expression showed substantially higher Rad6 levels in A2058, MelJuso, G361 and M14 melanoma lines compared to Malme-3 M and A375 cells, whereas it was negligible in normal HeMa-LP melanocytes (Figure 1A). Simultaneous analysis of β-catenin in the lysates showed 6 to 10-fold higher levels of high molecular weight forms of β-catenin in MelJuso, G361, M14 and A2058 cells compared to HeMa-LP cells (Figure 1A). A375 and Malme-3 M cells expressed ~ 1.5- to 2-fold higher levels of high molecular weight β-catenin compared to HeMa-LP cells (Figure 1A). Levels of the nascent 97 kDa β-catenin protein were similar or only marginally (1.5-fold) higher in melanoma cells compared to normal HeMa-LP melanocytes. These data show a positive association between Rad6 and modified β-catenin protein levels (Figure 1A). We next performed TOP/FOP Flash reporter assays to determine whether the increased levels of high molecular weight or modified forms of β-catenin protein in melanoma cell lines translate into higher β-catenin transcriptional activity. A2058 cells showed > 100-fold higher TOP/Flash activity compared to normal HeMa-LP cells, and MelJuso, M14 and G361 lines showed > 10-fold higher levels of TOP/Flash activity compared to HeMa-LP, Malme-3 and A375 cells (P < .0001; Figure 1B). These data suggest that melanoma lines expressing high molecular weight β-catenin have transcriptionally active β-catenin. Since canonical Wnt signaling is implicated in migration of melanocytes, we assessed the migratory/invasive potential of the melanoma lines. Metastatic MDA-MB-231 human breast cancer cells were used as a positive control. Consistent with the lack of β-catenin transcriptional activity, normal HeMa-LP melanocytes failed to migrate, whereas all melanoma lines migrated/invaded the Matrigel barrier (P < .001; Figure 1C). Interestingly, migratory potentials correlated with Rad6 and modified β-catenin protein levels.
Figure 1.
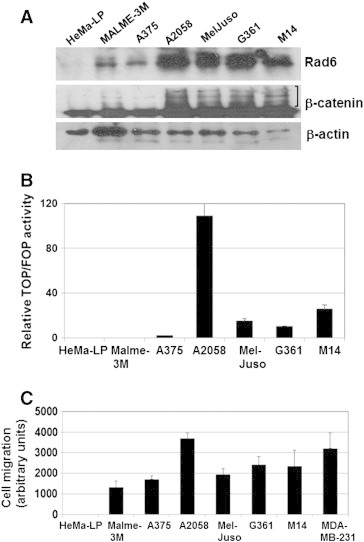
Rad6 and high molecular weight β-catenin forms are upregulated in melanoma lines. (A) Western blot analysis of Rad6 and β-catenin. High molecular weight species of β-catenin are indicated by a bracket. (B) transcriptional activity of β-catenin. Luciferase activity from TOP/Flash and FOP/Flash vectors were normalized against Renilla luciferase. Columns, mean of four independent experiments; bars, S.E. (P < .0001). (C) migration/invasion of the indicated cell models measured by Boyden chamber assay. Metastatic MDA-MB-231 breast cancer cells were included as a positive control. Results are expressed as mean ± S.E (P < .001) of replicate assays conducted in sextuplets.
To further evaluate the functionality of β-catenin transcriptional activity in melanoma lines, we analyzed the subcellular distributions of β-catenin transcriptional targets Rad6 and Mitf in the cytoplasmic and nuclear fractions of normal HeMa-LP and melanoma cells. Rad6 was detected in the cytoplasm of HeMa-LP and melanoma cells, albeit at much higher levels in A2058, Mel-Juso, G361 and Malme-3 M cells. Relative to the nuclear marker lamin A/C loading control, normal HeMa-LP cells had negligible nuclear Rad6, whereas Rad6 was detectable in the nuclei of all melanoma lines (Figure 2A, and C). Similar analysis of Mitf using a commonly used antibody that is not selective to specific isoforms showed strong expression of Mitf-M (55-60 kDa doublet indicated by open and closed circles in Figure 2A) and lower levels of Mitf-A (indicated by the triangle in Figure 2A) isoforms in the cytoplasm of normal HeMa-LP and melanoma lines (Figure 2A). This pattern of Mitf-M and Mitf-A immunoreactive bands detected by the clone C5 Mitf antibody is consistent with those described by Li et al. [39]. HeMa-LP cells showed only the Mitf-M isoform in the nucleus, whereas A2058 cells showed similar expression profiles of Mitf-M and Mitf-A in the cytoplasm and nucleus (Figure 2A and B). Interestingly, nuclear Mitf was negligible or very weakly detectable in A375, MelJuso and M14 cells, while G361 and Malme-3 M cells had detectable but lower levels of nuclear Mitf-M and Mitf-A compared to A2058 cells (Figure 2A). Consistent with elevated β-catenin transcriptional activity in melanoma cell lines, Rad6 was found to accumulate in both the cytoplasm and nucleus of melanoma cells compared to normal melanocytes. However, since strong expression of Mitf-M was detected in all cell lines including normal HeMa-LP cells regardless of TOP/Flash activity, these findings suggest that expression of Mitf-M is not dependent upon β-catenin activity.
Figure 2.
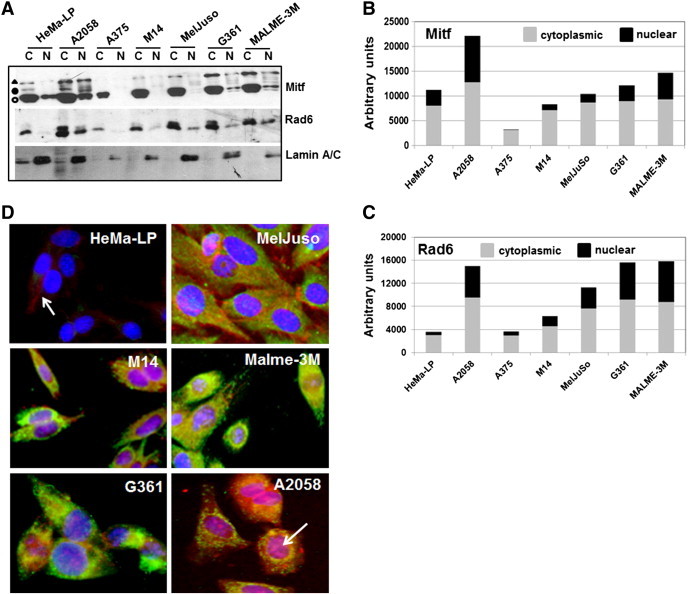
Expression of Rad6, β-catenin and Mitf proteins in melanoma lines. (A) detection of Rad6, Mitf and Lamin A/C proteins in the cytoplasmic and nuclear subfractions by western blotting. Mitf-M isoforms are indicated by open and closed circles, and Mitf-A is indicated by triangle. (B and C) Graphic representation of Mitf and Rad6 proteins levels, respectively, in the cytoplasmic and nuclear fractions quantified by NIH Imaging J Version 1.62 software. (D) Dual immunofluorescence analysis of Rad6 (green) and β-catenin (red). Note the strong expression of Rad6 in melanoma lines as compared to normal HeMa-LP cells. Also note that β-catenin staining is weak and localized to the cell membrane of HeMa-LP cells (arrow). Nuclear localization of β-catenin in A2058 cells is indicated by an arrow.
Dual immunofluorescence staining of Rad6 and β-catenin were performed to verify their presence and localization in normal HeMa-LP and melanoma cells. HeMa-LP cells showed negligible Rad6 immunoreactivity, and β-catenin staining was localized to the cell membranes (Figure 2D). In contrast, Rad6 and β-catenin staining was intense in the melanoma lines. Consistent with TOP/Flash reporter activity data, β-catenin was detected in the nuclei of all melanoma lines with strongest nuclear β-catenin immunoreactivity in M14 and A2058 melanoma cells. Intense Rad6 staining was detected in the cytoplasm that colocalized with β-catenin in the melanoma lines (Figure 2D).
Relevance of Rad6 in Clinical Melanoma
To analyze the potential role of Rad6 in melanoma development, we evaluated expression of Rad6 and the melanocyte differentiation antigen Melan-A in a melanoma tissue microarray by dual immunofluorescence staining. The numbers of Rad6 positive and Melan-A positive cells were scored, and Poisson regression analysis was applied to compare the percentage of cells costaining for Rad6 and Melan-A in nevi vs. primary cutaneous melanomas. The percent of Rad6 and Melan-A dual positive cells ranged from 0% to 43.5% in the nevi group, and from 51.4% to 98.2% in the melanoma group. Limiting Rad6 expression analysis to Melan-A positive cells could lead to underestimation of the number of Rad6 positive cells in the tissue specimens as Melan-A is not uniformly expressed in all nevi and melanomas. However, comparison of Melan- A expression in nevi and melanoma samples have been shown to have similar sensitivity and specificity values (75% to 92%, nevi vs. 95% to 100%, melanoma) [40]. Our data demonstrated that although the number of cells positive for Melan-A was not significantly different between the nevi and primary cutaneous melanoma groups (P = .5696), histological diagnosis of melanoma was significantly associated with the occurrence of Rad6/Melan-A dual positivity (P = .0029) with the odd ratio of 1.98 (95% confidence interval 1.6-2.46) compared to the nevi group. Also, compared to the nevi where only a few cell populations, if any, showed Rad6 staining (Figure 3A), Rad6 was abundantly expressed in malignant melanomas (Figure 3B). Similar analysis of Rad6 and β-catenin in nevi and malignant melanomas by dual immunofluorescence staining showed Rad6 and β-catenin costaining predominantly in melanoma specimens. In malignant melanomas, β-catenin staining was intense and widespread and colocalized with Rad6 (Figure 4). β-catenin was localized on the cell membrane and cytoplasm of nevi and melanomas, but was not found to localize in the nucleus. In rare cases of nevi that showed Rad6 expression, Rad6 was present in the cytoplasm, whereas in Rad6-positive melanomas, Rad6 was localized both in the cytoplasm and nucleus (Figure 4). These data suggest that up-regulation of Rad6 may play a role in the conversion of nevus to cutaneous melanoma. The positive relationship between Rad6 expression and melanoma development was further verified in superficial spreading malignant melanoma (SSMM) biopsies, the most common form of cutaneous melanoma accounting for ~ 70% of all diagnosed melanomas [41].
Figure 3.
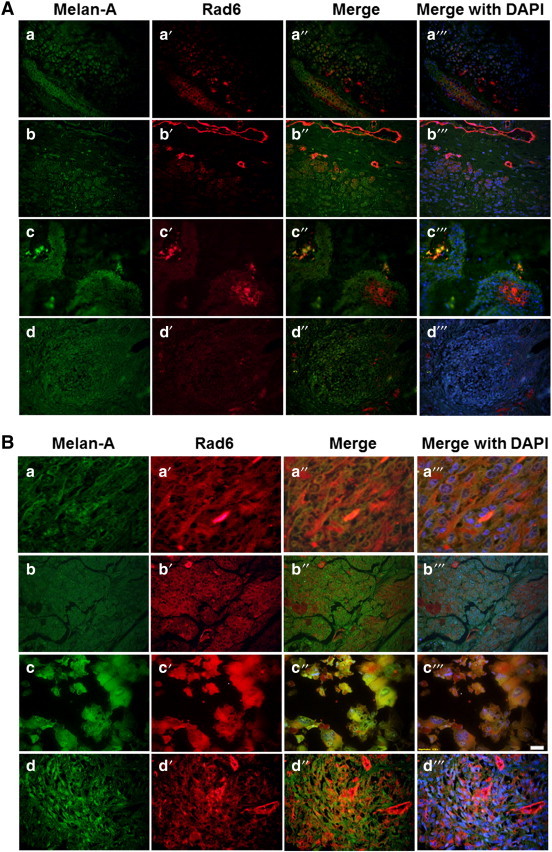
Representative pictures of Rad6 and Melan-A staining detected by double immunofluorescence staining of nevi (A) and primary melanomas (B) in melanoma tissue microarray. Original magnification, × 400.
Figure 4.
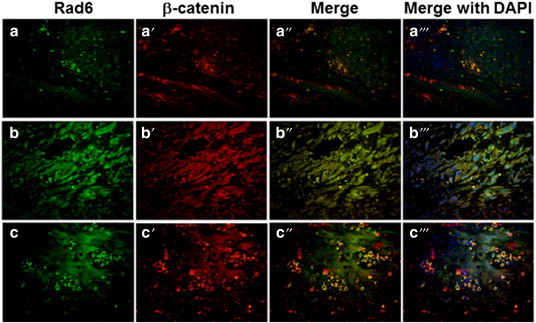
Representative pictures of Rad6 and β-catenin staining detected by double immunofluorescence staining of nevi and primary melanomas in the melanoma tissue microarray. Panels a-a”’, staining in nevus, and panels b-b”’ and c-c”’, staining in malignant melanomas. Original magnification, × 400.
Expression and distribution of Rad6 and Melan-A, or Rad6 and β-catenin in the normal adjacent and transformed areas of the same specimen of SSMM were evaluated by double immunofluorescence staining. Consistent with the maintenance of a stable ratio between melanocytes and keratinocytes in the normal skin, few melanocytes, as identified by Melan-A staining, were found in normal areas of the skin. Rad6 expression was undetectable in these normal regions (Figure 5A, panels a-a” and b-b”). Rad6 expression became noticeable in the neighboring areas of skin that showed increased numbers of (Melan-A positive) melanocytes (Figure 5A, panels c-c” and d-d”), and Rad6 was overexpressed and colocalized with Melan-A stained cells in tumor regions (Figure 5A, panels e-e”, f-f”). Similar analysis of Rad6 and β-catenin showed an inverse relationship between Rad6 and β-catenin in the normal areas of SSMM samples with strong β-catenin staining and negligible Rad6 (Figure 5B, panels a-a”), whereas both Rad6 and β-catenin staining were detected in the adjacent tumor areas (Figure 5B, panels b-b”). These data suggest that unlike β-catenin, Rad6 may contribute to the development of cutaneous melanoma.
Figure 5.
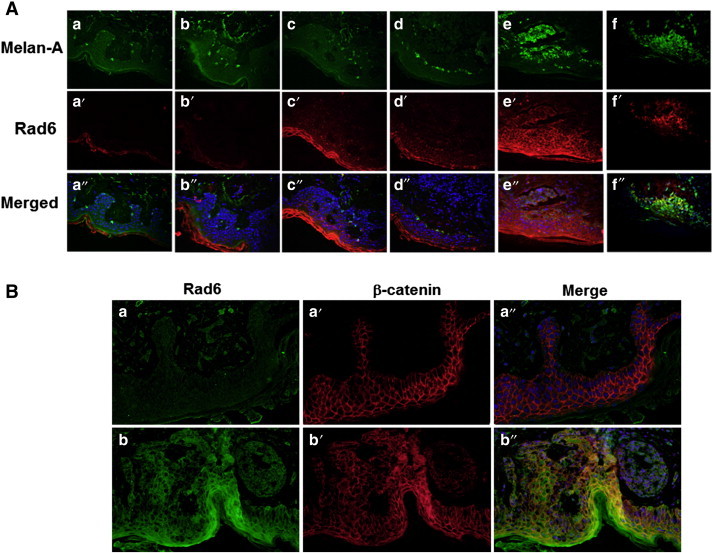
Representative pictures of Rad6 and Melan-A (A) and Rad6 and β-catenin (B) staining detected by double immunofluorescence staining of a representative superficial spreading malignant melanoma biopsy. (A) Note the absence of Rad6 in the normal areas (a-a”), and its appearance that coincides with increases in Melan-A positive cells (b-b” to f-f”). Also note the presence of Rad6/Melan-A dual positive cells in tumor areas (e-e” and f-f”). (B) Note the presence of β-catenin and absence of Rad6 in normal areas (a-a”) and coexpression of Rad6 and β-catenin in the tumor regions of SSMM (b-b”). Original magnification, × 400.
Discussion
A major finding of this study is the discovery of Rad6 as an early marker for cutaneous melanoma development. We show that Rad6, an ubiquitin conjugating enzyme, and activator of canonical Wnt/β-catenin signaling via β-catenin stabilizing modifications, plays an important role in melanoma development. Analysis of clinical melanoma and nevi cores in melanoma tissue microarray showed up-regulation of Rad6 expression in primary melanoma cases compared to nevi. The present data are supported by a detailed immunohistochemical study of Rad6 and β-catenin in archived nevi, primary, and metastatic melanoma samples from 90 patients that showed Rad6 expression is associated with primary and metastatic melanoma but not nevi, and that Rad6 that is overexpressed in > 95% of metastatic melanomas co-occurs with β-catenin in about half of metastatic melanomas [42].
In support of the clinical data, western blot and immunofluorescence analysis showed an inverse relationship between Rad6 and β-catenin in normal melanocytes, whereas primary and metastatic melanoma cell lines showed a direct correlation between levels of Rad6, high molecular weight β-catenin, β-catenin-mediated TOP/Flash reporter activity, and migratory potential. These data are consistent with the positive feedback loop between Rad6B gene expression and β-catenin stabilization/activation reported in breast cancer, wherein Rad6B, a transcriptional target of β-catenin, is induced by T-Cell factor/β-catenin [25], and Rad6B in turn stabilizes β-catenin by inducing K63-linked polyubiquitin modifications (high molecular weight β-catenin forms) that bestow β-catenin with elevated transcriptional activity and resistance to 26S proteasomal degradation [24]. While the Rad6/β-catenin connection is apparent in malignant melanoma cell lines, expression of Mitf, also a transcriptional target of β-catenin [11], does not show a similar association with the Wnt/β-catenin pathway as the melanocyte specific Mitf isoform, Mitf-M, is abundantly expressed in normal HeMa-LP melanocytes despite the absence of β-catenin mediated TOP/Flash activity. Mitf-M is readily detectable in the nuclei of normal HeMa-LP cells and A2058 metastatic melanoma cells, but is decreased or lost from the nuclei of MelJuso, M14, G361 and Malme-3 M cells. These data suggest that Mitf-M is necessary for the regulation of genes required for the maintenance and differentiation of melanocytes, as absence of Mitf-M in the nucleus is seen only in melanoma lines. The Mitf gene is amplified in some melanomas, and it has been suggested that Mitf can function as a melanoma oncogene [43]. Mitf is down-regulated in B-raf transformed murine melanocytes and B-raf overexpressing human melanocytes, and exogenous reexpression of Mitf inhibited the proliferation of these cells [44]. These data suggest a tumor-suppressive or differentiation-promoting role for Mitf in melanocytes. This role is consistent with the function of Mitf in regulating cell cycle arrest via activation of p21/WAF1 and p16Ink4a [45], [46]. Since the melanoma line A2058 shows abundant expression of Mitf-M and other Mitf isoforms in the nucleus, this suggests that Mitf can support both oncogenic and tumor suppressor functions. Cumulatively, these data suggest that Rad6 may be a more reliable marker than Mitf for melanoma development.
Double labeling analysis of Rad6 and Melan-A, and Rad6 and β-catenin in normal adjacent and transformed areas of the same SSMM specimens shed further light on the significance of Rad6 as a potential early marker for neoplastic conversion to melanoma. When melanocyte homeostasis is tightly regulated by keratinocytes, a process occurring in normal skin, Rad6 is undetectable. However, when homeostasis regulation is lost, as evidenced by increases in the number of Melan-A positive cells, Rad6 expression becomes noticeable. However, it is interesting to note that Rad6 expression is not initially localized in melanocytes, but rather expressed in the neighboring keratinocytes, prompting us to speculate that up-regulation of Rad6 in neighboring cells likely plays a role in the deregulation of melanocyte homeostasis and contributes to the risk of melanoma development. This supposition is supported not only by concurrent increases in the number of Melan-A positive cells, but also by increases in Melan-A/Rad6 double positive cells in tumor regions. Since the first detectable increase in Rad6 expression occurs in the neighboring keratinocytes that strongly express β-catenin prompts us to speculate that Rad6 gene expression may be induced by β-catenin, it’s transcriptional activator [25]. Double immunofluorescence staining of Rad6 and β-catenin in SSMM samples further confirmed the inverse and direct relationships between Rad6 and β-catenin expressions in the normal adjacent and tumor regions, respectively, of SSMM, suggesting that Rad6 could be a good marker for distinguishing benign nevi from melanoma.
Constantly elevated Rad6 expression in primary and metastatic melanomas suggests that Rad6 may play an active role during all phases of melanoma pathogenesis: initiation, maintenance and progression to metastatic disease. It remains to be determined, however, whether the melanoma transformation-inducing properties of Rad6 are solely transmitted through β-catenin or through the function of Rad6 as a postreplication DNA repair protein. The postreplication repair pathway enables completion of DNA replication blocked by damaging DNA lesions via error-free and error-prone bypass mechanisms [18], and the ubiquitin conjugating activity of Rad6 is critical to this process [47]. Since cells are challenged by environmental or endogenous processes that induce DNA damage, we posit that the activation of Rad6 postreplication repair pathway in the early phase of melanoma development may be necessary for ensuring completion of stalled DNA replication and hence cell survival. Because postreplication repair is often error prone or mutagenic, it is tempting to speculate that Rad6 may participate in melanocyte transformation by directly contributing to genomic alterations underlying melanoma pathogenesis.
In summary, our data suggest that Rad6 may serve as an early marker for melanoma development. The first detectable increase in Rad6 expression is correlated with melanocyte transformation, and is further augmented in malignant melanoma, there by implicating Rad6 as a novel anti-melanoma therapeutic target.
Acknowledgments
The authors thank Dr. Michael Tainsky for programmatic support of this project. This work was supported by U.S. Army Medical Research Acquisition W81XWH07-1-0562, NIH R21CA178117-01 (MPS), and startup funds from Wayne State University (KR).
Contributor Information
Karli Rosner, Email: krosner@med.wayne.edu.
Malathy P.V. Shekhar, Email: shekharm@karmanos.org.
References
- 1.Larson AR, Konat E, Alani RM. Melanoma biomarkers: current status and vision for the future. Nat Clin Pract Oncol. 2009;6:105–117. doi: 10.1038/ncponc1296. [DOI] [PubMed] [Google Scholar]
- 2.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 3.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta- catenin and GSK3 beta. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 4.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 5.Seidensticker MJ, Behrens J. Biochemical interactions in the wnt pathway. Biochim Biophys Acta. 2000;1495:168–182. doi: 10.1016/s0167-4889(99)00158-5. [DOI] [PubMed] [Google Scholar]
- 6.Kitagawa M, Hatakeyama S, Shirane M. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 9.Rimm DL, Caca K, Hu G, Harrison FB, Fearon E. Frequent nuclear/cytoplasmic localization of beta-catenin without exon 3 mutations in malignant melanoma. Am J Pathol. 1999;154:325–329. doi: 10.1016/s0002-9440(10)65278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He T-C, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 11.Widlund HR, Horstmann MA, Price ER, Cui J, Lessnick SL, Wu M, He X, Fisher DE. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia- associated transcription factor. J Cell Biol. 2002;158:1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, Fisher DE. Microphthalmia transcription factor: A sensitive and specific melanocyte marker for melanoma diagnosis. Am J Pathol. 1999;155:731–738. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 15.Udono T, Yasumoto K, Takeda K, Amae S, Watanabe K, Saito H, Fuse N, Tachibana M, Takahashi K, Tamai M. Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochim Biophys Acta. 2000;1491:205–219. doi: 10.1016/s0167-4781(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 16.Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, Aaronson SA, Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14:50–54. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- 17.Dorsky RI, Raible DW, Moon RT. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 2000;14:158–162. [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence CW. The Rad6 DNA repair pathway in Saccharomyces cerevisiae: What does it do, and how does it do it? BioEssays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds P, Weber S, Prakash L. Rad6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985;82:168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair Rad6 encodes a ubiquitin conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 21.Haynes RH, Kunz BA. Life cycle and inheritance. In: Strathern J, Jones E, Broach J, editors. The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1981. pp. 371–414. [Google Scholar]
- 22.Lawrence CW. Mutagenesis in Saccharomyces cerevisiae. Adv Genet. 1982;21:173–254. doi: 10.1016/s0065-2660(08)60299-0. [DOI] [PubMed] [Google Scholar]
- 23.Prakash S, Sung P, Prakash L. Structure and Function of RAD3, RAD6, and other DNA repair genes of Saccharomyces cerevisiae. In: Strauss PR, Wilson SH, editors. The eukaryotic nucleus. vol. 1. Telford Press; Caldwell, New Jersey: 1990. pp. 275–292. [Google Scholar]
- 24.Shekhar MP, Gerard B, Pauley RJ, Williams BO, Tait L. Rad6B is a positive regulator of beta-catenin stabilization. Cancer Res. 2008;68:1741–1750. doi: 10.1158/0008-5472.CAN-07-2111. [DOI] [PubMed] [Google Scholar]
- 25.Shekhar MP, Tait L, Gerard B. Essential role of T-cell factor/beta-catenin in regulation of Rad6B: a potential mechanism for Rad6B overexpression in breast cancer cells. Mol Cancer Res. 2006;4:729–745. doi: 10.1158/1541-7786.MCR-06-0136. [DOI] [PubMed] [Google Scholar]
- 26.Gerard B, Tait L, Nangia-Makker P, Shekhar MP. Rad6B acts downstream of Wnt signaling to stabilize β-catenin: Implications for a novel Wnt/β-catenin target. J Mol Signal. 2011;6:6–19. doi: 10.1186/1750-2187-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shekhar MP, Lyakhovich A, Visscher DW, Heng H, Kondrat N. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 2002;62:2115–2124. [PubMed] [Google Scholar]
- 28.Gerard B, Sanders MA, Visscher DW, Tait L, Shekhar MP. Lysine 394 is a novel Rad6B-induced ubiquitination site on beta-catenin. Biochim Biophys Acta. 2012;1823:1686–1696. doi: 10.1016/j.bbamcr.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9:119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- 30.Sanders MA, Brahemi G, Nangia-Makker P, Balan V, Morelli M, Kothayer H, Westwell AD, Shekhar MP. Novel inhibitors of Rad6 ubiquitin conjugating enzyme: design, synthesis, identification, and functional characterization. Mol Cancer Ther. 2013;12:373–383. doi: 10.1158/1535-7163.MCT-12-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koken MH, Smith EM, Jaspers-Dekker I, Oostra BA, Hagemeijer A, Bootsma D, Hoeijmakers JH. Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24–q25 and 5q23–q31. Genomics. 1992;12:447–453. doi: 10.1016/0888-7543(92)90433-s. [DOI] [PubMed] [Google Scholar]
- 32.Omholt K, Platz A, Ringborg U, Hansson J. Cytoplasmic and nuclear accumulation of beta-catenin is rarely caused by CTNNB1 exon 3 mutations in cutaneous malignant melanoma. Int J Cancer. 2001;92:839–842. doi: 10.1002/ijc.1270. [DOI] [PubMed] [Google Scholar]
- 33.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 34.Larue L, Delmas V. The WNT/Beta-catenin pathway in melanoma. Front Biosci. 2006;11:733–742. doi: 10.2741/1831. [DOI] [PubMed] [Google Scholar]
- 35.Kielhorn E, Provost E, Olsen D, D'Aquila TG, Smith BL, Camp RL, Rimm DL. Tissue microarray-based analysis shows phospho-beta-catenin expression in malignant melanoma is associated with poor outcome. Int J Cancer. 2003;103:652–656. doi: 10.1002/ijc.10893. [DOI] [PubMed] [Google Scholar]
- 36.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 37.Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL, Moon RT. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y, Nishikawa M, Suehara T, Takiguchi N, Takakura Y. Gene silencing of beta-catenin in melanoma cells retards their growth but promotes the formation of pulmonary metastasis in mice. Int J Cancer. 2008;123:2315–2320. doi: 10.1002/ijc.23812. [DOI] [PubMed] [Google Scholar]
- 39.Li KKC, Goodall J, Goding CR, Liao S-K, Wang C-H, Lin Y-C, Hiraga H, Nojima T, Nagashima K, Schaefer K-L. The melanocyte inducing factor MITF is stably expressed in cell lines from human clear cell sarcoma. Br J Cancer. 2003;89:1072–1078. doi: 10.1038/sj.bjc.6601212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohsie SJ, Sarantopoulos GP, Cochran AJ, Binder SW. Immunohistochemical characteristics of melanoma. J Cutan Pathol. 2008;35:433–444. doi: 10.1111/j.1600-0560.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 41.Forman SB, Ferringer TC, Peckham SJ, Dalton SR, Sasaki GT, Libow LF, Elston DM. Is superficial spreading melanoma still the most common form of malignant melanoma? J Am Acad Dermatol. 2008;58:1013–1020. doi: 10.1016/j.jaad.2007.10.650. [DOI] [PubMed] [Google Scholar]
- 42.Rosner K, Mehregan DR, Kirou E, Abrams J, Kim S, Campbell M, Frieder J, Lawrence K, Haynes B, Shekhar MP. Melanoma development and progression are associated with Rad6 upregulation and β-catenin relocation to the cell membrane. J Skin Cancer. 2014 doi: 10.1155/2014/439205. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF- stimulated melanocyte and melanoma cell proliferation. J Cell Biol. 2005;170:703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 46.Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung P, Prakash S, Prakash L. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc Natl Acad Sci U S A. 1990;87:2695–2699. doi: 10.1073/pnas.87.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]


