Abstract
Whether Cell block (CB) samples are applicable to detect anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1) and ret proto-oncogene (RET) fusion genes in lung adenocarcinoma is still unknown. In this study, 108 cytological samples that contained lung adenocarcinoma cells were collected, and made into CB. The CB samples all contained at least 30% lung adenocarcinoma cells. In these patients, 48 harbored EGFR mutation. Among the 50 EGFR wild type patients who detected fusion genes, 14 carried EML4-ALK fusion (28%), 2 had TPM3-ROS1 fusion (4%), and 3 harbored KIF5B-RET fusion (6%). No double fusions were found in one sample. Patients with fusion genes were younger than those without fusion genes (p = 0.032), but no significant difference was found in sex and smoking status (p > 0.05). In the thirty-five patients who received first-line chemotherapy, patients with fusion gene positive had disease control rate (DCR) (72.7% VS 50%, p > 0.05) and objective response rate (ORR) (9.1% VS 4.2%, p > 0.05) compared with those having fusion gene negative. The median progression free survival (mPFS) were 4.0 and 2.7 months in patients harbored fusion mutations and wild type, respectively (p > 0.05). We conclude that CB samples could be used to detect ALK, ROS1 and RET fusions in NSCLC. The frequency distribution of three fusion genes is higher in lung adenocarcinoma with wild-type EGFR, compared with unselected NSCLC patient population. Patients with fusion genes positive are younger than those with fusion gene negative, but they had no significantly different PFS in first-line chemotherapy.
Introduction
Lung cancer is the leading cause of cancer death worldwide [1], non–small-cell lung cancer (NSCLC) accounts for about 85%. Along with the discovery of somatic epidermal growth factor receptor (EGFR) mutations, NSCLC patients with activating EGFR mutations benefit from EGFR-TKI therapy [2], [3], [4]. Since then, targeted therapies according to gene mutations lead a new trend in tumor therapy. Subsequently, more driver mutations are found in NSCLC, including many fusion gene mutations, such as anaplastic lymphoma kinase (ALK), ROS1 and RET.
Echinoderm microtubule associated protein like 4 (EML4)-ALK is the first targetable fusion gene to be identified in NSCLC [5]. The fusion is found about 2-7% in lung cancer [5], [6], [7], [8]. Other genes which can fuse with ALK had also been found, including KIF5B and TFG [7], [9], [10]. In NSCLC never/light smokers without EGFR mutation the mutation frequency of EML4-ALK was 33% [11], and in lung adenocarcinoma patients with malignant pleural effusions having wild-type EGFR and measurable target lesions it was reported as 34% [12]. Many drugs that target EML4-ALK had been discovered, such as crizotinib, which was effective in ALK-rearranged NSCLC [13] and approved by US food and drug administration (FDA) in treating ALK-positive NSCLC. ROS1 was also reported to be a target of crizotinib [14], [15], but its frequency only ranges from 0.7-1.7% [13], [15], [16], [17] in lung adenocarcinoma. RET, as another fusion gene, is rarely detected in NSCLC, which is reported from 1-2% [18], [19], [20]. Several drugs (sunitinib, sorafenib, and vandetanib) that target RET fusions are effective [18], [21].
Molecular typing is essential for NSCLC patients to select the optimal treatment. Although tumor tissue is the most valuable specimen for gene mutation detection, it is not always available especially for advanced NSCLC patients that are old aged and have inoperable tumor. In advanced lung cancer patients, 50% has malignant pleural effusions and 80% of the effusions can find tumor cells in microscope [22], [23]. Therefore, this kind of cytological samples could be a surrogate to tumor tissues. In this study, CB samples were done and used to detect ALK, ROS1 and RET fusion genes, the relationship between clinicopathologic characteristics and the fusion genes were analyzed.
Method
Patients and CB Samples
108 patients with pleural, ascitic or pericardial effusions conducted EGFR mutation detection. They were all lung adenocarcinoa patients, in stage IV and had PS score 0-1. All patients had signed an informed consent for future molecular analyses. Patient follow-up was ended in 20th, December, 2013. The effusions (50 to 1200 ml) containing lung adenocarcinoma cells were collected from October 2012 to August 2013. Simply, the effusion was centrifuged at 2500 rpm for 3 minutes, the supernatant was removed and the precipitant was mixed with erythrocyte lysate for 10 minutes. After centrifuging at 2500 rpm for 3 minutes the precipitant was resuspended in normal saline solution and then was centrifuged again. The precipitant was packaged by mixing with warm agarose gel and had routinely dehydration before packaging in paraffin wax. Sections of 5 μm thick from the samples were used for hematoxylin and eosin staining and assessed by pathologists.
EGFR Mutations Detection
DNA was extracted from the 108 effusion samples or CB samples using tissue DNA kit and FFPE DNA kit (QIAGEN, Hilden, Germany) respectively. EGFR was examined using amplification refractory mutation system (ARMS) PCR method. The ARMS PCR procedure was as follows: 5 μl of 1 (effusion samples) or 2 ng/μl (CB samples) template DNA solutions was added to each reaction buffer and then [1] initial denaturation at 95°C for 5 min, [2] 15 cycles of 95°C 25 s, 64°C 20s, and 72°C 20s, [3] 31 cycles of 93°C 25 s, 60°C 35 s, and 72°C 20s was conducted before analyzing the results.
ALK, ROS1 and RET Fusion Gene Detection by ARMS PCR
CB samples were scraped into 1.5 mL tubes, and then total RNA was extracted using RNeasy FFPE kit (QIAGEN, Hilden, Germany). RNA was reversed to cDNA, added to reaction buffer and then ALK, ROS1 and RET fusion genes were detected using EML4-ALK, ROS1 and RET Fusion Gene Detection Kit (Amoydx, Xiamen, China) respectively by ARMS method as mentioned above. All the fusion positive samples were confirmed by DNA sequencing.
Statistical Analysis
The ORR, DCR, the relationship between fusion gene mutations and other clinical characteristics were evaluated by Pearson Chi-square test or Fisher’s exact test. Median PFS was analyzed by Kaplan–Meier method and compared between different groups using the log-rank test. The 2-sided significance level was set at P < 0.05. All data were analyzed using the Statistical Package for the Social Sciences version 17.0 software package (SPSS Inc., Chicago, Ill).
Results
The Quality of CB Samples
The CB samples were preserved between days to 10 months before cut into 5 μm thick sections, and then routinely stained by hematoxylin and eosin. Tumor cell content and pathological type were assessed by pathologists (Figure 1). All the samples were confirmed to be lung adenocarcinoma, and the tumor cell content of each specimen was more than 30%.
Figure 1.
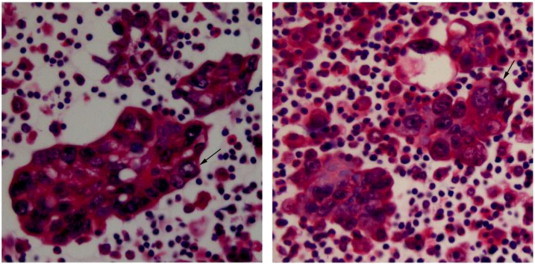
Cell block samples contain lung adenocarcinoma cells. CB samples of 5 μm thick sections from two patients were stained by hematoxylin and eosin. The lung adenocarcinoma cells in the pictures were marked by the black arrows.
ALK, ROS1 and RET Fusion Types in the Fusion Positive Patients
In the 108 patients, 48 (44%) had EGFR mutation. The characteristics of the 108 lung adenocarcinoma patients were listed in Table 1. They had no significant difference in age, sex and smoking status between patients with or without EGFR mutation. In the EGFR wild type patients 50 conducted fusion gene detection. Of these, 14 had ALK fusion (28%), 2 had ROS1 fusion (4%), and 3 had RET fusion (6%). PCR positive samples were all verified by DNA sequencing. The ALK fusions were: eight E(EML4) exon 13 with A(ALK) exon 20 fusions, four E20 with A20 fusions, one E18 with A20 fusion, and one E6 with A20 fusion. The ROS1 fusions were ROS1 exon 34 with TPM3 exon 8. The three RET fusions were all RET exon 12 with KIF5B exon n15.
Table 1.
EGFR Mutation in 108 Lung Adenocarcinoma Patients.
| Characteristic | Patients Without EGFR Mutation | Patients with EGFR Mutation | P Value |
|---|---|---|---|
| Age, years | |||
| < 65 | 41 | 38 | 0.080 |
| ≥ 65 | 19 | 10 | |
| Sex | |||
| Male | 35 | 29 | 0.153 |
| Female | 25 | 19 | |
| Smoking status | |||
| Never/light smokers | 36 | 34 | 0.082 |
| Smokers | 24 | 14 |
Light smokers: smoking less than 10 pack-years.
Clinicopathologic Characteristics of the Gene Fusion Positive Patients
The patients who harbored fusion gene mutation were listed in Table 2. In the EML4-ALK patients, 11 were under 60 and 8 were none or light smokers. The TPM3-ROS1 and two KIF5B-RET patients were under 60 years old and none-smokers, and one KIF5B-RET patient was a heavy smoker (30 pack-years) and under 60. There was no significant difference between the patients with and without any one of the fusion genes in sex, and smoking status (p > 0.05), but the patients with fusion gene mutations were younger than those without mutations (median age, 51 vs 61, p = 0.032).
Table 2.
Clinicopathologic Characteristics of Lung Adenocarcinoma Patients (n = 50).
| Characteristic | ALK Positive (n = 14) | ROS1 Positive (n = 2) | RET Positive (n = 3) | Either Positive (n = 19) | Negative Patients (n = 31) | P Value of All |
|---|---|---|---|---|---|---|
| Age, years | ||||||
| < 65 | 11 | 2 | 3 | 16 | 17 | 0.032 |
| ≥ 65 | 3 | 0 | 0 | 3 | 14 | |
| Sex | ||||||
| Male | 9 | 1 | 1 | 11 | 18 | 0.610 |
| Female | 5 | 1 | 2 | 8 | 13 | |
| Smoking status | ||||||
| Never/light smoker | 8 | 2 | 2 | 12 | 16 | 0.308 |
| Smoker | 6 | 0 | 1 | 7 | 15 |
Clinical Outcome of Patients With and Without the Fusion Genes
Thirty-five of the 50 patients received first-line chemotherapy in this hospital, including 29 carboplatin or cisplatin contained therapies, 2 single drug therapies and 4 TKI targeting EGFR therapies. In these patients, twenty-four did not carry any mutation of three fusion genes, eight were ALK fusion positive and three were RET fusion positive (Table 3). In the last follow-up, three patients did not get disease progression. ORR was 4.2% and 9.1% in patients without and with fusion gene mutation, respectively (p > 0.05); DCR was 50% and 72.7%, respectively (p > 0.05). The median PFS of the EML4-ALK positive patients was 4.2 (95% confidence interals, 1.890-6.510) months vs 2.8 (95% CI, 1.658-3.942) months (p = 0.706) in the EML4-ALK negative patients and in either one of three genes positive patients it was 4.0 (95% CI, 2.605-5.395) months vs 2.7 (95% CI, 1.551-3.849) months (p = 0.371) in the none-positive patients (Figure 2). Although there was no significant difference between the two cohorts, the results showed a trend that patients with fusion genes had a better chemotherapy response than those without any one of fusion genes in chemotherapy.
Table 3.
Response to First-line Chemotherapy in Lung Adenocarcinoma Patients (n = 35).
| Response | ALK Positive Patients (n = 8) | Ros1 Positive Patients (n = 0) | RET Positive Patients (n = 3) | Gene Wild Type Patients (n = 24) |
|---|---|---|---|---|
| PR | 0 | 0 | 1 | 1 |
| SD | 6 | 0 | 1 | 11 |
| PD | 2 | 0 | 1 | 12 |
Figure 2.
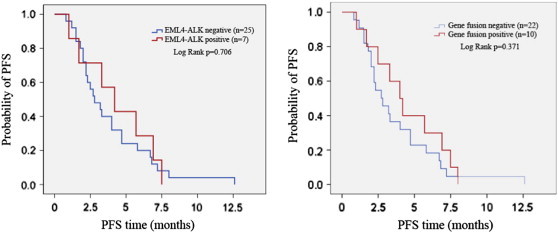
Comparison of PFS between patients with or without fusion genes. The change of PFS between patients with or without EML4-ALK mutation (left) and the change of PFS between patients with or without any fusion gene mutation (right) were shown in the figure.
Discussion
Cell block (CB) is a method to concentrate and preserve cells in fluid samples for long use. Compared with effusion smears, CB contains more cells to be identified and helps pathologists in decision making. It has been used routinely in pathological classification and also in gene detection. In certain cases it has an advantage to other conventional pathological methods [24]. In advanced-stage patients who cannot have their tissues dissected, CB samples could be an alternative selection. In this study, the authors detected ALK, ROS1 and RET fusion genes in EGFR wild type lung adenocarcinoma patients using cell block samples and analyzed the prevalence of the fusion genes and the relationship between clinicopathologic characteristics and fusion gene mutations.
Fluorescence in situ hybridization (FISH) is the primary method to detect ALK, ROS1 and RET fusions in NSCLC [14], [25], [26]. However, it is not wildly used in China due to its high spent, time consuming and also the interpretation of results. Immunohistochemistry (IHC) is another method to detect ALK fusion, but there is no standard procedure for all the labs and the same result could be explained differently by different pathologists. Soda showed us in his study that different technologies should apply to different samples, and multiplex RT-PCR was applicable for the fluid samples [27]. Here, we use a reverse-transcript polymerase chain reaction (RT-PCR) method-ARMS-to detect ALK, ROS1 and RET fusions in 50 CB samples. Wu [12] used RT-PCR and FISH to detect ALK fusion and they found a concordance rate of 85%, but they did not check cell block samples that were ALK fusion positive using FISH. Soda [27] reported in their research that PCR-based detection of EML4-ALK should have a higher analytic sensitivity compared with IHC or FISH. In this study, although we did not use FISH to conform the PCR results, we used DNA sequencing as a substitute. All the positive results using the PCR method were all conformed by DNA sequencing. We believe that the cell block samples could detect the three fusion genes using both RT-PCR and DNA sequencing.
We tested the quality of cell block samples from the points of malignant cell ratio and PCR controls, finding that they were qualified to do the gene detection. The fusion positive results were all validated by DNA sequencing and the specific variants were also given. The results indicate that cell block samples preserved at least 10 months could be used to detect fusion genes. EML4-ALK fusion gene detection using plural effusions had been reported by Wu et al [12]. They used RT-PCR and direct sequencing methods and found a 34% presence in EGFR wild type lung adenocarcinoma patients. Shaw et al. [19] got a 33% prevalence in never/light smokers in EGFR wild type lung adenocarcinoma patients using FISH method. Although Cai et.al [28] used 19 cell block samples and 35 fine-needle aspirates to detect EGFR, KRAS and ALK genes in primary and metastatic lung adenocarcinomas, they did not show whether the CB samples be used for ALK detection or not. As far as we know, there is no study that reports the three fusion genes detected specially using CB samples. In the 50 EGFR wild type lung adenocarcinoma patients, EML4-ALK had a prevalence of 28%, which was a little lower than the former data [11], [12]. Nonetheless, considering the small number of cases in our study, this slight difference should be reasonable. We had also examined ROS1 and RET fusion genes in the 50 samples. Compared with their frequency in NSCLC or lung adenocarcinoma (1-2%), they were 2-3 times higher in EGFR wild type lung adenocarcinoma. Therefore, gene screening on the basis of other genes may improve its detection rate.
No significant difference was found between patients with and without genes mutation in sex and smoking status (p > 0.05), but they as a whole had a significant difference in age (51 versus 61, p = 0.032). The patients with fusion genes are younger than those without mutations, which is in concordance with other reports [11], [18], [29], [30]. In the absence of EML4-ALK targeted therapy, patients have a similar prognosis whether ALK was fusion positive or not [7], but other data indicate that the prognosis is controversial [12], [31]. Here, we observed that patients with fusion genes had a clinical benefit in ORR, DCR and median PFS than those without mutations. Although they were not significantly different, which may due to the limitation of the sample numbers, the results showed a positive response in the patients harboring fusion genes. In addition, the new targeted medicines for ALK, ROS1 and RET have been come into the market or in clinical trials. For example, crizotinib was approved by US FDA in 2011 for the treatment of patients with locally advanced or metastatic ALK-positive NSCLC and it was available in the market of China since June 2013. Sunitinib, sorafenib and vandetanib could effectively inhibit RET positive lung cancer cells [21].
In this study, we demonstrated that CB samples could be an option to substitute tissues to detect ALK, ROS1 and RET fusion genes in lung cancer patients. Patients with fusion gene mutation may have a better clinical response than those without mutations, which needed to be confirmed by a large sample study.
Footnotes
Funding sources: This study was supported by grants from the National Natural Science Foundation of China (grant 81172101), the Key Project of the Science and Technology Commission of Shanghai Municipality (grant 11JC1411301 and 124119a8000) and Project of Shanghai Pulmonary Hospital (grant FK1207).
Conflicts: The authors declare no conflicts with other studies.
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Koivunen J.P., Mermel C., Zejnullahu K., Murphy C., Lifshits E., Holmes A.J., Choi H.G., Kim J., Chiang D., Thomas R. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi K., Choi Y.L., Soda M., Inamura K., Togashi Y., Hatano S., Enomoto M., Takada S., Yamashita Y., Satoh Y. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 8.Inamura K., Takeuchi K., Togashi Y., Nomura K., Ninomiya H., Okui M., Satoh Y., Okumura S., Nakagawa K., Soda M. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 9.Choi Y.L., Takeuchi K., Soda M., Inamura K., Togashi Y., Hatano S., Enomoto M., Hamada T., Haruta H., Watanabe H. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi K., Choi Y.L., Togashi Y., Soda M., Hatano S., Inamura K., Takada S., Ueno T., Yamashita Y., Satoh Y. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 11.Shaw A.T., Yeap B.Y., Mino-Kenudson M., Digumarthy S.R., Costa D.B., Heist R.S., Solomon B., Stubbs H., Admane S., McDermott U. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S.G., Kuo Y.W., Chang Y.L., Shih J.Y., Chen Y.H., Tsai M.F., Yu C.J., Yang C.H., Yang P.C. EML4-ALK translocation predicts better outcome in lung adenocarcinoma patients with wild-type EGFR. J Thorac Oncol. 2012;7:98–104. doi: 10.1097/JTO.0b013e3182370e30. [DOI] [PubMed] [Google Scholar]
- 13.Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.H., Dezube B.J., Janne P.A., Costa D.B. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2011;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergethon K., Shaw A.T., Ou S.H., Katayama R., Lovly C.M., McDonald N.T., Massion P.P., Siwak-Tapp C., Gonzalez A., Fang R. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovly C.M., Heuckmann J.M., de Stanchina E., Chen H., Thomas R.K., Liang C., Pao W. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res. 2011;71:4920–4931. doi: 10.1158/0008-5472.CAN-10-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimkunas V.M., Crosby K.E., Li D., Hu Y., Kelly M.E., Gu T.L., Mack J.S., Silver M.R., Zhou X., Haack H. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18:4449–4457. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- 17.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Kohno T., Ichikawa H., Totoki Y., Yasuda K., Hiramoto M., Nammo T., Sakamoto H., Tsuta K., Furuta K., Shimada Y. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi K., Soda M., Togashi Y., Suzuki R., Sakata S., Hatano S., Asaka R., Hamanaka W., Ninomiya H., Uehara H. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 20.Suehara Y., Arcila M., Wang L., Hasanovic A., Ang D., Ito T., Kimura Y., Drilon A., Guha U., Rusch V. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipson D., Capelletti M., Yelensky R., Otto G., Parker A., Jarosz M., Curran J.A., Balasubramanian S., Bloom T., Brennan K.W. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavalli P., Riboli B., Generali D., Passalacqua R., Bosio G. EGFR genotyping in pleural fluid specimens in NSCLC patients. Lung Cancer. 2006;54:265–266. doi: 10.1016/j.lungcan.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Kimura H., Fujiwara Y., Sone T., Kunitoh H., Tamura T., Kasahara K., Nishio K. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer. 2006;95:1390–1395. doi: 10.1038/sj.bjc.6603428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz-Santos J., Serra P., Andreo F., Llatjos M., Castella E., Monso E. Contribution of cell blocks obtained through endobronchial ultrasound-guided transbronchial needle aspiration to the diagnosis of lung cancer. BMC cancer. 2012;12:34. doi: 10.1186/1471-2407-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw A.T., Solomon B., Kenudson M.M. Crizotinib and testing for ALK. J Natl Compr Canc Netw. 2011;9:1335–1341. doi: 10.6004/jnccn.2011.0115. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki H., Shimizu S., Tani Y., Maekawa M., Okuda K., Yokota K., Shitara M., Hikosaka Y., Moriyama S., Yano M. RET expression and detection of KIF5B/RET gene rearrangements in Japanese lung cancer. Cancer Med. 2012;1:68–75. doi: 10.1002/cam4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soda M., Isobe K., Inoue A., Maemondo M., Oizumi S., Fujita Y., Gemma A., Yamashita Y., Ueno T., Takeuchi K. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res. 2012;18:5682–5689. doi: 10.1158/1078-0432.CCR-11-2947. [DOI] [PubMed] [Google Scholar]
- 28.Cai G., Wong R., Chhieng D., Levy G.H., Gettinger S.N., Herbst R.S., Puchalski J.T., Homer R.J., Hui P. Identification of EGFR mutation, KRAS mutation, and ALK gene rearrangement in cytological specimens of primary and metastatic lung adenocarcinoma. Cancer Cytopathol. 2013;121:500–507. doi: 10.1002/cncy.21288. [DOI] [PubMed] [Google Scholar]
- 29.Cai W., Su C., Li X., Fan L., Zheng L., Fei K., Zhou C. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer. 2013;119:1486–1494. doi: 10.1002/cncr.27940. [DOI] [PubMed] [Google Scholar]
- 30.Rodig S.J., Mino-Kenudson M., Dacic S., Yeap B.Y., Shaw A., Barletta J.A., Stubbs H., Law K., Lindeman N., Mark E. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scagliotti G., Stahel R.A., Rosell R., Thatcher N., Soria J.C. ALK translocation and crizotinib in non-small cell lung cancer: an evolving paradigm in oncology drug development. Eur J Cancer. 2012;48:961–973. doi: 10.1016/j.ejca.2012.02.001. [DOI] [PubMed] [Google Scholar]


