Abstract
The protein kinase mTOR (mechanistic target of rapamycin) in complex 1 (mTORC1) promotes cell growth and proliferation in response to anabolic stimuli, including growth factors and nutrients. Growth factors activate mTORC1 by stimulating the kinase Akt, which phosphorylates and inhibits the tuberous sclerosis complex (TSC; which is comprised of TSC1, TSC2, and TBC1D7), thereby stimulating the mTORC1 activator Rheb. Here, we identified the mechanism through which REDD1 (regulated in DNA damage and development 1) represses the mTORC1 signaling pathway. We found that REDD1 promoted the protein phosphatase 2A (PP2A)-dependent dephosphorylation of Akt at Thr308 but not at Ser473. Consistent with previous studies showing that phosphorylation of Akt on Thr308, but not Ser473, is necessary for phosphorylation of TSC2, we observed a REDD1-dependent reduction in the phosphorylation of TSC2 and subsequently in the activity of Rheb. REDD1 and PP2A coimmunoprecipitated with Akt from wild-type but not REDD1-knockout mouse embryonic fibroblasts, suggesting that REDD1 may act as a targeting protein for the catalytic subunit of PP2A. Furthermore, binding to both Akt and PP2A was essential for REDD1 to repress signaling to mTORC1. Overall, the results demonstrate that REDD1 acts not just as a repressor of mTORC1, but also as a constant modulator of the phosphorylation of Akt in response to growth factors and nutrients.
Introduction
The serine-threonine protein kinase mechanistic target of rapamycin (mTOR) is a master regulator of the cellular signaling response to growth factor and nutrient sufficiency (1). The kinase exists in two interdependent complexes with distinct functions. mTOR complex 1 (mTORC1) regulates processes such as protein synthesis, de novo lipogenesis, and autophagy, whereas mTOR complex 2 (mTORC2) modulates cell survival and migration (2). To balance these processes with the energy and nutrient demands of a cell, two convergent signaling pathways have evolved to regulate mTORC1 activation: growth factors such as insulin signal to mTORC1 through one pathway and amino acids such as leucine signal through the other (3). Insulin and amino acids activate mTORC1 in a cooperative manner, because signaling inputs from both pathways are necessary for maximal phosphorylation of the two best-studied mTORC1 substrates, 70-kDa ribosomal S6 kinase 1 (p70S6K1) and eukaryotic initiation factor 4E binding protein 1 (4E-BP1) (3-5).
Insulin activates mTORC1 primarily through the PI3K/Akt signaling pathway, leading to the phosphorylation and inhibition of tuberous sclerosis complex 2 (TSC2). TSC2 acts in a complex with TSC1 and Tre2-Bub2-Cdc16 domain family member 7 (TBC1D7) as a GTPase activating protein (GAP) for Ras homologue enriched in brain (Rheb) (6). Direct binding of Rheb-GTP, but not Rheb-GDP, with mTORC1 results in its activation (7). On the other hand, amino acids activate mTORC1 through a TSC2-independent pathway that is mediated by a heterodimeric complex consisting of either Ras-related GTP binding (Rag) A or B (RagA/B) and Rag C or D (RagC/D) (8). Deprivation of either complete amino acids or leucine alone suppresses mTORC1 signaling (9); however, expression of a dominant-active mutant of RagA/B is sufficient to maintain mTORC1 activity in cells incubated in amino acid deficient medium (8).
Various upstream repressors of mTORC1 signaling have been identified, including a 25 kDa protein regulated in DNA damage and development 1 (REDD1; also known as DDIT4). REDD1 was initially identified as a gene induced by hypoxia and other stresses (10-13), but we have demonstrated that it is also induced by nutrient (14) and serum deprivation (15) in association with repressed mTORC1 signaling. Conversely, the enhanced protein synthesis and mTORC1 signaling that occurs in skeletal muscle after a bout of resistance exercise is associated with reduced expression of both REDD1 and its homolog REDD2 (16). REDD1 is a ubiquitous protein of low abundance in adult tissues; however during development, changes in its abundance are dynamic and tissue-specific (17). Thus, REDD1 likely serves as a key regulator of mTORC1 activation under various conditions, and not just as a stress response protein.
The mechanism through which REDD1 acts to repress mTORC1 signaling has been actively investigated for almost a decade, and the results of those studies have led to development of a model in which REDD1 acts to promote the GAP activity of TSC2 toward Rheb, leading to accumulation of Rheb•GDP and subsequent repression of mTORC1 signaling. However, this has not been shown experimentally. Furthermore, the mechanism through which REDD1 might act to stimulate TSC2 activity is unclear, with the single proposal that REDD1 activates TSC2 by sequestration of 14-3-3 leading to stabilization of the TSC2 protein (18) being questioned in a later study (19). Moreover, if REDD1 activates TSC2 GAP activity, it would be expected that overexpression of its substrate, Rheb, might overcome the repressive effects of REDD1 on mTORC1 signaling. However, although ectopic expression of Rheb attenuates REDD1-induced repression of mTORC1 in one report (12), another study demonstrates that REDD1 overexpression does not prevent the repression (19). Thus, the mechanism of REDD1 action remains unresolved.
The present study was designed to define the mechanism through which REDD1 acts to repress mTORC1 signaling. Using a cell line stably expressing REDD1 under an inducible promoter and mouse embryo fibroblasts (MEFs) deficient in REDD1 expression (REDD1−/−), we performed experiments that provide support for a model in which REDD1 promotes the association of protein phosphatase (PP) 2A with Akt, leading to dephosphorylation of the kinase on Thr308, a subsequent reduction in Akt-mediated phosphorylation of TSC2, and a decrease in the proportion of Rheb in the active GTP-bound state.
Results
Increased REDD1 abundance represses mTORC1 in a concentration dependent fashion
To study REDD1-mediated mTORC1 repression, we generated a HEK293 cell line with tetracycline-inducible HA-tagged REDD1, so that its abundance could be rapidly induced without subjecting cells to a stress such as hypoxia or irradiation. Upon doxycycline administration, HA-REDD1 protein abundance was increased, reaching a maximum by 4 h, which was maintained for at least 6 h (Fig 1A). In the absence of REDD1 induction, the presence of growth factors and nutrients in complete cell culture medium maintained the activation state of mTORC1. However, even in the presence of serum and amino acids, increased REDD1 abundance was sufficient to repress mTORC1 signaling to p70S6K1 and 4E-BP1 in a concentration dependent fashion (Fig 1A), an effect that was absent in HEK293 cells lacking inducible REDD1 (Fig 1B). Moreover, when cells were treated with cycloheximide to inhibit protein synthesis, REDD1 abundance was rapidly attenuated and mTORC1 signaling recovered (Fig 1C). These results demonstrate that REDD1 represses the activation state of mTORC1 in response to signaling inputs provided by growth factors and nutrients present in cell culture medium.
Figure 1. REDD1 expression represses mTORC1 signaling in cells maintained in complete medium.
HEK293 Tet-On HA-REDD1 cells (A) and HEK293 Tet-On control cells (B) were treated with doxycycline (Dox) for the time periods indicated. (C) HEK293 Tet-On HA-REDD1 cells were treated with Dox for 6 h followed by cycloheximide for up to 30 min. Abundance of HA-tagged and total REDD1, p70S6K1, 4E-BP1, GAPH, phosphorylation of p70S6K1 on Thr389 and 4E-BP1 on Ser65 were assessed in whole cell lysates by Western blot analysis. Blots shown in panels A-C are representative of results for two experiments (N=2); within an experiment, two independent samples were analyzed.
REDD1 acts to repress mTORC1 following activation by expression of Rags, but not following activation by expression of Rheb
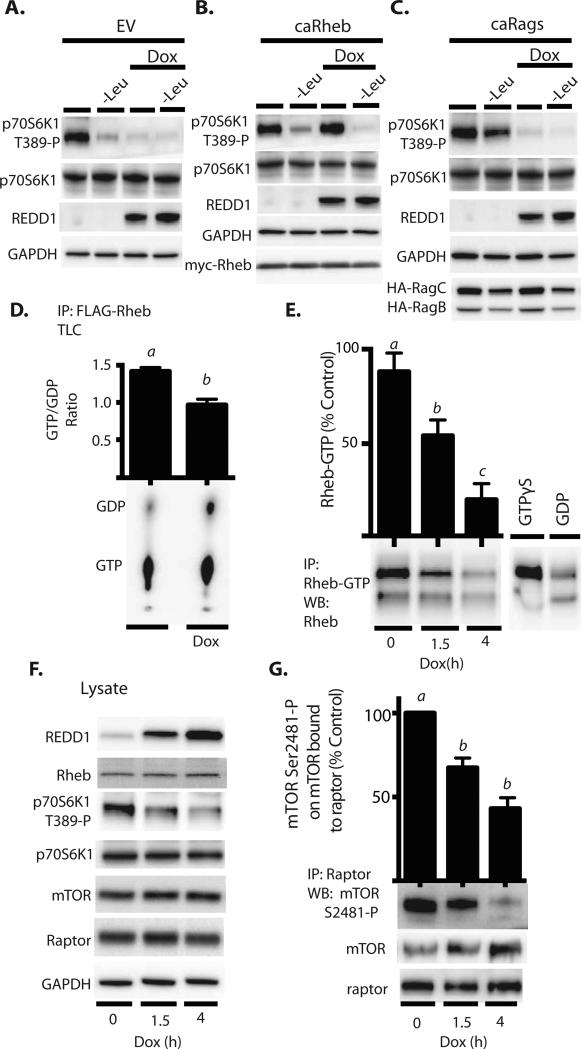
A previous report (20) suggested that mTORC1 is activated in response to cycloheximide due to the accumulation of intracellular amino acids. Furthermore, this effect is attenuated in cells lacking REDD1, suggesting an integral role for REDD1 in the cycloheximide-induced activation of mTORC1 (21). The most proximal activators of mTORC1 are the small GTPases Rheb and Rags. Amino acids signal through the Rag complex and insulin through Rheb to coordinate activation of mTORC1 (3). To assess the role of REDD1 in governing the activation state of mTORC1, HEK293 cells were transfected with a control plasmid, a plasmid expressing a constitutively active Rheb variant that preferentially binds GTP (caRheb), or a combination of plasmids expressing a variant of RagB that preferentially binds GTP and a variant of RagC that preferentially binds GDP (caRags) which dimerize to form a constitutively active RagBC complex (8). Both deprivation of the single amino acid leucine and REDD1 induction attenuated the mTORC1-mediated phosphorylation of p70S6K1 on Thr389 (Fig 2A). Consistent with our previous findings (3), ectopic expression of caRheb was not sufficient to prevent the reduction in the phosphorylation of p70S6K1 caused by leucine deprivation (Fig 2B). On the other hand, when cells were maintained in complete growth medium, expression of caRheb was sufficient to prevent the REDD1-mediated inhibition of p70S6K1 phosphorylation (Fig 2B, compare lanes 1 and 3). In contrast, whereas expression of caRags was largely sufficient to maintain mTORC1 signaling in leucine deficient medium, it was insufficient to prevent repression of mTORC1 by REDD1 (Fig 2C). These findings are consistent with the conclusion that REDD1 acts upstream of Rheb, but not of Rags, to repress mTORC1.
Figure 2. REDD1 expression attenuates Rheb-GTP loading and mTORC1 autophosphorylation.
HEK293 Tet-On HA-REDD1 cells were transfected with either an empty vector control plasmid (EV) (A), a plasmid expressing constitutively active (caRheb) (B), or plasmids expressing caRags (C). REDD1 expression was induced with doxycycline (Dox) in cells maintained in either complete growth medium or medium lacking leucine (-Leu) as indicated in the figure. (D) Cells expressing FLAG-Rheb were labeled with [32P]phosphate, FLAG-Rheb was immunoprecipitated, and the amounts of GTP and GDP in the immunoprecipitate were determined by thin layer chromatography (TLC). (E) Rheb-GTP loading was also assessed by immunoprecipitation using an antibody that selectively recognizes Rheb-GTP. Samples incubated with GTPγS or GDP were used as positive and negative controls, respectively. (F) Western blot analysis of whole cell lysates after doxycycline administration. (G) Western blot analysis of autophosphorylation of mTOR on Ser2481 in raptor immunoprecipitates. Blots or phosphorimages shown in panels A-D and E-G are representative of results for two (N=2) or three experiments (N=3), respectively; within an experiment, three independent samples were analyzed. Results shown in (D), (E), and (G) represent means ± SEM.
REDD1 acts to govern Rheb-GTP loading
Because REDD1 represses mTORC1 signaling upstream of Rheb, and Rheb-GTP (but not Rheb-GDP) activates mTORC1, we next sought to evaluate whether REDD1 acts to govern Rheb-GTP loading. To investigate how REDD1 affects Rheb GTP loading, we determined the nucleotide-binding status of Rheb in HEK293 cells labeled with 32P-phosphate. Induction of REDD1 decreased the GTP/GDP ratio of Rheb (Fig 2D), indicating an increase in the GTPase activity of Rheb. To further test the effects of REDD1 on Rheb GTP loading, Rheb was immunoprecipitated with a configuration-specific monoclonal antibody that selectively recognizes Rheb-GTP after induction of REDD1 expression. In cells maintained in complete growth medium, Rheb GTP loading was high, with approximately 90% of Rheb associated with GTP (Fig 2D). This observation is consistent with a previous report (22) that the proportion of Rheb bound to GTP is high in cells maintained in normal growth medium. Following induction of REDD1, the proportion of Rheb bound to GTP was reduced to 57% after 1.5 h and 25% after 4 h of doxycycline treatment (Fig 2D), concomitant with reduced phosphorylation of p70S6K1 on Thr389 (Fig 2E). Autophosphorylation of mTOR on Ser2481 within the mTORC1 complex indicates activation of the complex (23). To compare the reduction in Rheb-GTP binding with mTORC1 catalytic activity, we measured the autophosphorylation of mTOR on Ser2481 in raptor immunoprecipitates following induction of REDD1 expression. In a manner that paralleled Rheb-GTP binding, induction of REDD1 expression reduced the phosphorylation of mTOR on Ser2481 to 67% after 1.5 h and 43% after 4 h (Fig 2F and G). Overall, these data demonstrate that REDD1 represses mTORC1 by attenuating Rheb-GTP loading.
REDD1 co-immunoprecipitates with TSC2 and 14-3-3
A previous study suggested that REDD1 activates TSC2, which acts as a GAP for Rheb, by sequestering the protein 14-3-3 and decreasing its inhibitory association with TSC2 (18). To assess whether ectopic expression of REDD1 promoted dissociation of 14-3-3 from TSC2, FLAG-tagged TSC2 (Fig S1A), endogenous TSC2 (Fig S1B), and endogenous 14-3-3 (Fig S1C) were immunoprecipitated from cells incubated in the presence or absence of doxycycline treatment to induce REDD1 expression. Consistently, we observed co-immunoprecipitation of REDD1 with both TSC1/2 and 14-3-3, suggesting that all three proteins are potentially part of a multi-subunit complex. However, neither the abundance of 14-3-3 nor that of TSC2 in the immunoprecipitate fraction was consistently altered in response to increased REDD1 abundance (Fig S1).
Phosphorylation of TSC2 is enhanced in the absence of REDD1
To further investigate the mechanism whereby REDD1 governs the activation state of mTORC1, serum-starved REDD1−/− and REDD1+/+ MEFs were treated with insulin. Consistent with the repressive role of REDD1 in complete medium observed above, stimulation of p70S6K1 phosphorylation on Thr389 was enhanced in response to insulin in REDD1−/− MEFs as compared to REDD1+/+ MEFs (Fig 3A-B). Notably, the maximal insulin-induced phosphorylation of p70S6K1 on Thr389 was enhanced by the absence of REDD1 (Fig. 3B). Consistent with the effect of REDD1 on Rheb GTP loading, phosphorylation of TSC2 on Thr1462 and Ser939 was not only increased in REDD1−/− MEFs as compared to REDD1+/+ MEFs after serum deprivation, but insulin-stimulated phosphorylation of these residues was also enhanced in cells lacking REDD1 (Fig 3A, C-D). Conversely, doxycycline-induced REDD1 expression in HEK293 Tet-on HA-REDD1 cells led to decreased phosphorylation of TSC2 (Fig. 3E) These finding are consistent with a model in which REDD1 acts to repress the inhibitory phosphorylation of TSC2 in response to insulin, and because Akt mediates phosphorylation of TSC2 on Thr1462 and Ser939 (24), the results suggest that REDD1 may act to repress the kinase activity of Akt toward the GAP.
Figure 3. REDD1 expression differentially alters the phosphorylation of Akt on Thr308 and Ser473.
(A-D, F-I) REDD1+/+ and REDD1−/− MEFs were serum starved and subsequently treated with the indicated concentrations of insulin. Relative phosphorylation of various proteins in the AktmTORC1 signaling pathway was assessed in whole cell lysates by Western blot analysis. Results represent mean phosphorylation relative to protein abundance ± SEM for (B) to (D) and (F) to (I). Dose response curves for REDD1−/− MEFs were significantly different (p<0.01) than REDD1+/+ MEFs for (B) to (D), (F), (G), and (I). (E) HEK293 Tet-On control cells (Control) and HEK293 Tet-On HA-REDD1 cells (HA-REDD1) were treated with doxycycline (Dox) as indicated. Phosphorylation of Akt on Thr308 and Ser473, TSC2 on Thr1462, PDK1 on Ser241, S6K1 on Ser229, as well as the abundance of each respective protein, HA-REDD1, and GAPDH was examined by Western blot analysis. (J) The relative phosphorylation of Akt on Thr308 and Ser473 in HEK293 Tet-On HA-REDD1 cells following treatment with doxycycline (Dox) as indicated. Blots in panel (A) and (E) are representative of results for three experiments (N=3); within an experiment, two independent samples were analyzed. Results represented as means ± SEM are shown in (J). *p<0.05 compared to 1 h.
REDD1 represses site-specific phosphorylation of Akt at Thr308
To provide further evidence that REDD1 acts to repress Akt activity, we also evaluated the phosphorylation of two other Akt substrates, GSK3 and FOXO1/3. Similar to TSC2, phosphorylation of both GSK3 on Ser21/9 and FOXO1/3 on Thr24/32 was enhanced in serum-deprived REDD1−/− MEFs as compared to REDD1+/+ MEFs (Fig 3A). In addition, insulin-induced phosphorylation of both proteins was higher in REDD1−/− MEFs than in REDD1+/+ MEFs (Fig 3F and G). However, repressed activation of Akt did not initially seem consistent with reports from both our laboratory (15) and others (25) that demonstrate increased phosphorylation of Akt at Ser473 in response to REDD1. To further assess this apparent discrepancy, phosphorylation of Akt on both Ser473 and Thr308 was measured because differential phosphorylation of these residues alters the substrate specificity of the kinase (26). In accordance with our previous findings (15), insulin-induced phosphorylation of Akt on Ser473 was slightly higher in REDD1+/+ MEFs than in REDD1−/− MEFs (Fig 3H). Surprisingly, the effect of insulin on the phosphorylation of Akt at Thr308 was distinctly different compared to that of Ser473. Specifically, phosphorylation of Akt at Thr308 was higher in REDD1−/− MEFs than in REDD1+/+ MEFs (Fig 3I). Furthermore, increased REDD1 abundance specifically attenuated phosphorylation of Akt on Thr308 (Fig 3E and S2A) but not on Akt Ser473 in HEK293 Tet-on HA-REDD1 cells, whereas parental cells lacking HA-REDD1 did not show obvious changes in the phosphorylation of either phosphorylation site (Fig 3E and S2B). REDD1 expression reduced the ratio of Akt phosphorylation at Thr308 to Ser473 by 60% (Fig 3J). Because PDK1 phosphorylates Akt at Thr308 (27), we also evaluated activation of this upstream kinase. However, REDD1 induction did not alter the phosphorylation of PDK1 on Ser241, an autophosphorylation site in the activation loop of the kinase, or the phosphorylation of another PDK1 substrate, p70S6K1, on Ser229 (Fig 3E). Together these findings are consistent with a model in which REDD1 inhibits activation of mTORC1 by specifically impairing the phosphorylation of Akt at Thr308, an effect that is independent of changes in PDK1 activity.
PP2A co-immunoprecipitates with Akt in the presence of REDD1
Because the changes in the phosphorylation of Akt on Thr308 could not be explained by modulation of PDK1 activity, we tested the alternative hypothesis that REDD1 promotes the dephosphorylation of Akt on Thr308 by modulating protein phosphatase activity toward the residue. The site-specific dephosphorylation of Akt at Thr308 by protein phosphatase 2A (PP2A) has been previously demonstrated both in cells and with recombinant proteins (28). HA-tagged REDD1 from doxycycline-treated cells coimmunoprecipitated with both Akt and the catalytic subunit of PP2A (PP2Ac) but not with ERK or PP1 (Fig 4A).
Figure 4. PP2A co-immunoprecipitates with Akt in the presence of REDD1.
(A) Lysates from HEK293 Tet-On HA-REDD1 cells incubated with or without doxycycline (Dox) were immunoprecipitated using anti-HA-agarose followed by Western blot analysis. (B) HEK293 Tet-On HA-REDD1 cells expressing caAkt were incubated with or without Dox and phosphorylation of caAkt on Thr308 was assessed by Western blot analysis in three experiments (N=3); within an experiment, three independent samples were analyzed. (C) REDD1+/+ and REDD1−/− MEFs expressing HA-tagged caAkt were serum deprived and HA immunoprecipitates and cell lysates were subject to Western blot analysis. (D) Endogenous Akt was immunoprecipitated following treatment with thapsigargin (TG) and immunoprecipitates and lysates subjected to Western blot analysis. (E & G) HEK293 Tet-On cells were transiently transfected with empty pTREtight vector (EV) or pTREtight plasmid encoding wild-type (WT), P139A, K219A/Y222A (KYAA), or E68A REDD1 as indicated. HA-tag immunoprecipitates and lysates were subjected to Western blot analysis. Blots shown in panels A, C-E, and G are representative of results for two experiments (N=2); within an experiment, two independent samples were analyzed. (F) Alignment of REDD1 amino acid sequences with a conserved region on the PP2AB regulatory subunits required for interaction with the core enzyme.
To further explore the role of REDD1 in regulating phosphorylation of Akt on Thr308, a constitutively active variant of Akt (caAkt) was employed. The caAkt variant bears a myristoylation signal that targets the protein to the plasma membrane, promoting its phosphorylation on Thr308 by PDK1 and Ser473 by mTORC2 (29). Ectopic expression of caAkt is not sufficient to maintain mTORC1 signaling in cells subjected to hypoxia, a condition that potently increases REDD1 abundance (18). Thus we questioned whether or not REDD1-mediated recruitment of PP2A and dephosphorylation of Thr308 might be sufficient to inhibit the downstream effect of Akt that is “constitutively activated” by targeting to the plasma membrane. Indeed, increased REDD1 abundance not only repressed the phosphorylation of p70S6K1 on Thr389 in the presence of caAkt, but also resulted in a 29% decrease in the phosphorylation of caAkt at Thr308 (Fig 4B).
To gain further evidence of REDD1-mediated targeting of PP2A to Akt, immunoprecipitates of HA-tagged caAkt overexpressed in serum-starved REDD1−/− and REDD1+/+ MEFs were probed for REDD1 and PP2A. Whereas REDD1 and PP2A were both detected in the anti-HA-tag immunoprecipitate from REDD1+/+ MEFs, neither protein was detected in immunoprecipitates from REDD1−/− MEFs (Fig 4C). REDD1 was also necessary for the interaction of PP2A with Akt in response to endoplasmic reticulum stress. Induction of ER stress in response to thapsigargin treatment was confirmed by increased abundance of the transcription factor ATF4 (activating transcription factor 4) in treated compared to control cells (Fig. 4D). Moreover, induction of ER stress by thapsigargin promoted the co-immunoprecipitation of PP2A with Akt in REDD1+/+ MEFs, but not REDD1−/− MEFs (Fig 4D).
If REDD1 acts to repress mTORC1 signaling by targeting PP2A to Akt to promote dephosphorylation of Thr308, then REDD1 variants that do not repress mTORC1 signaling are unlikely to either promote the interaction of the phosphatase with Akt or to cause dephosphorylation of Thr308. A form of REDD1 with an Ala substitution at Pro139 does not inhibit mTORC1 (19). Structure/function studies suggest that Pro139 is located in a surface patch on the protein that is critical for function (19). This “functional hotspot” also includes Lys219 and Tyr222, and simultaneous alanine substitutions at these residues (KYAA) completely abrogates the ability of REDD1 to repress mTORC1 signaling (19). To test the possibility that these critical residues participate in REDD1-mediated recruitment of PP2A to Akt, HEK293 Tet-On cells were transiently transfected with empty vector, wild-type HA-REDD1, or either the P139A HA-REDD1 or KYAA HA-REDD1 variants. In confirmation of the previous study (19), expression of wild type REDD1 suppressed mTORC1 signaling whereas expression of the two variants had no effect (Fig 4E). Notably, unlike wild-type REDD1, the P139A and KYAA variants also failed to repress phosphorylation of Akt at Thr308 (Fig 4E). Moreover, whereas Akt and PP2A coimmunoprecipitated with wild-type HA-REDD1, the association of both proteins with the P139A REDD1 variant was reduced (Fig 4E). Furthermore, although PP2A was present in KYAA HAREDD1 immunoprecipitates, little or no Akt was detected.
Structural studies of PP2A have identified a conserved loop in the PP2AB regulatory subunit that is required for their interaction with the core enzyme (30). Intriguingly, a number of these residues, including an obligatory glutamic acid that is required for dephosphorylation of PP2A/B-targeted substrates (31), align with residues in REDD1 (Fig 4F), raising the possibility that REDD1 functions in a manner analogous to a regulatory B-subunit of PP2A in the recruitment of the core enzyme to Akt. To evaluate this possibility, Glu68 was mutated to alanine to assess the role of this residue in REDD1-mediated mTORC1 inhibition. As previously observed with the P139A and KYAA REDD1 variants, REDD1 E68A failed to attenuate phosphorylation of either p70S6K1 at Thr389 or Akt at Thr308 (Fig 4G). Further, PP2A failed to co-immunoprecipitate with the E68A variant, although Akt was still detected in the immunoprecipitate. Overall, these results are consistent with a model in which binding both Akt and PP2A is essential for REDD1 signaling to mTORC1.
Discussion
Our results support a model in which REDD1 acts to repress mTORC1 signaling by promoting PP2A-dependent dephosphorylation of Akt on Thr308, leading to reduced phosphorylation of TSC2 and subsequently to reduced Rheb GTP loading (Fig S3). A previous study (28) demonstrated the preferential dephosphorylation of Akt on Thr308 by PP2A, with little or no effect on phosphorylation of Ser473, and our results corroborate that finding. We found that ectopic expression of REDD1 led to reduced phosphorylation of Akt on Thr308, whereas in REDD1-deficient MEFs, insulin-stimulated Akt phosphorylation on Thr308 was enhanced. A key difference between the present and previous study (28) is that the earlier study did not assess the possible involvement of REDD1 in the differential dephosphorylation by PP2A. We also extend the findings of Kuo et al. (28) showing that ectopic expression of REDD1 results in increased association of both Akt and the catalytic subunit of PP2A with REDD1, suggesting the formation of a multiprotein complex consisting of REDD1, Akt, and PP2A.
The activity of Akt is regulated by the phosphorylation and dephosphorylation of multiple residues. In response to growth factors, the activation loop of Akt is phosphorylated at Thr308 by PDK1 (27), whereas the C-terminal hydrophobic motif is phosphorylated by mTORC2 at Ser473 (32). Since phosphorylation of both Thr308 and Ser473 produces higher in vitro kinase activity (33), it is often assumed that phosphorylation of both residues is required for Akt activation. Indeed because insulin-induced phosphorylation of Thr308 occurs prior to Ser473 (34), phosphorylation of Ser473 is often used alone as an indicator of Akt activity. However, this assumption often leads to confounding conclusions regarding the activation state of Akt. For example, in response to various stimuli, Akt phosphorylates numerous downstream substrates, including GSK3, FoxO transcription factors, TSC2, PRAS40, and Bad (35). However, defective phosphorylation of Akt at Ser473 only affects a subset of Akt targets, including FoxO transcription factors, whereas TSC2 phosphorylation is unaffected (26). Moreover, phosphorylation of Thr308, but not Ser473, correlates with Akt protein kinase activity toward TSC2, PRAS40, and TBC1D4 (36). Our finding that the selective dephosphorylation of Akt on Thr308 that occurred in response to increased REDD1 expression was associated with decreased phosphorylation of at least three Akt substrates (TSC2, GSK-3, and FOXO1/3) provides further support for this conclusion.
PP2A is a major cellular serine/threonine phosphatase and exists in both a dimeric form (referred to as the core enzyme) consisting of a catalytic (C) and a scaffold subunit (A), and a trimeric form (referred to as the holoenzyme) which in addition to the A and C subunits, also includes a B regulatory subunit (37). Four diverse gene families account for approximately 15 different PP2AB subunits (38). While the different family members and isoforms of PP2AB bind to the same recognition sequences of PP2AA, they possess distinct gene sequences. Akt directly interacts with the PP2A-B55 holoenzyme (28). We showed that only the catalytic, subunit (PP2AC) interacted with REDD1. Thus, REDD1 may not only promote a stable interaction of the phosphatase with Akt, leading to preferential dephosphorylation at Thr308, but may also function as a regulatory, B-subunit of PP2A.
Based on studies showing that mTORC1 signaling is not repressed by REDD1 in cells deficient for TSC2 (39), it has been assumed that REDD1 activates TSC2 GAP activity and consequently decreases the proportion of Rheb in the GTP-bound form. However, we showed that Rheb GTP loading was inversely proportional to the amount of REDD1 expressed. Notably, mTORC1 activity, assessed either by the autophosphorylation of mTOR on Ser2481 in raptor immunoprecipitates or by changes in phosphorylation of the mTORC1 substrate p70S6K1, was also inversely proportional to REDD1 expression, but conversely, was directly proportional to the amount of Rheb bound to GTP. We also showed that REDD1-induced inhibition of mTORC1 was attenuated by ectopic expression of a Rheb variant that preferentially binds to GTP, but not by a combination of constitutively active RagB and RagC. Overall, our results suggest that REDD1 acts to repress mTORC1 signaling through activation of Rheb GTPase activity. A caveat to this conclusion is that REDD1-induced inhibition of mTORC1 has previously been shown to be incompletely prevented by Rheb overexpression (19), although that study used wild-type Rheb, rather than caRheb, and consequently its GTP loading status would have been sensitive to TSC2 activation state. Thus, as long as the total amount of Rheb did not exceed the capacity of TSC2 to stimulate its GTPase activity, then repression of mTORC1 signaling by REDD1 expression would still be detected. It is noteworthy that, in agreement with our results, a previous study in which caRheb was overexpressed (26) showed that REDD1 did not repress mTORC1 signaling.
One model that has been proposed to explain how the TSC complex acts to repress mTORC1 involves the dissociation of the active TSC1•TSC2 complex as a result of TSC2 phosphorylation by Akt and subsequent sequestration of TSC2 by 14-3-3 proteins (18). In support of that model, various studies have shown that Akt phosphorylates TSC2 on multiple residues (40) and in some studies phosphorylation of TSC2 is associated with increased association of the protein with 14-3-3 proteins (41-43). We found that changes in REDD1 abundance were inversely associated with alterations in phosphorylation of both Akt and TSC2. This finding might seem to provide a missing piece to the puzzle: By inhibiting Akt-mediated phosphorylation of TSC2, REDD1 would promote dissociation of TSC2 from 14-3-3 proteins, thereby allowing it to bind to TSC1 to form the active TSC complex. However, we did not observe either a decrease in TSC2 association with 14-3-3 proteins or an increase in association of TSC2 with TSC1 in response to increased REDD1 abundance, suggesting that REDD1 may work through additional mechanisms to regulate TSC2 function. In this regard, Menon et al. (44) have shown that Akt does not promote release of TSC2 from TSC1, but instead activates mTORC1 by causing dissociation of the TSC holocomplex from the lysosomal membrane where Rheb is located. Moreover, other studies (45, 46) suggest that Akt-mediated phosphorylation of TSC2 is not responsible for its interaction with 14-3-3 proteins, but that instead phosphorylation of the protein by MAP kinase on Ser1210 is essential for 14-3-3 binding. Thus, although it is clear that TSC2 binds to 14-3-3 proteins, whether or not REDD1 also binds to them is unclear. Indeed, structural analysis of the REDD1 protein suggests that the 14-3-3 binding motif in REDD1 not conserved and immunoprecipitation studies suggest that REDD1 does not interact directly with 14-3-3 proteins (19). Our findings neither prove nor disprove the model in which REDD1 promotes TSC1•TSC2 complex assembly by sequestering 14-3-3 proteins. However, the presence of REDD1 in both TSC2 and 14-3-3 immunoprecipitates suggests that the association of REDD1 and TSC2 with 14-3-3 proteins is more complex than originally anticipated.
Our results establish REDD1 as a dominant governor of mTORC1 activity that attenuates Rheb-GTP loading such that phosphorylation of p70S6K1 and 4E-BP1 phosphorylation are impaired in cells cultured in complete cell culture medium. Mechanistically, REDD1 promotes dephosphorylation of Akt at Thr308 and represses TSC2 activation through the recruitment of PP2A to the kinase (Fig S3). Thus, REDD1 not only governs mTORC1 activity in response to growth factors and nutrients, but also modulates Akt phosphorylation in a manner that potentially alters the substrate specificity of this critical kinase.
Materials and Methods
Materials
Protease inhibitor mixture was purchased from Sigma, and ECL Western blot detection reagent from Pierce. Horseradish peroxidase-conjugate, anti-phospho-S6K1 (Thr389), and goat anti-rabbit IgG antibodies were purchased from Bethyl Laboratories. Anti-REDD1 antibody was purchased from Proteintech, anti-phospho-p70S6K1 Ser229 antibody was purchased from Abcam, anti-PP2A was purchased from Transduction Laboratories, and anti-GAPDH antibody was purchased from Santa Cruz Biotechnology. Anti-PPP1CA antibody was purchased from Antibody Verify and all other antibodies were purchased from Cell Signaling Technology. Cell culture medium lacking leucine, histidine, and pyruvate was a custom formulation purchased from Atlanta Biologicals; histidine was added to the medium prior to use.
Construction of pTREtight HA-REDD1 plasmid
A cDNA clone (MGC-12610) encoding human REDD1 was purchased from ATCC (Manassas, VA) and used as a template for PCR. Primers (sense: 5’- CGC GAA TTC GGA TGC CTA GCC TTT GGG AC-3’ and antisense: 5’-CTC ATT GAG GAG TGT TGA CTC GAG GGA TCC CGC-3’) were used to generate a product that was digested and ligated into the EcoR1/Xho1 restriction sites of a mammalian expression plasmid (pCMV; Clontech) that encoded REDD1 with an N-terminal HA epitope. The resultant plasmid, pCMV-HA-REDD1 was used as a template in a PCR reaction with primers (sense: 5’-CTC GGT ACC ATG TAC CCA TAC GAT GTT C-3’ and antisense: 5’-CGC GGA TCC TCA ACA CTC CTC AAT GAG -3’) to generate the HA-REDD1 insert that was digested and cloned into the Kpn1/BamH1 restriction sites of a doxycycline responsive plasmid, pTREtight-HA-REDD1 (Clontech).
Derivation of HEK293 Tet-On HA-REDD1 Advanced Stable Cell Line
HEK293 TetON cells (Clontech) were transfected in a 60 mm dish according to the manufacturer's recommendations using 5 μl Lipofectamine 2000 with 0.5 μg of pTREtight HA-REDD1 together with 0.025 μg of a linearized hygromycin selection marker. After 5 hours, the same volume of high glucose Dulbecco's modified eagle medium (DMEM, Invitrogen) with 20% FBS was added to avoid lifting cells from the plates. Transfected cells were passaged 48 h after the start of transfection and replated at a series of low densities in DMEM with 10% FBS and maintained under selective pressure of G418 (100 μg/ml) and hygromycin B (200 μg/ml). Surviving colonies that formed were selected using cloning cylinders and frozen stocks were established.
Cell culture
Parental HEK293 Tet-On cells or HEK293 Tet-On HA-REDD1 were maintained at 37 °C and 5% CO2 in DMEM supplemented with Tet System Approved FBS (Clontech) and placed under selective pressure using G418 and hygromycin B as described above. Where indicated cell culture medium was supplemented with 1 μg/μl doxycycline (Clontech) to induce HA-REDD1 protein expression and 1 mM cycloheximinde to block protein synthesis as indicated. Wild-type MEF and REDD1 knockout MEF (kindly provided by Dr. Leif Ellison, Massachusetts General Hospital Cancer Center) were maintained similarly, but DMEM was supplemented with 1% penicillin/streptomycin (Invitrogen) and 10% heat-inactivated fetal bovine serum (FBS, Atlas Biologicals). Transfections were performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. The following plasmids were used in transfections: pRK5-Myc-Rheb-S16H (kindly provided by Dr. Kun-Liang Guan, University of California, San Diego); pRK5-HA-GST-RagC-S75L, pRK5-HA-GST-RagB-Q99L (purchased from Addgene; deposited by Dr. David Sabatini, Massachusetts Institute of Technology); pRK7-FLAG-TSC2 (kindly provided by Dr. John Blenis, Harvard Medical School); and pCMV5-HA-caAkt (purchased from Addgene; submitted to Addgene by Dr. Mien-Chie Hung). A pRK5 empty vector was used as a control plasmid. Cells were deprived of serum and/or leucine for 2 h and then treated with insulin (Novolin) for 30 min as indicated. In other experiments, cells were administered 100 nM thapsigargin (Sigma) for 4 h. Cells were harvested in 1 X SDS sample buffer for analysis of total cell lysate by SDS-PAGE and Western blot analysis as previously described (47). Site-directed mutagenesis was performed on pTREtight HA-REDD1 plasmid using Quick-Change Lightning (Agilent Technologies) and the following primers: 5'-CCTACAGCGAGGCGTGCGGCCTG-3' and 5'-CAG GCC GCA CGC CTC GCT GTA GG-3' to mutate Pro139 to Ala; 5'-GCT TCC GAG TCA TCA AGG CGA AGC TGT ACA GCT CGG-3' and 5'-CCG AGC TGT ACA GCT TCG CCT TGA TGA CTC GGA AGC-3' to mutate Lys219 to Ala; 5'-AGC AGC TGT TCC GAG CTG GCC AGC TTC TTC TTG ATG AC-3'; 5'-GTC ATC AAG AAG AAG CTG GCC AGC TCG GAA CAG CTG CT-3' to mutate Tyr222 to Ala; and 5'-GTA AGC CGT GTC TGC CTC CGG CCC GAA-3' and 5'-TTC GGG CCG GAG GCA GAC ACG GCT TAC-3' to mutate Glu68 to Ala. All plasmids were sequenced to confirm mutations and then transiently transfected into HEK293 Tet-On cells using Lipofectamine 2000 as described above.
Immunoprecipitations
Immunoprecipitations were performed on 1000 × g supernatant fractions of cell lysates. Raptor immunoprecipitations were performed as previously described (47). Akt immunoprecipitation was performed with 10 μl Protein G Plus/Protein A-Agarose (EMD Millipore) and 4 μg of anti-Akt/PH domain antibody (Upstate). TSC2 (48) and 14-3-3 (49) immunoprecipitations were performed as previously described. For anti-HA and anti-FLAG immunoprecipitations, 10 μl of either anti-HA-agarose affinity resin (Sigma) or EZview Red anti-FLAG M2 affinity gel (Sigma) were used. Beads were washed with CHAPS lysis buffer (40 mM HEPES pH 7.2, 0.3% CHAPS, 1 mM EDTA, 50 mM NaF, 40 mM NaCl, 50 mM β-glycerophosphate) and blocked with CHAPS lysis buffer containing 1% BSA. Cells were harvested in CHAPS lysis buffer supplemented with 10 mM sodium pyrophosphate, 1 mM benzamidine, 200 mM sodium vanadate, and 10 μl/ml protease inhibitor mixture and lysed for 30 min at 4 °C. Cell supernatants were collected by centrifuging lysates 3 min at 1000 × g and incubated with the appropriate affinity resin for 2 h. Affinity resins were washed (3X) with either cold CHAPS lysis buffer or RIPA wash buffer (25mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), resuspended in 1X SDS sample buffer, and boiled for 5 min. The immunoprecipitates were subjected to Western blot analysis.
Measurement of Rheb GTP loading status
Rheb GTP loading was assessed as previously described (50) with the following modifications. HEK293 Tet-On HA-REDD1 cells were cultured in 10 cm dishes, transfected with pRK7-FLAG-Rheb (kindly provided by Dr. John Blenis, Harvard Medical School). Forty-eight h after transfection cells were placed in phosphate-free DMEM (Gibco) supplemented with 10% FBS for 90 min and treated with doxycycline to induce HA-REDD1. Cells were labeled with 25 μCi of [32P]phosphate/mL for 4 h. Cells were collected in lysis buffer (0.5% NP-30, 50 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 1 mM benzamidine, 200 mM sodium vanadate, and 10 l/ml protease inhibitor mixture). FLAG-Rheb was immunoprecipitated with anti-FLAG M2 affinity gel and the Rheb-bound nucleotides were eluted with elution buffer (2 mM EDTA, 0.2% SDS, 1 mM GDP, 1 mM GTP) at 68°C. GTP and GDP were resolved by thin layer chromatography and visualized by a Typhoon 9400 Phosphorimager (GE Healthcare). Rheb-GTP loading was also assessed using a RheB Activation Assay Kit (NewEast Biosciences) according to the manufacturer's instructions.
Statistical analysis
Data are presented as means ± SEM. Statistical analysis was performed using GraphPad Prism and R. The quantitative graphics and statistical assessments were largely derived from 3 biological replicates per group with each biological replicate being the average of 2-3 technical replicates. Values used to normalize/calibrate the results in each assay were appropriately excluded from the statistical analysis. Data in Fig 2E and 2G were analyzed overall with an ANOVA and trend test and pairwise comparisons were conducted with Tukey's test for multiple comparisons. The dose response curves for REDD1−/− and REDD1+/+ MEFs in Fig 3 were analyzed using a linear regression model after controlling for insulin dose. In Figs 2D, 3J, 4B, and S2, a two-sample t test comparison compared controls to other groups. p < 0.05 was considered statistically significant.
Supplementary Material
Fig. S1. Effect of REDD1 on the interaction of 14-3-3 protein with TSC2.
Fig. S2: REDD1 promotes the dephosphorylation of Akt on Thr308.
Fig. S3. Regulation of the phosphorylation of Akt at Thr308 and the TSC2-TBC complex by REDD1.
Acknowledgments
Funding: This work was supported by NIH grants DK15658 and DK13499 (to LSJ), UL1 TR000127 (to AB), and DK094141 (to SRK), as well as EY023612 and the American Diabetes Association Pathway to Stop Diabetes Grant 1-14-INI-04 (to MDD). Acknowledgments: The authors thank Dr. Leif Ellisen (Massachusetts General Hospital Cancer Center) for kindly providing REDD1 wild type and knockout MEFs as well as Holly Lacko, Chen Yang, and Tony Martin for technical assistance in performance of the studies described herein. The authors also thank Vincent Chau for advice with regards to manuscript preparation.
Footnotes
Author Contributions: M.D.D. and C.S.C. designed experiments, researched data, wrote, and edited the manuscript. L.S.J. and S.R.K. designed experiments, contributed to discussion, and reviewed/edited the manuscript. A.B. performed statistical analysis on the data. Data availability: An MTA is required by The Pennsylvania State University College of Medicine for the cell lines and expression plasmids generated by the Kimball laboratory.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dennis MD, Baum JI, Kimball SR, Jefferson LS. Mechanisms involved in the coordinate regulation of mTORC1 by insulin and amino acids. J Biol Chem. 2011;286:8287–8296. doi: 10.1074/jbc.M110.209171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter JN, Fox TE, Kester M, Jefferson LS, Kimball SR. Phosphatidic acid mediates activation of mTORC1 through the ERK signaling pathway. Am J Physiol Cell Physiol. 2010;299:C335–344. doi: 10.1152/ajpcell.00039.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest. 1998;101:1519–1529. doi: 10.1172/JCI1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 8.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 10.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 11.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XH, Ha CT, Fu D, Xiao M. REDD1 protects osteoblast cells from gamma radiation-induced premature senescence. PLoS One. 2012;7:e36604. doi: 10.1371/journal.pone.0036604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGhee NK, Jefferson LS, Kimball SR. Elevated corticosterone associated with food deprivation upregulates expression in rat skeletal muscle of the mTORC1 repressor, REDD1. J Nutr. 2009;139:828–834. doi: 10.3945/jn.108.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis MD, McGhee NK, Jefferson LS, Kimball SR. Regulated in DNA damage and development 1 (REDD1) promotes cell survival during serum deprivation by sustaining repression of signaling through the mechanistic target of rapamycin in complex 1 (mTORC1). Cell Signal. 2013;25:2709–2716. doi: 10.1016/j.cellsig.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond MJ, Fujita S, Abe T, Dreyer HC, Volpi E, Rasmussen BB. Human muscle gene expression following resistance exercise and blood flow restriction. Med Sci Sports Exerc. 2008;40:691–698. doi: 10.1249/MSS.0b013e318160ff84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q, Zou X, Lu L, Li Y, Liu Y, Zhou J, Duan C. The stress-response gene redd1 regulates dorsoventral patterning by antagonizing Wnt/beta-catenin activity in zebrafish. PLoS One. 2012;7:e52674. doi: 10.1371/journal.pone.0052674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vega-Rubin-de-Celis S, Abdallah Z, Kinch L, Grishin NV, Brugarolas J, Zhang X. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry. 2010;49:2491–2501. doi: 10.1021/bi902135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003;372:555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimball SR, Do AN, Kutzler L, Cavener DR, Jefferson LS. Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. J Biol Chem. 2008;283:3465–3475. doi: 10.1074/jbc.M706643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 25.Jin HO, Hong SE, Kim JH, Choi HN, Kim K, An S, Choe TB, Hwang CS, Lee JH, Kim JI, Kim HA, Kim EK, Noh WC, Hong YJ, Hong SI, Lee JK, Park IC. Sustained overexpression of Redd1 leads to Akt activation involved in cell survival. Cancer Lett. 2013;336:319–324. doi: 10.1016/j.canlet.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 26.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 28.Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–1892. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- 29.Warfel NA, Niederst M, Newton AC. Disruption of the interface between the pleckstrin homology (PH) and kinase domains of Akt protein is sufficient for hydrophobic motif site phosphorylation in the absence of mTORC2. J Biol Chem. 2011;286:39122–39129. doi: 10.1074/jbc.M111.278747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 31.Saraf A, Oberg EA, Strack S. Molecular determinants for PP2A substrate specificity: charged residues mediate dephosphorylation of tyrosine hydroxylase by the PP2A/B' regulatory subunit. Biochemistry. 2010;49:986–995. doi: 10.1021/bi902160t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 33.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. Embo J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 34.Humphrey SJ, Yang G, Yang P, Fazakerley DJ, Stockli J, Yang JY, James DE. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013;17:1009–1020. doi: 10.1016/j.cmet.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent EE, Elder DJ, Thomas EC, Phillips L, Morgan C, Pawade J, Sohail M, May MT, Hetzel MR, Tavare JM. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer. 2011;104:1755–1761. doi: 10.1038/bjc.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seshacharyulu P, Pandey P, Datta K, Batra SK. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013;335:9–18. doi: 10.1016/j.canlet.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slupe AM, Merrill RA, Strack S. Determinants for Substrate Specificity of Protein Phosphatase 2A. Enzyme Res. 2011;2011:398751. doi: 10.4061/2011/398751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Inoki K, Yeung R, Guan KL. Regulation of TSC2 by 14-3-3 binding. J Biol Chem. 2002;277:44593–44596. doi: 10.1074/jbc.C200510200. [DOI] [PubMed] [Google Scholar]
- 42.Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shumway SD, Li Y, Xiong Y. 14-3-3beta binds to and negatively regulates the tuberous sclerosis complex 2 (TSC2) tumor suppressor gene product, tuberin. J Biol Chem. 2003;278:2089–2092. doi: 10.1074/jbc.C200499200. [DOI] [PubMed] [Google Scholar]
- 44.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Inoki K, Vacratsis P, Guan KL. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J Biol Chem. 2003;278:13663–13671. doi: 10.1074/jbc.M300862200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dennis MD, Kimball SR, Jefferson LS. Mechanistic target of rapamycin in complex 1 (mTORC1)-mediated phosphorylation is governed by competition between substrates for interaction with raptor. J Biol Chem. 2012;288:10–19. doi: 10.1074/jbc.M112.402461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson DL, Bolster DR, Kimball SR, Jefferson LS. Time course changes in signaling pathways and protein synthesis in C2C12 myotubes following AMPK activation by AICAR. Am J Physiol Endocrinol Metab. 2006;291:E80–89. doi: 10.1152/ajpendo.00566.2005. [DOI] [PubMed] [Google Scholar]
- 49.Miyazaki M, Esser KA. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am J Physiol Cell Physiol. 2009;296:C583–592. doi: 10.1152/ajpcell.00464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Inoki K, Guan KL. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol. 2004;24:7965–7975. doi: 10.1128/MCB.24.18.7965-7975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Effect of REDD1 on the interaction of 14-3-3 protein with TSC2.
Fig. S2: REDD1 promotes the dephosphorylation of Akt on Thr308.
Fig. S3. Regulation of the phosphorylation of Akt at Thr308 and the TSC2-TBC complex by REDD1.