ABSTRACT
Succinate:quinone oxidoreductase (Sdh) is a membrane-bound complex that couples the oxidation of succinate to fumarate in the cytoplasm to the reduction of quinone to quinol in the membrane. Mycobacterial species harbor genes for two putative sdh operons, but the individual roles of these two operons are unknown. In this communication, we show that Mycobacterium smegmatis mc2155 expresses two succinate dehydrogenases designated Sdh1 and Sdh2. Sdh1 is encoded by a five-gene operon (MSMEG_0416-MSMEG_0420), and Sdh2 is encoded by a four-gene operon (MSMEG_1672-MSMEG_1669). These two operons are differentially expressed in response to carbon limitation, hypoxia, and fumarate, as monitored by sdh promoter-lacZ fusions. While deletion of the sdh1 operon did not yield any growth phenotypes on succinate or other nonfermentable carbon sources, the sdh2 operon could be deleted only in a merodiploid background, demonstrating that Sdh2 is essential for growth. Sdh activity and succinate-dependent proton pumping were detected in cells grown aerobically, as well as under hypoxia. Fumarate reductase activity was absent under these conditions, indicating that neither Sdh1 nor Sdh2 could catalyze the reverse reaction. Sdh activity was inhibited by the Sdh inhibitor 3-nitroproprionate (3NP), and treatment with 3NP dissipated the membrane potential of wild-type or Δsdh1 mutant cells under hypoxia but not that of cells grown aerobically. These data imply that Sdh2 is the generator of the membrane potential under hypoxia, an essential role for the cell.
IMPORTANCE
Complex II or succinate dehydrogenase (Sdh) is a major respiratory enzyme that couples the oxidation of succinate to fumarate in the cytoplasm to the reduction of quinone to quinol in the membrane. Mycobacterial species harbor genes for two putative sdh operons, sdh1 and sdh2, but the individual roles of these two operons are unknown. In this communication, we show that sdh1 and sdh2 are differentially expressed in response to energy limitation, oxygen tension, and alternative electron acceptor availability, suggesting distinct functional cellular roles. Sdh2 was essential for growth and generation of the membrane potential in hypoxic cells. Given the essentiality of succinate dehydrogenase and oxidative phosphorylation in the growth cycle of Mycobacterium tuberculosis, the potential exists to develop new antituberculosis agents against the mycobacterial succinate dehydrogenase. This enzyme has been proposed as a potential target for the development of new chemotherapeutic agents against intracellular parasites and mitochondrion-associated disease.
INTRODUCTION
The genus Mycobacterium comprises a group of obligately aerobic bacteria that have adapted to inhabit a wide range of intracellular and extracellular environments. A fundamental feature of this adaptation is the ability to respire and generate energy from variable sources or to sustain metabolism in the absence of growth. To achieve this, mycobacteria use a respiratory chain that consists of two types of NADH dehydrogenase (types I and II), multiple succinate dehydrogenases/fumarate reductases (FRDs), a menaquinol (MQH2)-cytochrome c oxidoreductase termed the bc1 complex, and two terminal respiratory oxidases, an aa3-type cytochrome c oxidase (encoded by ctaBCDE) belonging to the heme-copper respiratory oxidase family and a cytochrome bd-type MQH2 oxidase (cydABCD) (1–5). The regulator(s) and molecular signals that control the expression of these complexes in response to environmental change remain unknown.
Succinate dehydrogenase forms complex II of the respiratory chain and couples oxidative phosphorylation to central carbon metabolism by being an integral part of the citric acid cycle (6). Succinate dehydrogenase couples the oxidation of succinate to fumarate in the cytoplasm to the reduction of quinone to quinol in the membrane. The reverse reaction can be catalyzed by fumarate reductase, which is generally found in anaerobic or facultative anaerobes that utilize low-potential quinols (MQH2 Eo′ = −74 mV) to reduce fumarate as the final step in the anaerobic electron transport chain (7–9). Succinate dehydrogenase and fumarate reductase have the same overall structural architecture (10, 11), and the reaction catalyzed (i.e., succinate oxidation or fumarate reduction) cannot be predicted solely on the basis of the primary amino acid sequence of the enzyme subunits. In general, increased reaction rates and catalytic efficiency in a particular direction reflect whether the enzyme is a succinate dehydrogenase or a fumarate reductase (12, 13).
Both succinate dehydrogenase and fumarate reductase consist of a large soluble domain and a smaller membrane-bound domain (membrane anchor) (10, 11). The soluble domain contains two hydrophilic subunits designated A and B. Flavoprotein subunit A contains a covalently bound flavin adenine dinucleotide (FAD) cofactor and the catalytic substrate-binding site. Subunit B is a small iron-sulfur protein containing three distinct iron-sulfur clusters that mediate electron transfer between the flavin and quinone catalytic sites in the membrane-bound domain. Subunits A and B have high sequence similarity between different bacterial species. The smaller membrane-bound domain varies between bacterial species consisting of one large or two small hydrophobic subunits designated C or C and D. Variation also occurs in the number of heme b groups (none, one, or two) and the type of quinone used (menaquinone or ubiquinone) (14).
Most mycobacterial genomes harbor two annotated succinate dehydrogenases, designated Sdh1 and Sdh2 (15). The oxidation of succinate to fumarate (Eo′ = +30 mV) and reduction of ubiquinone to ubiquinol (Eo′ = +113 mV) is a thermodynamically favorable reaction (ΔG° = −15 kJ/mol). In mycobacteria, the oxidation of succinate to fumarate (Eo′ = +30 mV) is coupled to menaquinone reduction to MQH2 (Eo′ = −74 mV), an endergonic reaction under standard conditions (ΔG° = +21 kJ/mol) (16). How this thermodynamically unfavorable reaction is driven in mycobacteria is not known. In M. smegmatis, two putative operons for succinate dehydrogenase are present, Sdh1 (MSMEG_0416-MSMEG_0420) and Sdh2 (MSMEG_1672-MSMEG_1669) (15). These M. smegmatis sdh operons are contiguous with the two annotated sdh operons of M. tuberculosis designated Sdh1 (Rv0249c-Rv0247c) and Sdh2 (Rv3316-Rv3319). M. smegmatis does not harbor genes for fumarate reductase, making it a genetically tractable model to dissect the roles of the individual sdh operons. Here, we report that the sdh1 and sdh2 operons of M. smegmatis are differentially expressed in response to carbon limitation, hypoxia, and fumarate. Sdh1 was nonessential for growth, but Sdh2 was essential and generates the membrane potential under hypoxia.
RESULTS
M. smegmatis expresses two distinct succinate dehydrogenase operons
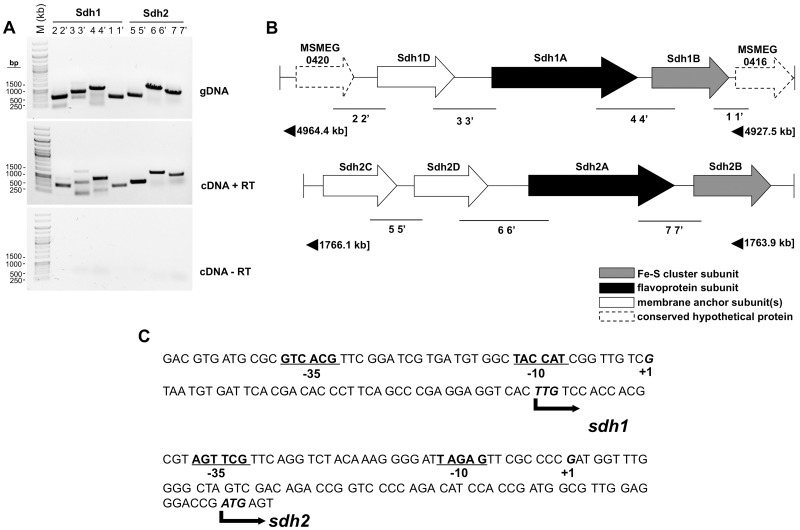
The operon structure of the two putative succinate dehydrogenases in M. smegmatis was determined by reverse transcriptase PCR (RT-PCR) with appropriate controls (Fig. 1A). On the basis of these data, we confirmed the operons as sdh1 (MSMEG_0420-MSMEG_0416) (Fig. 1B, top panel) and sdh2 (MSMEG_1672-MSMEG_1669) (Fig. 1B, bottom panel). BLAST searches based on the predicted translation products determined that the operon structure of sdh2 is similar to that of the canonical Escherichia coli succinate:quinone oxidoreductase (SQR) enzyme in that it contains a putative catalytic flavoprotein subunit (Sdh2A), a soluble iron-sulfur cluster protein (Sdh2B), and two integral membrane subunits (Sdh2C and Sdh2D) (Fig. 1B). sdh1, on the other hand, is operonic with two unknown hypothetical proteins (MSMEG_0420 and MSMEG_0416) and has an integral membrane protein that appears to be a fusion of the expected two membrane subunits (Sdh1D) (Fig. 1B). These differences between the Sdh1 and Sdh2 operon structures suggest that they may play distinct cellular and energetic roles.
FIG 1 .
Transcriptional organization and start site mapping of sdh1 and sdh2 gene clusters in M. smegmatis. (A) Gels showing RT-PCR products encompassing the start, end, and intergenic region of two adjacent genes. Numbers above lanes correspond to amplified regions indicated in panel B. Each primer pair (see Table S2 in the supplemental material) was tested with the genomic DNA (gDNA) (top gel), cDNA synthesized with RT (cDNA + RT) (middle gel), and a control reaction mixture excluding RT (bottom gel) (cDNA-RT). Lane M, 1-kb DNA ladder molecular size marker. (B) Schematic view of the sdh1 (MSMEG_0416-MSMEG_0420) and sdh2 (MSMEG_1672-MSMEG_1669; sdh2C-sdh2B) gene clusters, including the predicted function of each subunit. (C) Upstream nucleotide sequence illustration showing the TSSs of sdh1 and sdh2, located 39 and 62 bp upstream from the TTG and ATG codons (labeled +1 and in bold), respectively, identified by 5′ RACE. Bold and underlined −10 and −35, promoter elements.
Sdh1 and Sdh2 protein sequence alignments show differences in heme-binding and catalytic residues.
Using the functional classification of Hågerhåll (17), the M. smegmatis Sdh1 and Sdh2 enzymes are succinate:menaquinone oxidoreductases and belong to subclass 3. These enzymes are characterized by the oxidation of succinate coupled to the reduction of a low-potential quinone (menaquinone) in the respiratory chain. On the basis of their membrane-bound domain (subunit C or subunits C and D) and heme content, succinate dehydrogenases can be classified into five different types (18–20). According to this classification, Sdh2, with its two heme groups and two small hydrophobic subunits (subunits C and D) (Fig. 1B), can be classified as type A, previously reported in some extremophiles (21). Sdh1, with its large single hydrophobic subunit, C (Sdh1D) (Fig. 1B), can be classified as a type B enzyme; type B enzymes are found in a wide variety of microorganisms (reviewed in reference 14). Given that the operon structure of Sdh1 is similar to that of SQR subclasses known to have different numbers of hemes involved in quinone interactions (22), we made protein sequence alignments of Sdh1D and Sdh2D to representative SQR/MQH2:fumarate oxidoreductase (QFR) sequences with experimentally determined numbers of hemes and also with the SQR- and QFR-encoding genes found in M. tuberculosis and other members of the family Corynebacterineae (see Fig. S1 and S2 in the supplemental material). We aligned only Sdh1D with other fused subunits and Sdh2D with other nonfused subunits to avoid erroneous alignments. It has been previously determined that conserved histidines are required for the binding of heme molecules proximal and distal to the cytoplasmic membrane interface (23) and so used this to make predictions of the heme contents of Sdh1 and Sdh2.
Sdh2D and Sdh1D appeared to have histidines that aligned in a pattern consistent with the presence of both a proximal and a distal heme (see Fig. S1 and S2). In addition, other closely related Corynebacterineae species (Gordonia, Nocardia, and Rhodococcus) displayed high sequence identity with Sdh1 homologues, including the conserved histidine positions. This prediction suggests that Sdh1 and Sdh2 may have structural specializations to facilitate interactions with menaquinones, relative to bacteria containing differing types of quinone. Alignments of Sdh2C and other C subunits gave the same result as Sdh2D alignments (data not shown). We aligned mycobacterial flavoprotein SdhA subunits with those of E. coli. It has been previously shown that differences in the FAD-binding site correspond to either efficient SQR or efficient QFR activity (13). Correspondingly, the known fumarate reductase in M. tuberculosis has the glutamate required for efficient E. coli QFR activity (see Fig. S3), as does the mycobacterial Sdh1. Sdh2 has a glutamine that is required for efficient SQR activity in E. coli. These analyses point to biochemical differences in M. smegmatis Sdh1 and Sdh2 that may relate to the physiological function of each enzyme and the conditions under which they operate.
sdh-lacZ transcriptional fusions are differentially regulated in response to energy limitation, hypoxia, and fumarate.
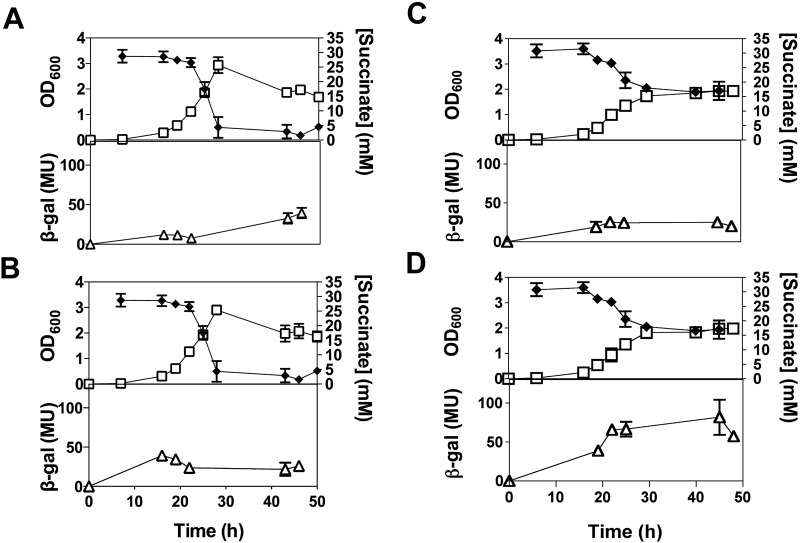
To facilitate the design of sdh-lacZ fusions, the transcriptional start sites (TSSs) of the sdh1 and sdh2 operons were determined by 5′ rapid amplification of cDNA ends (RACE) (Fig. 1C). Using this information, we constructed sdh1- and sdh2-lacZ transcriptional promoter fusions and studied operon promoter expression in response to carbon limitation (succinate) and oxygen limitation (hypoxia) (Fig. 2). When cells were grown aerobically (normoxia) with succinate (30 mM) as the sole carbon and energy source, sdh1-lacZ expression increased in response to the depletion of succinate (Fig. 2A). The expression of sdh2-lacZ remained relatively unchanged throughout the growth cycle (Fig. 2B). Under hypoxia (oxygen limitation), with succinate-replete conditions, the expression of sdh1-lacZ remained relatively constant during the growth cycle (Fig. 2C), while sdh2-lacZ expression increased in response to hypoxia (Fig. 2D).
FIG 2 .
Levels of sdh1-lacZ and sdh2-lacZ expression in M. smegmatis. M. smegmatis cells were grown in HdB minimal medium supplemented with 30 mM succinate under normoxic (conical flasks) or hypoxic (stoppered serum vials) conditions. Hypoxic conditions were achieved at approximately 25 to 30 h, as indicated by methylene blue decolorization. The graphs show how sdh1-lacZ expression under normoxic (A) and hypoxic (C) conditions and sdh2-lacZ expression under normoxic (B) and hypoxic (D) conditions affect OD600 (open squares), the succinate concentration (solid diamonds), and β-galactosidase (β-gal) activity (in Miller units [MU]; open triangles). The experiments were carried out in biological triplicate, and the results shown are mean values and standard deviations. The empty vector pJEM15 was used as a control and gave no detectable activity (data not shown).
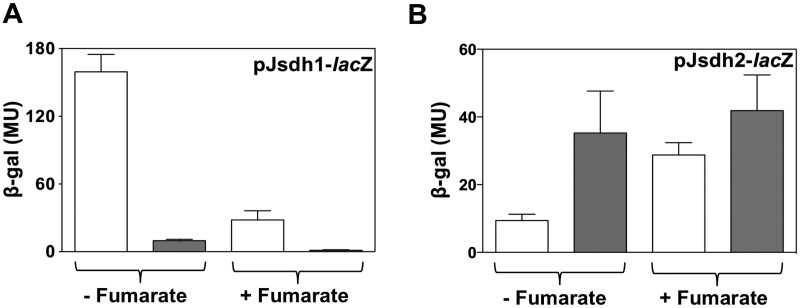
Fumarate reductase and succinate dehydrogenase are closely related enzymes, and the preferred reaction catalyzed cannot be predicted on the basis of the amino acid sequence alone. The effect of fumarate on sdh expression was investigated under normoxic and hypoxic conditions (Fig. 3). When cells were grown on glycerol, sdh1-lacZ expression was repressed by the addition of fumarate under both normoxic and hypoxic conditions (Fig. 3A). In contrast, the expression of sdh2-lacZ was increased by the addition of fumarate under both normoxic and hypoxic conditions (Fig. 3B).
FIG 3 .
Effect of fumarate on sdh1-lacZ and sdh2-lacZ expression. M. smegmatis cells were grown in HdB minimal medium supplemented with 0.2% glycerol with (+) or without (−) 30 mM fumarate under normoxic (open bars) or hypoxic (gray bars) conditions. Panels: A, sdh1-lacZ expression; B, sdh2-lacZ expression. The experiments were carried out in biological triplicate, and the results shown are mean values and standard deviations. MU, Miller units; β-gal, β-galactosidase.
The sdh2 operon is essential for M. smegmatis growth.
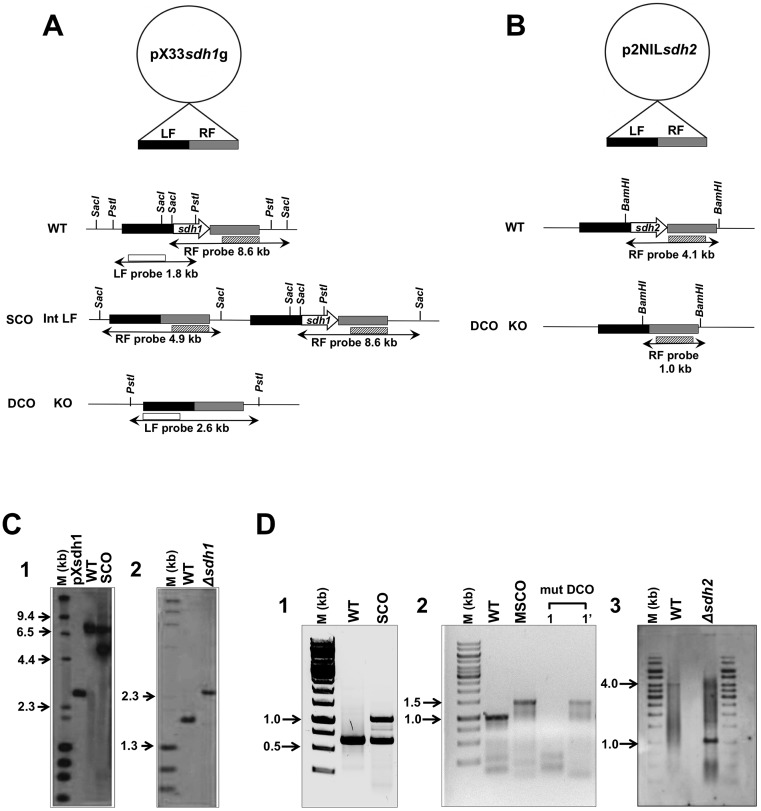
To dissect the roles of Sdh1 and Sdh2, we attempted to create gene deletions of both sdh operons. For deletion of sdh1, a markerless deletion construct to replace the entire sdh1 operon was made and introduced into M. smegmatis as described in the legend to Fig. 4. Single-crossover (SCO) and double-crossover (DCO) events were screened by PCR and Southern hybridization (Fig. 4C). The increased size of the left flank product in a DCO event indicated successful deletion of the sdh1 operon, thereby showing that this operon is nonessential for the growth of M. smegmatis.
FIG 4 .
Construction of markerless sdh1 and sdh2 operon deletion mutants of M. smegmatis. Schematic diagrams of the vectors pX33sdh1g (A) and p2NILsdh2 (B) with the left flank (LF) and right flank (RF) of the sdh1 and sdh2 operons, integration of the suicide vector into the chromosome, and subsequent deletion of the sdh1 and sdh2 operons are shown. Relevant restriction sites: SacI for SCO event and PstI for DCO event screens for the sdh1 operon; BamHI for the DCO event screen for the sdh2 operon. Fragment sizes detected by Southern hybridization are indicated. KO, knockout. Southern blot analysis of pX33sdh1g vector integration due to a SCO event (C1) yielding the 4.9-kb fragment (lane SCO). The WT M. smegmatis chromosomal DNA, yielding an 8.6-kb fragment, was used as a positive control (lane WT). Plasmid pXsdh1 was also used as a control (lane pXsdh1). Confirmation of sdh1 operon nonessentiality (C2). The probe used to perform the hybridization contained the 1.8-kb WT and 2.6-kb deletion mutant sdh1 operon (Δsdh1) restriction fragments, respectively. Lanes M, HindIII-digested λ DNA molecular size marker (D1). Identification of sdh2 SCO strain by colony PCR. SCO strains were generated by homologous recombination of p2Nbk-sdh2 with a chromosomal copy of the sdh2 operon. Chromosomal DNA of M. smegmatis, yielding the 0.6-kb fragment (lane WT), was used as a positive control; the p2NILsdh2 vector integration due to a SCO event yielded the 1.0-kb fragment (lane SCO) (D2). Colony PCR of the generated DCO strain. Lanes 1 and 1′ represent samples from the same cell line. The potential DCO cell lines were screened for both the WT copy (designated 1) and the sdh2 operon deletion mutant (designated 1′). The lengths of the expected PCR products for the WT and deletion mutant sdh2 operons were 0.9 and 1.4 kb, respectively. A merodiploid SCO (MSCO) strain was used as a positive control (D3). A Southern blot analysis of the mutant (mut) DCO strain is shown. The probe used to perform the hybridization contained the 4.1-kb WT and 1.0-kb deletion mutant sdh2 operon (Δsdh2) restriction fragments, respectively. Lanes M, 1-kb DNA ladder molecular size marker. Drawings are not to scale.
Using a similar strategy (Fig. 4B), Δsdh2 mutants could not be isolated. A total of 82 potential DCO events were screened, and 74 were found to be wild-type (WT) DCOs while the other 8 proved to be spontaneous sucrose-resistant SCO strains. Screening was also performed with medium supplementation with either succinate or fumarate (1 to 30 mM concentrations of each were tested); however, this could not rescue the lethal sdh2 phenotype. The fact that we could not isolate an sdh2 deletion mutant suggested that the sdh2 operon might be essential for M. smegmatis growth. To confirm the essentiality of sdh2, we constructed a merodiploid strain carrying an additional copy of the sdh2 operon expressed from its native promoter by using the mycobacteriophage L5-based integrating pUHA267 vector, which integrates into the attB chromosomal site of the genome. We integrated this complementing vector into the SCO strain and screened for DCO events in this merodiploid background. Of 26 potential DCOs screened at the native locus, 25 were of the WT and one sdh2 deletion mutant was isolated, which was confirmed by the smaller size of a left flank product than that of the WT in a Southern blot assay (Fig. 4D). This shows that the sdh2 operon can be deleted at its native locus but only if a functional copy of the sdh2 operon is supplied elsewhere on the chromosome; therefore, we conclude that the sdh2 operon is essential for the growth of M. smegmatis.
We analyzed the Δsdh1 mutant for a growth phenotype over a wide range of succinate concentrations (Fig. 5A and B). No significant differences in growth rate or yield (final optical density at 600 nm [OD600]) were observed. We rationalized that this was due to upregulation (compensation) of Sdh2 in the Δsdh1 background. To test this hypothesis, we measured the expression of sdh2-lacZ in a Δsdh1 mutant background (Fig. 5C and D) and compared it to sdh2-lacZ expression in WT cells (Fig. 2B and D). The expression of sdh2-lacZ was elevated in the Δsdh1 mutant background relative to that in the WT background under both normoxic and hypoxic growth conditions (compared Fig. 5C and D with 2B and D), suggesting that Sdh2 was compensating for the loss of Sdh1 in M. smegmatis during growth on succinate.
FIG 5 .
Growth and sdh2-lacZ expression of Δsdh1 mutant. Shown are the growth curves (A) and plotted growth rates (B) of the WT (solid lines and filled bars) and the Δsdh1 deletion mutant (dotted lines and open bars) in HdB minimal medium under normoxic conditions with succinate at 30 mM (closed circles), 15 mM (closed squares), 5 mM (closed triangles), and 1 mM (inverted closed triangles). The specific growth rate was calculated between the two time points indicated in panel A. (C and D) Expression levels of sdh2-lacZ in the M. smegmatis Δsdh1 mutant grown under normoxic (C) or hypoxic (D) conditions in HdB minimal medium supplemented with 30 mM succinate. (D) Hypoxic conditions were achieved at approximately 30 h, as indicated by methylene blue decolorization. OD600 (open squares), succinate concentration (solid diamonds), β-galactosidase (β-gal) activity (in Miller units [MU]; open triangles) are shown. The experiments were carried out in biological triplicate, and the results shown are mean values and standard deviations.
Enzyme activities of the Sdh1 and Sdh2 complexes in M. smegmatis.
In order to understand the biochemical differences between WT M. smegmatis and the Δsdh1 mutant; we prepared inverted (inside-out) membrane vesicles (IMVs) of both WT and Δsdh1 mutant cells grown under both normoxic and hypoxic conditions on succinate (Hartman’s de Bont [HdB] minimal medium). We assayed for Sdh activity by using the dye-based phenazine ethosulfate (PES)–2,6-dichlorophenolindophenol (DCIP) assay (24) with appropriate E. coli controls (positive and negative) to validate our enzyme assays (Fig. 6A and B). IMVs of E. coli exhibited high rates of succinate dehydrogenase activity and succinate-dependent oxygen consumption (Fig. 6A and B). Both enzyme activities were essentially absent from an E. coli ΔfrdABCD ΔsdhABCD double mutant (strain RP437; see Table S1 in the supplemental material) and well above WT levels in E. coli strains overexpressing either the SQR operon (pFAS, SdhABCD+) or the QFR operon (pH3, FrdABCD+) (Fig. 6A and B). IMVs prepared from M. smegmatis cells grown aerobically had similar kinetics of succinate oxidation, regardless of the presence or absence of Sdh1 (Fig. 6C). IMVs prepared from Δsdh1 mutant cells grown under hypoxic conditions exhibited a 2-fold lower apparent Vmax than WT cells (Fig. 6D; see Table S3). The apparent Km values for succinate were consistently lower in the Δsdh1 mutant than in the isogenic WT parent, suggesting a higher affinity for succinate oxidation in the Δsdh1 genetic background (see Table S3).
FIG 6 .
Kinetic characterization of succinate oxidation by IMVs of WT and Δsdh1 mutant E. coli and M. smegmatis. Succinate dehydrogenase activities in IMVs of WT and Δsdh1 mutant E. coli and M. smegmatis strains were measured with the PES-DCIP assay (A) or a Clark O2 electrode-based method (B). IMVs of E. coli were prepared from cells grown aerobically in LB medium at 37°C. MG1655, E. coli WT; RP437, E. coli ΔfrdABCD ΔsdhABCD double mutant; pFAS, E. coli overexpressing the sdhABCD operon; pH3, E. coli overexpressing the frdABCD operon. (C and D) Rates of succinate oxidation by WT and Δsdh1 mutant M. smegmatis IMVs prepared from cells grown under normoxic (C) and hypoxic (D) conditions in HdB minimal medium supplemented with 30 mM succinate. Data were analyzed by nonlinear least-squares regression of the Michaelis-Menten equation with GraphPad Prism 6, and Vmax and Km data are summarized in Table S3 in the supplemental material. WT, solid squares; SDH2 (Δsdh1 deletion mutant strain), solid triangles. Error bars indicate the standard error of a biological triplicate.
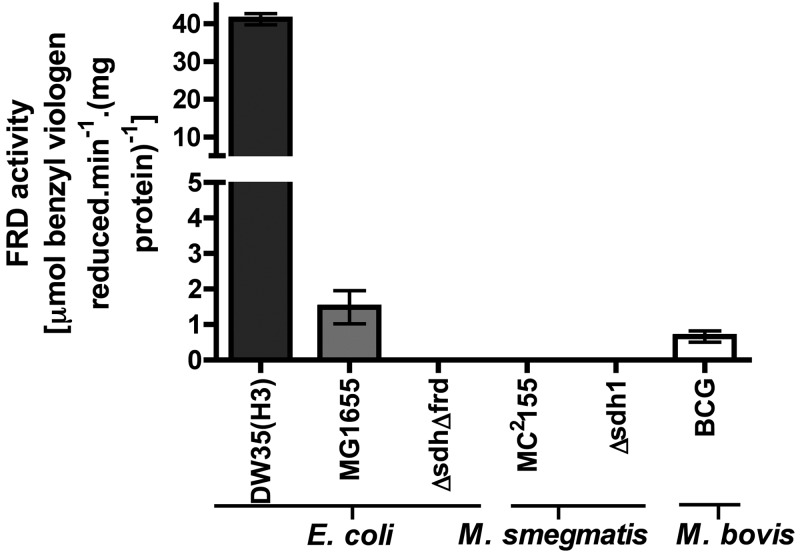
To determine if the M. smegmatis Sdh enzymes are capable of fumarate reduction, we assayed for fumarate reductase activity in mycobacterial IMVs. A direct assay for fumarate reductase activity in mycobacteria has not been previously reported in the literature. We used a benzyl viologen (BV) assay that has been described for use with the E. coli QFR enzyme (13). We validated that the assay was correctly working by detecting fumarate reductase activity in E. coli IMVs that were either WT or overexpressing the E. coli QFR (DW35 pH3 FrdABCD+) (Fig. 7). Consistently, an E. coli ΔfrdABCD ΔsdhABCD double mutant (strain RP437) gave no detectable activity. Furthermore, we were able to detect fumarate reductase activity in IMVs prepared from M. bovis BCG (Fig. 7). However, we were unable to detect any fumarate reductase activity in IMVs prepared from WT or Δsdh1 mutant M. smegmatis grown under any of the conditions used in this study (Fig. 7).
FIG 7 .
Fumarate reductase activities in E. coli, M. bovis BCG, and M. smegmatis IMVs. Fumarate reductase (FRD) activities (in µmol of BV oxidized min−1 mg of protein−1) were calculated with BV as the electron donor (0.2 mM) under anaerobic conditions. No fumarate reductase activity could be detected in IMVs prepared from M. smegmatis cells grown under either normoxic or hypoxic conditions on HdB minimal medium supplemented with 30 mM succinate. IMVs of E. coli and M. bovis BCG were prepared from cells grown aerobically in LB and 7H9 media with ADS at 37°C. DW35(H3), E. coli overexpressing the frdABCD operon; MG1655, E. coli WT; E. coli ΔfrdABCD ΔsdhABCD double mutant (RP437). Fumarate (5 mM) was present in each assay. Error bars indicate the standard error of a technical triplicate.
Sdh2 is the generator of the membrane potential under hypoxia.
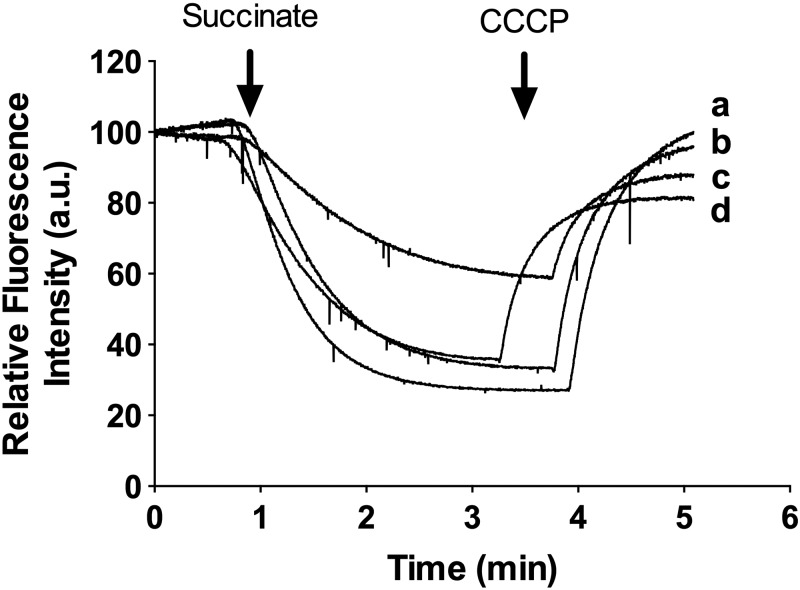
To understand the importance of succinate oxidation in M. smegmatis, we determined the contributions of succinate oxidation to the membrane potential (Δψ) and proton gradient (ΔpH). Succinate oxidation coupled to proton translocation was measured by using acridine orange (AO) quenching in IMVs prepared from M. smegmatis cells grown under normoxia or hypoxia on succinate (Fig. 8). In these experiments, succinate was provided as the electron donor for the electron transport chain. Succinate oxidation to fumarate results in the generation of MQH2, which is then used by either the proton-pumping MQH2-cytochrome c oxidoreductase bc1-aa3-type cytochrome c oxidase (bc1-aa3 pathway) and/or non-proton-translocating cytochrome bd-type MQH2 oxidase (1, 2). Succinate oxidation by either WT or Δsdh1 mutant IMVs resulted in significant proton pumping (AO quenching), which was reversed by the protonophore m-chlorophenylhydrazone (CCCP), demonstrating that succinate oxidation was coupled to proton pumping by the bc1-aa3 pathway (Fig. 8). Lower levels of succinate-driven proton pumping (quenching) were observed in WT cells grown under hypoxia (Fig. 8, trace c) than in the Δsdh1 mutant grown under the same conditions (Fig. 8, trace d).
FIG 8 .
Succinate-driven proton translocation in IMVs of M. smegmatis (WT versus Δsdh1 mutant). Quenching of AO fluorescence in IMVs was initiated with 5 mM succinate, and at the indicated time points, the uncoupler CCCP at 50 µM was added to collapse the proton gradient (reversal of AO fluorescence). IMVs were prepared from M. smegmatis cells grown under normoxic (traces a and b) or hypoxic (traces c and d) conditions in HdB minimal medium supplemented with 30 mM succinate. Traces were normalized to a starting value of 100 arbitrary units (a.u.). Experiments are representative of a technical triplicate. Traces: a, WT cells grown under normoxic conditions; b, Δsdh1 mutant cells grown under normoxic conditions; c, WT cells grown under hypoxic conditions; d, Δsdh1 mutant cells grown under hypoxic conditions.
Succinate dehydrogenase activity in M. tuberculosis is inhibited by 3-nitroproprionate (3NP) (25), which is a complex II-specific suicide inhibitor (26). We validated this as an inhibitor of Sdh activity in M. smegmatis (see Fig. S4 in the supplemental material) and used it to study the effect of 3NP treatment on the generation of the electrical potential across the cell membrane (ΔΨ) in whole cells. When WT or Δsdh1 mutant cells were grown under normoxia in the presence of 3NP (400 µM), the growth rate and final OD600 were the same as those of untreated cells (Fig. 9A and B). The same strains grown under hypoxia in the presence of 3NP showed some growth inhibition, and the final OD600 was always lower in the 3NP-treated cells, particularly for the Δsdh1 mutant (Fig. 9C and D). To determine the effect of 3NP on succinate dehydrogenase inhibition and electron transport chain activity, we measured the Δψ under these conditions. Under normoxic growth conditions, the Δψ was comparable between WT and Δsdh1 mutant cells (170 mV) and 3NP treatment had no significant effect on the Δψ (Fig. 9E, cells sampled at 48 h) (P value, >0.05; 95% confidence interval [CI], one-way analysis of variance [ANOVA]), suggesting that generation of the membrane potential under these conditions was not dependent on succinate or was a minor contributor. Under hypoxic conditions, the Δψ was significantly dissipated by 3NP (P value, <0.001 for WT untreated versus 3NP-treated cells; P value, <0.01 for Δsdh1 untreated versus 3NP-treated cells, 95% CI, one-way ANOVA) in both the WT and Δsdh1 mutant. Taken together, these data suggest that succinate oxidation (Sdh activity) was responsible for generation of the Δψ under hypoxia but not under normoxia and was not dependent on the presence of the sdh1 operon for this activity.
FIG 9 .
Effect of 3NP on the growth and membrane potential of the WT and the Δsdh1 mutant. Cells (WT, squares; Δsdh1 mutant, triangles) were grown under normoxic (A and B) or hypoxic (C and D) conditions in HdB minimal medium with 30 mM succinate in the absence or presence of 400 µM 3NP. The dotted vertical line (approximately 40 h) indicates the point at which methylene blue was decolorized and hence hypoxic conditions were achieved. At 48 h of growth, the membrane potential from normoxic (E) and hypoxic (F) cells was determined as described in Materials and Methods. Error bars represent the standard deviation (A to D) or the standard error (E and F) of a biological triplicate. Ethanol was used as the vehicle in all of the experiments.
DISCUSSION
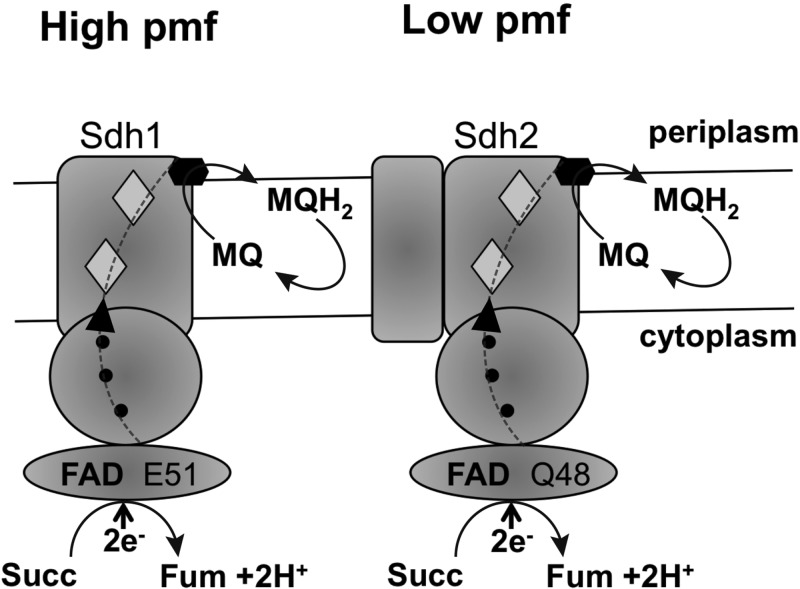
Members of the genus Mycobacterium harbor two copies of the succinate dehydrogenase operon designated sdh1 and sdh2, but the reasons for this remain unclear. In this communication, we report that the sdh operons of M. smegmatis are differentially expressed in response to energy and oxygen limitations and fumarate, suggesting distinct cellular and energetic roles. The sdh1 operon was upregulated in response to energy limitation (normoxic conditions, high proton motive force) and downregulated in response to hypoxia and fumarate (Fig. 10). Sdh1 was dispensable for growth, and no phenotype for the Δsdh1 mutant was identified. In contrast, Sdh2 was downregulated in response to energy limitation and upregulated in response to hypoxia (low proton motive force) and fumarate (Fig. 10). The sdh expression data obtained with sdh-lacZ fusions were consistent with microarray data obtained for the sdh1 and sdh2 genes, when bacteria were grown in continuous culture in response to the growth rate and hypoxia (15). Exogenous fumarate addition induced sdh2 expression, even though Sdh2 was without measured fumarate reductase activity. One possible reason for this increased sdh2 expression is that fumarate accumulation signals hypoxia to M. smegmatis. The fact that fumarate addition increased sdh2 expression under both normoxic and hypoxic conditions supports this hypothesis.
FIG 10 .
Schematic diagram outlining the subunit structures and proposed functions of Sdh1 and Sdh2 in M. smegmatis. Sdh1 is expressed under normoxia (high proton motive force [pmf]), and Sdh2 is expressed under hypoxia (low pmf). Both enzymes catalyze the oxidation of succinate (Succ) to fumarate (Fum) and the two-electron (2e−, dotted line) reduction of menaquinone (MQ) to MQH2. Sdh1 is composed of three subunits (A, B, and D, type A enzyme), and Sdh2 is a four-subunit enzyme (A to D, type B enzyme). Shaded diamonds represent proposed proximal and distal hemes in the membrane anchor domain. Solid circles indicate proposed iron-sulfur clusters in subunit B. The solid hexagon is a proposed menaquinone-binding site.
Sdh2 was essential for growth, even in an Sdh1+ background, demonstrating that Sdh1 was unable to compensate for Sdh2 activity in M. smegmatis. The essentiality of Sdh enzymes for mycobacterial growth has previously been reported in a transposon site hybridization (TraSH) screening of M. tuberculosis that showed that Sdh1, but not Sdh2, is essential for optimal growth under aerobic conditions on standard laboratory medium (27). In contrast, TraSH screening selecting for M. tuberculosis mutants that continue to replicate under hypoxic conditions found that sdh2 mutants were overrepresented, suggesting that Sdh2 has a pivotal role in the transition of M. tuberculosis from aerobic to hypoxic conditions (28). The reasons for this essentiality, despite the presence of one intact copy of an sdh operon, has not been investigated.
The M. smegmatis Sdh1 and Sdh2 enzymes are succinate:menaquinone oxidoreductases. The oxidation of succinate to fumarate (Eo′ = +30 mV) coupled to menaquinone reduction to MQH2 (Eo′ = −74 mV) is an endergonic reaction under standard conditions (ΔG° = +21 kJ/mol) (16), and therefore, these bacteria face a thermodynamic problem. Bacillus species also use succinate:menaquinone oxidoreductases, and in Bacillus subtilis and Bacillus licheniformis, the endergonic succinate dehydrogenase reaction is driven by the proton motive force (29–31). The succinate-menaquinone reductases generally contain two heme b groups in the membrane anchor (14) (Fig. 10). These hemes are referred to as the proximal heme (bp), located close to the negative side of the membrane (near the hydrophilic subunits), and the distal heme (bD), close to the positive side of the membrane near the menaquinone-binding site. The hemes differ in redox potential, with heme bp having a high potential (e.g., Eo′ = +42 mV in B. subtilis) and heme bD having a low potential (e.g., Eo′ = −131 mV in B. subtilis) (19, 22, 30, 32). Mutations in the axial ligands (His-28 and His-113) for heme bD in B. subtilis lead to impaired electron transfer from succinate to menaquinone, demonstrating that heme bD is essential for electron transfer to menaquinone (32). In mycobacterial Sdh, a similar pair of histidines are found at His-65 to His-113 (Sdh1, M. smegmatis numbering) and His-65 to His-107 (Sdh2, M. smegmatis numbering) for potential heme b ligation, suggesting that this adaptation is conserved in the mycobacterial Sdh enzymes. Neither the Sdh1 nor the Sdh2 enzyme was able to catalyze the reduction of fumarate to succinate under the conditions tested. Our data support a model in which Sdh1 is adapted to high redox potentials (normoxic conditions) and Sdh2 is adapted to low redox potentials (hypoxic conditions) (Fig. 10). Such adaptations have been linked to the FAD cofactor of SQR/QFR in E. coli. The redox potentials of the FAD in SQR and QFR are −138 and −122 mV, respectively (13). Analysis of the amino acids of SQR and QFR involved in flavin binding has shown that in E. coli FrdA, Glu-49 is located close to the FAD cofactor and in E. coli SdhA, a Gln-50 residue is found at this position (13). Replacement of these residues with the residue found in the complementary enzyme (i.e., FrdA E49Q or SdhA Q50E) results in the enzyme becoming more efficient in the opposite direction (13). In mycobacterial Sdh1, Sdh2, and fumarate reductase enzymes, these same residues are conserved: SdhA1 Glu-51, SdhA2 Gln-48, and M. tuberculosis Frd Glu-51 (see Fig. S3 in the supplemental material). These observations raise the possibility that the reduction potential of the FAD is different in M. smegmatis Sdh1 and Sdh2, consistent with the enzymes functioning under different redox potentials. On the basis of E. coli data, one might speculate that M. smegmatis Sdh1 is more akin to QFR than Sdh2, which resembles SQR of E. coli.
The reason for the essentiality of Sdh2 in M. smegmatis remains unknown. Our data demonstrate an essential role for Sdh2 in generating the membrane potential of M. smegmatis under hypoxia but not under normoxic conditions, suggesting that succinate is the preferred respiratory electron donor at low oxygen tensions in M. smegmatis. The succinate dehydrogenase inhibitor 3NP was without effect on M. smegmatis cells growing aerobically but caused some growth inhibition under hypoxia, suggesting that blocking succinate dehydrogenase has different implications for cells growing under normoxia or hypoxia. These data suggest different carbon flux through succinate under the two conditions and the essentiality of succinate dehydrogenase in membrane potential generation under hypoxia. We propose an energetic scheme for M. smegmatis in which Sdh2-mediated succinate oxidation generates a reduced MQH2 pool that saturates the proton-pumping bc1-aa3 supercomplex, leading to the generation of the membrane potential and sustaining ATP synthesis under hypoxia. A similar mechanism has been proposed in M. tuberculosis, but the Sdh responsible for this process is not known (25). Rhee and coworkers (25) have proposed that succinate accumulation in M. tuberculosis during hypoxia plays an essential role in fueling Sdh activity to sustain the membrane potential, ATP synthesis, and anaplerosis. If oxygen is completely lacking, excess succinate may be secreted to sustain the membrane potential or stored to enable immediate resumption of growth when electron acceptors become available, akin to a metabolic battery (25). In our experiments, the membrane potential was still generated in hypoxic M. smegmatis cells even when the external succinate concentration was high (15 mM), indicating that the primary mechanism of membrane potential generation involved succinate oxidation coupled to proton pumping by the bcc-aa3 complex and not succinate secretion per se. Our findings and those of others (25) are consistent with mitochondrial studies where respiratory succinate dehydrogenase (complex II) is the primary generator of the membrane potential and ATP levels under hypoxic conditions and is more efficient than complex I under these conditions (33). The use of succinate and Sdh2 by M. smegmatis to generate the membrane potential may explain why the proton-pumping NADH oxidoreductase complex I (nuo operon, NDH-1) is downregulated under hypoxic conditions in M. smegmatis (15). The use of Sdh2 in favor of NDH-1 under these conditions may well prevent the excess production of reactive oxygen species (ROS) mediated by complex I, a major contributor to ROS production in bacteria and mitochondria.
M. smegmatis lacks genes for a canonical fumarate reductase, and we could not detect fumarate reductase activity in cells grown under either normoxic or hypoxic conditions. We could readily detect fumarate reductase activity in M. bovis BCG cells. It has been proposed that fumarate may be an important endogenous electron acceptor for energy production and maintenance of redox balance (oxidation of NADH to NAD+) in hypoxic nonreplicating mycobacteria (34) or used as a mechanism to generate succinate as an excreted end product for maintenance of the membrane potential (35). Interestingly, the use of fumarate as an electron acceptor in E. coli requires complex I, and expression of the nuo operon is stimulated by the presence of fumarate (7). This stands in direct contrast to M. smegmatis, where the nuo operon seems to be silent under hypoxic conditions (15).
Complex II plays an important role in the cellular physiology of eukaryotes and prokaryotes, and its malfunction has been associated with human disease (36, 37). The enzyme has been proposed as a potential target for the development of new chemotherapeutic agents against intracellular parasites (38) and mitochondrion-associated disease (39). Given the essentiality of succinate dehydrogenase and the importance of fumarate reductase in mycobacterial pathogens, the potential exists to develop new antituberculosis agents against these two respiratory complexes.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All of the strains and plasmids used in this study are listed in Table S1 in the supplemental material. Plasmid propagation was carried out with E. coli strains grown in Luria-Bertani (LB) medium at 37°C with agitation (200 rpm) or on LB 1.5% agar plates. M. smegmatis strain mc2155 (40) and derived strains were routinely grown at 37°C with agitation (200 rpm) in HdB minimal medium (41) supplemented with 0.05% Tween 80 (Sigma-Aldrich). Where specified, 25 mM glycerol, 30 mM succinate, or 30 mM fumarate was used as the sole carbon and energy source. For allelic-exchange mutagenesis experiments, M. smegmatis mc2155 was grown in Lemco medium with or without the addition of 15 g/liter Bacto agar as described previously (42). Kanamycin was added at 20 µg/ml for M. smegmatis and at 50 µg/ml for E. coli, and hygromycin B was added at 50 µg/ml for M. smegmatis and at 200 µg/ml for E. coli. Gentamicin was added at 5 µg/ml for M. smegmatis and at 20 µg/ml for E. coli. For sucrose selection, 10% (wt/vol) sucrose was included. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at 40 µg/ml. OD600s of culture samples diluted in saline (0.85% NaCl) were measured in cuvettes with a 1-cm path length in a Jenway 6300 spectrophotometer.
Small-scale batch growth of M. smegmatis was carried out aerobically (normoxic) in either 125- or 250-ml conical flasks or under hypoxic conditions in 120-ml sealed serum vials (butyl rubber stoppered, O2 impermeable). Oxygen depletion under hypoxia was confirmed by decolorization of methylene blue added (1.5 µM final concentration) (in a separate replicate) as previously described (43). Large-scale batch growth in either HdB containing 30 mM succinate (M. smegmatis) or LB (E. coli) for IMV preparations were carried out either aerobically in 2-liter conical flasks or under hypoxia in 2-liter Schott bottles (butyl rubber stoppered, O2 impermeable). A 600-ml volume of medium was used in the Schott bottles, which is the same headspace/medium volume ratio as in serum vials. M. bovis BCG was maintained according to established protocols used for M. tuberculosis (44). Large-scale growth was carried out in 2-liter conical flasks of 7H9 medium supplemented with only the albumin and dextrose components (ADS) of oleic acid-albumin-dextrose-catalase.
RNA extraction and RT-PCR.
Total RNA was extracted with TRIZOL reagent according to the manufacturer’s instructions. Cell lysis was achieved by three cycles of bead beating in a Mini-Beadbeater (Biospec) at 5,000 rpm for 30 s. DNA was removed from the RNA preparation by treatment with 2 U of RNase-free DNase with the TURBO DNA-free kit (Ambion) according to the manufacturer’s instructions. The quality of the RNA was checked on a 1.2% agarose gel, and the concentration was determined with a NanoDrop ND-1000 spectrophotometer. RT-PCR was performed with the SuperScript III RT kit (Invitrogen) according to the manufacturer’s instructions for the RT step and the Phusion High-Fidelity PCR kit for the PCR step. The RT reactions were carried out with 1 µg of RNA template and gene-specific primers (listed in Table S2 in the supplemental material) that bind within the sdh1 or sdh2 operon, followed by PCR with the same set of primers, and 5 µl of the resulting RT reaction mixture were used.
DNA manipulation and cloning of constructs.
All molecular biology techniques were carried out according to standard procedures (45). Amino acid sequences for the succinate dehydrogenases/fumarate reductases included in the analysis were obtained from the NCBI protein database and aligned with the Clustal Omega (46) server hosted at EMBL-EBI. Restriction or DNA-modifying enzymes and other molecular biology reagents were obtained from Roche Diagnostics or New England BioLabs. Genomic DNA of M. smegmatis was isolated as described previously (47). To create a markerless deletion of the sdh1 operon, which corresponds to the MSMEG_0420-to-MSMEG_0416 gene region, a 1,017-bp fragment flanking MSMEG_ 0420 upstream and a 916-bp fragment flanking MSMEG_0416 downstream were amplified. The two products were fused by overlap extension PCR (48), cloned into the SpeI site of pPR23-derived (49) vector pX33 (47), creating pXsdh1g, and transformed into M. smegmatis mc2155. The same procedure was used to create a markerless deletion of the sdh2 operon, which corresponds to the MSMEG_1672-to-MSMEG_1669 gene region; a 1,230-bp fragment flanking MSMEG_1669 upstream and a 1,031-bp fragment flanking MSMEG_1672 downstream were amplified. The two products were fused by overlap extension PCR and cloned into the HindIII site of the p2NIL vector (50), creating p2NILsdh2k, and transformed into M. smegmatis mc2155. To make the complementing construct for M. smegmatis WT sdh2, the complete operon, together with the native promoter 352 bp upstream of sdh2, was PCR amplified. Thereafter, the 3.7-kb PCR product was cloned into the HindIII site of the integrative E. coli-Mycobacterium shuttle vector pUHA267 (51), creating plasmid pUHA267sdh2h (for the primers used, see Table S2 in the supplemental material). All constructs were verified by restriction digestion and sequencing.
Generation of M. smegmatis sdh1 and sdh2 deletion mutant strains.
Allelic replacement of the sdh1 operon was carried out essentially as previously described (49). In brief, a culture of M. smegmatis mc2155 carrying pXsdh1g was grown at 28°C to an OD600 of approximately 0.6 to 0.8 and then plated onto LB medium with 0.05% Tween 80 (LBT) solid medium containing gentamicin, which selected for integration of the plasmid via a SCO event. Thereafter, colonies that formed a yellow product when exposed to 250 mM catechol were screened by Southern hybridization analysis for correct integration of the construct. One integrant was chosen and grown in 5 ml of LBT medium containing gentamicin, at 37°C. Aliquots of this culture were then plated onto low-salt LBT plates containing sucrose and incubated at 40°C to select for a DCO event resulting in loss of the plasmid and deletion of the sdh1 operon. Colonies that did not form the yellow product after exposure to catechol were screened by Southern hybridization analysis for correct deletion of the sdh1 operon.
For deletion of the sdh2 operon, a two-step deletion strategy similar to that described by Parish et al. (50, 52) was used. A 5-µg UV-pretreated (53) p2NILsdh2k plasmid sample was electroporated into competent M. smegmatis, and then SCO transformants were selected on medium containing kanamycin and X-Gal. Merodiploid strains were constructed by electroporation of the SCO strain with the complementing plasmid pUHA267sdh2h, followed by isolation of kanamycin- and hygromycin-resistant transformants. DCOs were generated in the WT and merodiploid backgrounds by streaking cells onto plates lacking antibiotics. DCO selection was performed on medium containing sucrose and X-Gal. Candidate clones (Hygr, Kans, and white color) were first screened by colony PCR with gene-specific screening primers (see Table S2 in the supplemental material) and then confirmed by Southern blotting to determine whether the WT or deletion mutant allele was present at the targeted chromosomal location. Southern blot hybridization analysis was carried out with the Amersham Gene Images AlkPhos Direct Labeling and detection system by using CDP-Star detection reagent (GE Healthcare) according to the manufacturer’s instructions. Restriction digestion of the genomic DNA was performed with PstI, resulting in 1.8- and 2.6-kb fragments in the cases of WT and sdh1 deletion strains, respectively, and with BamHI, resulting in 1.0- and 4.1-kb fragments in the cases of the WT and sdh2 deletion strains, respectively. The probe used for sdh1 was the 1.0-kb left flank fragment, and for sdh2, a 0.8-kb fragment was separately PCR amplified (for the primers used, see Table S2).
Mapping of sdh1 and sdh2 TSSs and construction of sdh1-lacZ and sdh2-lacZ transcriptional fusions.
The TSSs of the sdh1 and sdh2 operons were mapped by 5′ RACE by using the components of a 3′/5′ RACE kit according to the manufacturer’s instructions and as previously described (54), with the primers listed in Table S2 in the supplemental material.
To study sdh1 and sdh2 expression, transcriptional fusions of sdh1 and sdh2 operon promoters were cloned into pJEM15 (55), a plasmid containing a promoterless lacZ gene. PCR products of 607 and 654 bp encompassing the sdh1 (−424 to +183) and sdh2 (−453 to +201) promoter regions relative to the TSSs were amplified, respectively, with the primers listed in Table S2. Thereafter, the products were cloned into the BamHI and SphI sites of pJEM15, creating plasmids pJsdh1 and pJsdh2 (see Table S1). All of the promoter regions amplified by PCR were confirmed by DNA sequencing. The plasmids were transformed into M. smegmatis mc2155 by electroporation. The transcriptional activities of the sdh1 and sdh2 operons were determined with the β-galactosidase assay as described previously (47).
Preparation of IMVs.
Cell suspensions from large-scale batch growth (~2-liter total volume) were pooled and centrifuged at 7,000 rpm for 15 min. The pellet was resuspended in lysis buffer (50 ml of 50 mM morpholinepropanesulfonic acid [MOPS; pH 7.5] buffer with 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT], 1 Roche protease inhibitor tablet, 5 mM MgCl2, 0.05% Tween 80, 2.5 mg/ml lysozyme, 2.5 mg DNase I). The cell suspension was homogenized four times with a homogenizer and incubated for 30 min on ice. The cells were broken by six passages through a precooled French pressure cell at 20,000 lb/in2 (AMINCO). The lysate was centrifuged at 10,000 rpm at 4°C for 10 min to remove unbroken cells. The pellet was discarded. This step was repeated with this clarified lysate to ensure that no unbroken cells were carried over. The clarified lysate was centrifuged at 150,000 × g for 45 min at 4°C to harvest IMVs. The resulting membrane pellet was resuspended in lysis buffer that did not contain lysozyme or DNase I. Aliquots were used immediately or snap-frozen after 10% glycerol addition and stored at −80°C. Protein concentration was determined by using the bicinchoninic acid (BCA) reaction (Sigma) with bovine serum albumin as the standard.
Determination of succinate concentrations and succinate dehydrogenase and fumarate reductase assays.
Cell suspensions sampled during growth were centrifuged at 13,000 × g for 1 min, and supernatant samples were stored at −20°C until used for analysis. Succinate concentrations were estimated with the Megazyme Succinate Acid Assay kit according to the manufacturer’s instructions. Succinate dehydrogenase enzymes as isolated are partially deactivated by tightly bound oxaloacetate at their active sites, and thus, it is necessary to activate the enzyme to fully express their succino-oxidase activity (56, 57). To activate the enzyme, the membrane vesicles were incubated at 37°C for 5 min in 50 mM MOPS (pH 7.5) with 2 mM KCN and the desired amount of succinate. Activated membranes were then added, and the reaction was initiated by the addition of 660 µM PES and 50 µM DCIP. The reduction of DCIP was followed by measuring the absorbance at 600 nm of DCIP (ε600 = 19.1 mM−1 cm−1) with a Varian Cary 50 spectrophotometer. Additional supplementation with fumarate or 3NP was performed as described in Results.
Fumarate reduction activities of IMVs were determined by measuring the fumarate-dependent oxidation of BV. This method has been previously described for assays of E. coli QFR and SQR (13). The assay mixture contained 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 5 mM fumarate (pH 7.5), 0.2 mM BV, 60 µg/ml glucose oxidase, 4 µg/ml catalase, and 20 mM glucose. The Tris-HCl was boiled and then cooled under nitrogen, with the addition of DTT, to remove oxygen from the buffer before the addition of the other assay components. Glucose oxidase, catalase, and glucose were added to maintain anaerobiosis. Assay mixtures were prepared in 1.5-ml screw-top cuvettes under a flow of nitrogen. The desired amount of membrane vesicles was added, and then the assay was immediately initiated by the addition of a stoichiometric amount of sodium dithionite. The progress of the reaction was monitored by the decrease in BV absorbance at 602 nm (ε602 = 9.6 mM−1 cm−1) (13). Rates were corrected for baseline activity, which was estimated in technical triplicate for each organism and membrane concentration used. Fumarate reductase activity was calculated as μmol of BV oxidized min−1 mg of protein−1.
Measurement of oxygen consumption rates.
A model 10 Clark-type oxygen electrode (Rank Brothers Ltd., Cambridge, England) linked to a PicoLog ADC-20 data logger was used to measure O2 consumption in IMVs. IMVs were diluted to 0.5 mg/ml in 50 mM MOPS (pH 7.5) and supplemented with 5 mM succinate, with stirring, to initiate O2 consumption. The electrode was calibrated daily by flushing 0.1 M K-phosphate buffer with gaseous N2 for 5 min (oxygen solubility of 220 nmol/ml). Rates were normalized to the protein concentration of cell pellets estimated by using the BCA reaction (Sigma), which were lysed with 1 M NaOH with heating at 95°C and subsequently neutralized with 1 M HCl.
Membrane potential determination and proton-pumping assays.
Determination of ΔΨ was performed as previously described (58). Cells (1 ml, grown under normoxic or hypoxic conditions, Fig. 9) were transferred to glass tubes (in triplicate) containing [3H]methyltriphenylphosphonium iodide (TPP+) (30 to 60 Ci mmol−1) ([3H]TPP+, 2.4 nM final concentration). After incubation for 10 min at 37°C, the cultures were centrifuged through 0.35 ml of silicone oil (BDH Laboratory Supplies, Poole, England) in 1.5-ml microcentrifuge tubes (13,000 × g, 5 min, 22°C) and 20-µl samples of supernatant were removed. The tubes and contents were frozen (−20°C), and cell pellets were removed with dog nail clippers. The supernatant and cell pellets were dissolved in 2 ml of scintillation fluid (Amersham), and the amount of [3H]TPP+ taken up by the cells was determined with a Tri-Carb liquid scintillation analyzer (PerkinElmer). The silicone oil mixture was a 40% mixture of phthalic acid bis(2-ethyl-hexyl ester) and 60% silicone oil (40% part mixture of DC200/200 silicone oil and 60% DC 550). Silicone oils were left overnight at room temperature to equilibrate. The intracellular volume (3.45 ± 0.59 µl mg of protein−1) was estimated from the difference between the partitioning of 3H2O and [14C]taurine. The ΔΨ was calculated from the uptake of [3H]TPP+ according to the Nernst relationship. Nonspecific TPP+ binding was estimated from cells that had been treated with valinomycin and nigericin (10 µM each) for 25 min (58).
Proton translocation into IMVs was determined by measuring the quenching of the fluorescent probe AO with a Cary Eclipse Fluorescence spectrophotometer. IMVs (0.5 mg/ml) were preincubated at 30°C in a weak buffer system consisting of 10 mM HEPES (pH 7.5), 100 mM KOH, and 5 mM MgCl2 containing 5 µM AO for 2 min with constant stirring. The reaction was then initiated by adding 5 mM succinate as an electron donor. Once the fluorescence quenching reached a steady state, the proton gradient was collapsed by the addition of the uncoupler CCCP (50 µM). The excitation and emission wavelengths were 493 and 530 nm, respectively.
SUPPLEMENTAL MATERIAL
Clustal Omega alignment of SQR and QFR D subunits. Enzymes with known heme contents are in square brackets, and their respective subclasses are in parentheses; they have been previously reviewed by Lancaster (14). Histidines involved in the binding of a distal heme are highlighted with teal, while those involved in proximal heme binding are shown in dark blue. These assignments are based mainly on a similar analysis performed by Bott and Niebisch (23). In regions with >50% similarity, black shading indicates identical residues and gray shading indicates similar residues. Download
Clustal Omega alignment of SQR and QFR fused membrane subunits. Enzymes with known heme contents are in square brackets, and their respective subclasses are in parentheses; they have been previously reviewed by Lancaster (14). Histidines involved in the binding of a distal heme are highlighted with teal, while those involved in proximal heme binding are shown in dark blue. These assignments are based mainly on a similar analysis performed by Bott and Niebisch (23). In regions with >50% similarity, black shading indicates identical residues and gray shading indicates similar residues. Download
Clustal Omega alignment of SQR and QFR A subunits. Glutamine and glutamate, at the position with an asterisk, have previously been shown to be required for efficient SQR and QFR activity in E. coli, respectively (13). The glutamines are highlighted in pink, while the glutamates are highlighted in cyan. In regions with >50% similarity, black shading indicates identical residues and gray shading indicates similar residues. Download
Inhibition of Sdh activity in IMVs by 3NP. WT and Δsdh1 mutant M. smegmatis IMVs were preincubated with 3NP as described in Materials and Methods. IMVs were prepared from cells grown aerobically (A) or under hypoxic conditions (B) in HdB minimal medium supplemented with 30 mM succinate. Error bars represent the standard error of biological triplicates. The concentration of succinate in each assay was 500 µM. Ethanol was used as the vehicle in all of the experiments. Download
Bacterial strains and plasmids used in this study.
Primers used in this study.
Kinetic parameters of succinate oxidation in IMVs prepared from WT and Δsdh1 mutant M. smegmatis grown under normoxic or hypoxic conditions.
Supplemental material references. Download
ACKNOWLEDGMENTS
I.P. was funded by a Health Sciences Career Development Postdoctoral Fellowships, University of Otago. K.H. is supported by the Otago Medical Research Foundation. M.B. was financially supported by a Marsden Grant from the Royal Society of New Zealand. G.M.C. was supported by a James Cook Fellowship from the Royal Society of New Zealand and the Health Research Council of New Zealand. W.R.J., an HHMI investigator, is funded by NIH/NIAID AI26170 and R01AI097548-01A1.
We thank Gary Cecchini and Elena Maklashina for supplying E. coli bacterial strains and their helpful comments regarding Sdh and Frd assays.
Footnotes
Citation Pecsi I, Hards K, Ekanayaka N, Berney M, Hartman T, Jacobs WR, Jr, Cook GM. 2014. Essentiality of succinate dehydrogenase in Mycobacterium smegmatis and its role in the generation of the membrane potential under hypoxia. mBio 5(4):e01093-14. doi:10.1128/mBio.01093-14.
REFERENCES
- 1. Cook GM, Berney M, Gebhard S, Heinemann M, Cox RA, Danilchanka O, Niederweis M. 2009. Physiology of mycobacteria. Adv. Microb. Physiol. 55:81–182, 318–189. 10.1016/S0065-2911(09)05502-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boshoff HI, Barry CE., III 2005. Tuberculosis—metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3:70–80. 10.1038/nrmicro1065 [DOI] [PubMed] [Google Scholar]
- 3. Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. U. S. A. 102:4548–4553. 10.1073/pnas.0500469102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, Mizrahi V. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183:7076–7086. 10.1128/JB.183.24.7076-7086.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsoso LG, Kana BD, Crellin PK, Lea-Smith DJ, Pelosi A, Powell D, Dawes SS, Rubin H, Coppel RL, Mizrahi V. 2005. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J. Bacteriol. 187:6300–6308. 10.1128/JB.187.18.6300-6308.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cecchini G. 2013. Respiratory complex II: role in cellular physiology and disease. Biochim. Biophys. Acta 1827:541–542. 10.1016/j.bbabio.2013.02.010 [DOI] [PubMed] [Google Scholar]
- 7. Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217–234. 10.1016/S0005-2728(97)00034-0 [DOI] [PubMed] [Google Scholar]
- 8. Unden G, Schirawski J. 1997. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol. Microbiol. 25:205–210. 10.1046/j.1365-2958.1997.4731841.x [DOI] [PubMed] [Google Scholar]
- 9. Kröger A, Biel S, Simon J, Gross R, Unden G, Lancaster CR. 2002. Fumarate respiration of Wolinella succinogenes: enzymology, energetics and coupling mechanism. Biochim. Biophys. Acta 1553:23–38. 10.1016/S0005-2728(01)00234-1 [DOI] [PubMed] [Google Scholar]
- 10. Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC. 2002. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J. Biol. Chem. 277:16124–16130. 10.1074/jbc.M200815200 [DOI] [PubMed] [Google Scholar]
- 11. Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S. 2003. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299:700–704. 10.1126/science.1079605 [DOI] [PubMed] [Google Scholar]
- 12. Iverson TM. 2013. Catalytic mechanisms of complex II enzymes: a structural perspective. Biochim. Biophys. Acta 1827:648–657. 10.1016/j.bbabio.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maklashina E, Iverson TM, Sher Y, Kotlyar V, Andréll J, Mirza O, Hudson JM, Armstrong FA, Rothery RA, Weiner JH, Cecchini G. 2006. Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain. J. Biol. Chem. 281:11357–11365. 10.1074/jbc.M512544200 [DOI] [PubMed] [Google Scholar]
- 14. Lancaster CR. 2013. The di-heme family of respiratory complex II enzymes. Biochim. Biophys. Acta 1827:679–687. 10.1016/j.bbabio.2013.02.012 [DOI] [PubMed] [Google Scholar]
- 15. Berney M, Cook GM. 2010. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5(1):e8614. 10.1371/journal.pone.0008614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hågerhåll C. 1997. Succinate:quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320:107–141. 10.1016/S0005-2728(97)00019-4 [DOI] [PubMed] [Google Scholar]
- 18. Lancaster CR. 2001. Succinate:quinone oxidoreductases—what can we learn from Wolinella succinogenes quinol:fumarate reductase? FEBS Lett. 504:133–141. 10.1016/S0014-5793(01)02706-5 [DOI] [PubMed] [Google Scholar]
- 19. Hågerhåll C, Aasa R, von Wachenfeldt C, Hederstedt L. 1992. Two hemes in Bacillus subtilis succinate:menaquinone oxidoreductase (complex II). Biochemistry 31:7411–7421. 10.1021/bi00147a028 [DOI] [PubMed] [Google Scholar]
- 20. Lancaster CR, Kröger A. 2000. Succinate:quinone oxidoreductases: new insights from X-ray crystal structures. Biochim. Biophys. Acta 1459:422–431. 10.1016/S0005-2728(00)00180-8 [DOI] [PubMed] [Google Scholar]
- 21. Schafer G, Anemuller S, Moll R. 2002. Archaeal complex II: ‘classical’ and ‘non-classical’ succinate:quinone reductases with unusual features. Biochim. Biophys. Acta 1553:57–73. 10.1016/S0005-2728(01)00232-8 [DOI] [PubMed] [Google Scholar]
- 22. Hågerhåll C, Hederstedt L. 1996. A structural model for the membrane-integral domain of succinate:quinone oxidoreductases. FEBS Lett. 389:25–31. 10.1016/0014-5793(96)00529-7 [DOI] [PubMed] [Google Scholar]
- 23. Bott M, Niebisch A. 2003. The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104:129–153. 10.1016/S0168-1656(03)00144-5 [DOI] [PubMed] [Google Scholar]
- 24. Nachlas MM, Margulies SI, Seligman AM. 1960. A colorimetric method for the estimation of succinic dehydrogenase activity. J. Biol. Chem. 235:499–503 [PubMed] [Google Scholar]
- 25. Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 110:6554–6559. 10.1073/pnas.1219375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA. 2006. 3-Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 281:5965–5972. 10.1074/jbc.M511270200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7(9):e1002251. 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baek SH, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9(5):e1001065. 10.1371/journal.pbio.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schirawski J, Unden G. 1998. Menaquinone-dependent succinate dehydrogenase of bacteria catalyzes reversed electron transport driven by the proton potential. Eur. J. Biochem. 257:210–215 [DOI] [PubMed] [Google Scholar]
- 30. Lemma E, Unden G, Kröger A. 1990. Menaquinone is an obligatory component of the chain catalyzing succinate respiration in Bacillus subtilis. Arch. Microbiol. 155:62–67. 10.1007/BF00291276 [DOI] [PubMed] [Google Scholar]
- 31. Madej MG, Nasiri HR, Hilgendorff NS, Schwalbe H, Unden G, Lancaster CR, August RV, Re V, Recei M, October V, Goethe-universita JW, Gutenberg-universita J. 2006. Experimental evidence for proton motive force-dependent catalysis by the diheme-containing succinate:menaquinone oxidoreductase from the Gram-positive bacterium Bacillus licheniformis. Biochemistry 45:15049–15055. 10.1021/bi0618161 [DOI] [PubMed] [Google Scholar]
- 32. Matsson M, Tolstoy D, Aasa R, Hederstedt L. 2000. The distal heme center in Bacillus subtilis succinate:quinone reductase is crucial for electron transfer to menaquinone. Biochemistry 39:8617–8624. 10.1021/bi000271m [DOI] [PubMed] [Google Scholar]
- 33. Hawkins BJ, Levin MD, Doonan PJ, Petrenko NB, Davis CW, Patel VV, Madesh M. 2010. Mitochondrial complex II prevents hypoxic but not calcium- and proapoptotic Bcl-2 protein-induced mitochondrial membrane potential loss. J. Biol. Chem. 285:26494–26505. 10.1074/jbc.M110.143164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rao SP, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 105:11945–11950. 10.1073/pnas.0711697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE, III, Boshoff HI. 2011. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 7(10):e1002287. 10.1371/journal.ppat.1002287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. King A, Selak MA, Gottlieb E. 2006. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 25:4675–4682. 10.1038/sj.onc.1209594 [DOI] [PubMed] [Google Scholar]
- 37. Hoekstra AS, Bayley JP. 2013. The role of complex II in disease. Biochim. Biophys. Acta 1827:543–551. 10.1016/j.bbabio.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 38. Harada S, Inaoka DK, Ohmori J, Kita K. 2013. Diversity of parasite complex II. Biochim. Biophys. Acta 1827:658–667. 10.1016/j.bbabio.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 39. Kluckova K, Bezawork-Geleta A, Rohlena J, Dong L, Neuzil J. 2013. Mitochondrial complex II, a novel target for anti-cancer agents. Biochim. Biophys. Acta 1827:552–564. 10.1016/j.bbabio.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 40. Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911–1919. 10.1111/j.1365-2958.1990.tb02040.x [DOI] [PubMed] [Google Scholar]
- 41. Smeulders MJ, Keer J, Speight RA, Williams HD. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parish T. 2003. Starvation survival response of Mycobacterium tuberculosis. J. Bacteriol. 185:6702–6706. 10.1128/JB.185.22.6702-6706.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berney M, Weimar MR, Heikal A, Cook GM. 2012. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol. Microbiol. 84:664–681. 10.1111/j.1365-2958.2012.08053.x [DOI] [PubMed] [Google Scholar]
- 44. Larsen MH, Biermann K, Jacobs WR. 2007. Laboratory maintenance of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. Chapter 10:Unit 10A.1. 10.1002/9780471729259.mc10a01s6 [DOI] [PubMed] [Google Scholar]
- 45. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gebhard S, Tran SL, Cook GM. 2006. The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology 152:3453–3465. 10.1099/mic.0.29201-0 [DOI] [PubMed] [Google Scholar]
- 48. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 49. Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 94:10955–10960. 10.1073/pnas.94.20.10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975 [DOI] [PubMed] [Google Scholar]
- 51. Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. U. S. A. 88:3111–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parish T, Stoker NG. 2000. glnE is an essential gene in Mycobacterium tuberculosis. J. Bacteriol. 182:5715–5720. 10.1128/JB.182.20.5715-5720.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Triccas JA, Parish T, Britton WJ, Gicquel B. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 167:151–156. 10.1111/j.1574-6968.1998.tb13221.x [DOI] [PubMed] [Google Scholar]
- 54. Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. 2009. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 390:353–367. 10.1016/j.jmb.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 55. Timm J, Lim EM, Gicquel B. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ackrell BA, Cochran B, Cecchini G. 1989. Interactions of oxaloacetate with Escherichia coli fumarate reductase. Arch. Biochem. Biophys. 268:26–34. 10.1016/0003-9861(89)90561-4 [DOI] [PubMed] [Google Scholar]
- 57. Ackrell BA, Kearney EB, Singer TP. 1978. Mammalian succinate dehydrogenase. Methods Enzymol. 53:466–483. 10.1016/S0076-6879(78)53050-4 [DOI] [PubMed] [Google Scholar]
- 58. Rao M, Streur TL, Aldwell FE, Cook GM. 2001. Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147:1017–1024 [DOI] [PubMed] [Google Scholar]
- 59. Schulz S, Iglesias-Cans M, Krah A, Yildiz O, Leone V, Matthies D, Cook GM, Faraldo-Gómez JD, Meier T. 2013. A new type of Na(+)-driven ATP synthase membrane rotor with a two-carboxylate ion-coupling motif. PLoS Biol. 11(6):e1001596–e1001596. 10.1371/journal.pbio.1001596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clustal Omega alignment of SQR and QFR D subunits. Enzymes with known heme contents are in square brackets, and their respective subclasses are in parentheses; they have been previously reviewed by Lancaster (14). Histidines involved in the binding of a distal heme are highlighted with teal, while those involved in proximal heme binding are shown in dark blue. These assignments are based mainly on a similar analysis performed by Bott and Niebisch (23). In regions with >50% similarity, black shading indicates identical residues and gray shading indicates similar residues. Download
Clustal Omega alignment of SQR and QFR fused membrane subunits. Enzymes with known heme contents are in square brackets, and their respective subclasses are in parentheses; they have been previously reviewed by Lancaster (14). Histidines involved in the binding of a distal heme are highlighted with teal, while those involved in proximal heme binding are shown in dark blue. These assignments are based mainly on a similar analysis performed by Bott and Niebisch (23). In regions with >50% similarity, black shading indicates identical residues and gray shading indicates similar residues. Download
Clustal Omega alignment of SQR and QFR A subunits. Glutamine and glutamate, at the position with an asterisk, have previously been shown to be required for efficient SQR and QFR activity in E. coli, respectively (13). The glutamines are highlighted in pink, while the glutamates are highlighted in cyan. In regions with >50% similarity, black shading indicates identical residues and gray shading indicates similar residues. Download
Inhibition of Sdh activity in IMVs by 3NP. WT and Δsdh1 mutant M. smegmatis IMVs were preincubated with 3NP as described in Materials and Methods. IMVs were prepared from cells grown aerobically (A) or under hypoxic conditions (B) in HdB minimal medium supplemented with 30 mM succinate. Error bars represent the standard error of biological triplicates. The concentration of succinate in each assay was 500 µM. Ethanol was used as the vehicle in all of the experiments. Download
Bacterial strains and plasmids used in this study.
Primers used in this study.
Kinetic parameters of succinate oxidation in IMVs prepared from WT and Δsdh1 mutant M. smegmatis grown under normoxic or hypoxic conditions.
Supplemental material references. Download