ABSTRACT
In March 2013, three fatal human cases of infection with influenza A virus (H7N9) were reported in China. Since then, human cases have been accumulating. Given the public health importance of this virus, we performed a pathogenicity study of the H7N9 virus in the cynomolgus macaque model, focusing on clinical aspects of disease, radiographic, histological, and gene expression profile changes in the upper and lower respiratory tracts, and changes in systemic cytokine and chemokine profiles during infection. Cynomolgus macaques developed transient, mild to severe disease with radiographic evidence of pulmonary infiltration. Virus replicated in the upper as well as lower respiratory tract, with sustained replication in the upper respiratory tract until the end of the experiment at 6 days after inoculation. Virus shedding occurred mainly via the throat. Histopathological changes in the lungs were similar to those observed in humans, albeit less severe, with diffuse alveolar damage, infiltration of polymorphonuclear cells, formation of hyaline membranes, pneumocyte hyperplasia, and fibroproliferative changes. Analysis of gene expression profiles in lung lesions identified pathways involved in tissue damage during H7N9 infection as well as leads for development of therapeutics targeting host responses rather than virus replication. Overall, H7N9 infection was not as severe in cynomolgus macaques as in humans, supporting the possible role of underlying medical complications in disease severity as discussed for human H7N9 infection (H. N. Gao et al., N. Engl. J. Med. 368:2277–2285, 2013, doi:10.1056/NEJMoa1305584).
IMPORTANCE
Influenza A virus H7N9 emerged early in 2013, and human cases have continued to emerge since then. Although H7N9 virus-induced disease in humans is often very severe and even lethal, the majority of reported H7N9 cases occurred in older people and people with underlying medical conditions. To better understand the pathogenicity of this virus, healthy cynomolgus macaques were inoculated with influenza A virus H7N9. Cynomolgus macaques were used as a model because the receptor distribution for H7N9 virus in macaques was recently shown to be more similar to that in humans than that of other frequently used animal models. From comparison with previous studies, we conclude that the emerging H7N9 influenza virus was more pathogenic in cynomolgus macaques than seasonal influenza A viruses and most isolates of the pandemic H1N1 virus but less pathogenic than the 1918 Spanish influenza virus or highly pathogenic avian influenza (HPAI) H5N1 virus.
INTRODUCTION
In March 2013, three fatal human cases of infection with influenza A virus (H7N9) were reported in China (1). More cases were rapidly recognized in China; although no cases were reported in the summer of 2013, the number of cases started accumulating rapidly again in the fall of that same year (2). Common clinical symptoms in patients with H7N9 infection are fever and cough with lymphocytopenia; the disease progresses to an acute respiratory distress syndrome, often with a fatal outcome (3). Perimortem lung biopsies show diffuse alveolar damage, with infiltration of polymorphonuclear cells, formation of hyaline membranes, and fibroproliferative changes (4).
Although influenza A viruses of the H7 subtype have caused human infections in the past (reviewed in reference 5), most notably during an outbreak of highly pathogenic avian influenza virus H7N7 in the Netherlands in 2003 that resulted in 89 human cases (6), this is the first time that an H7 subtype virus has caused so many severe cases. The H7N9 virus is a reassortant consisting of Eurasian wild bird H7 and N9 viruses with H9N2 viruses circulating in poultry in China (1, 7, 8). An amino acid substitution in HA, Q226L, present in some H7N9 strains, increases the binding of HA of these viruses to α2,6-linked sialic acids (9). However, this single amino acid change does not seem to be sufficient for efficient human-to-human transmission.
The newly emerged H7N9 virus poses a significant pandemic threat, due to the rapid accumulation of cases in several distinct regions of China and the ability of the H7N9 viruses to be transmitted, albeit inefficiently, via respiratory droplets or aerosols in the ferret model (10–13). In order to allow a better estimation of the effect of a potential H7N9 pandemic, we studied the A/Anhui/1/2013 strain of this virus in the cynomolgus macaque model. This model was chosen because it most closely reflects the human physiology and the development of pneumonia, cytokine, and chemokine responses (14), and the pattern of attachment of H7N9 virus to respiratory tissues of macaques was recently shown to be more similar to that in humans than that in other frequently used animal models (15). Upon inoculation with influenza virus A/Anhui/1/2013, cynomolgus macaques developed transient, moderate disease with virus replication in the upper and lower respiratory tracts. The emerging H7N9 influenza virus was more pathogenic than seasonal influenza A virus and most isolates of the pandemic H1N1 virus but not as pathogenic as the 1918 Spanish influenza virus or highly pathogenic avian influenza (HPAI) H5N1 virus in cynomolgus macaques.
RESULTS
Clinical signs in cynomolgus macaques inoculated with A/Anhui/1/2013.
Eight 5-year-old cynomolgus macaques (4 male and 4 female) were inoculated with 7 × 106 50% tissue culture infectious doses (TCID50) of A/Anhui/1/2013 (H7N9) via a combination of ocular, oral, intranasal, and intratracheal inoculation as described previously (16). Animals started to show signs of disease at 1 day postinoculation (dpi). Clinical disease peaked at 3 and 4 dpi (Fig. 1A) and was moderate. Six out of 8 animals showed obvious respiratory signs, such as increased respiration rates, abdominal breathing, and coughing (see Table S1 in the supplemental material). Nasal discharge and cough were noted in only one animal. Hematologic and blood chemical analyses did not reveal any abnormalities during the course of infection (data not shown).
FIG 1 .
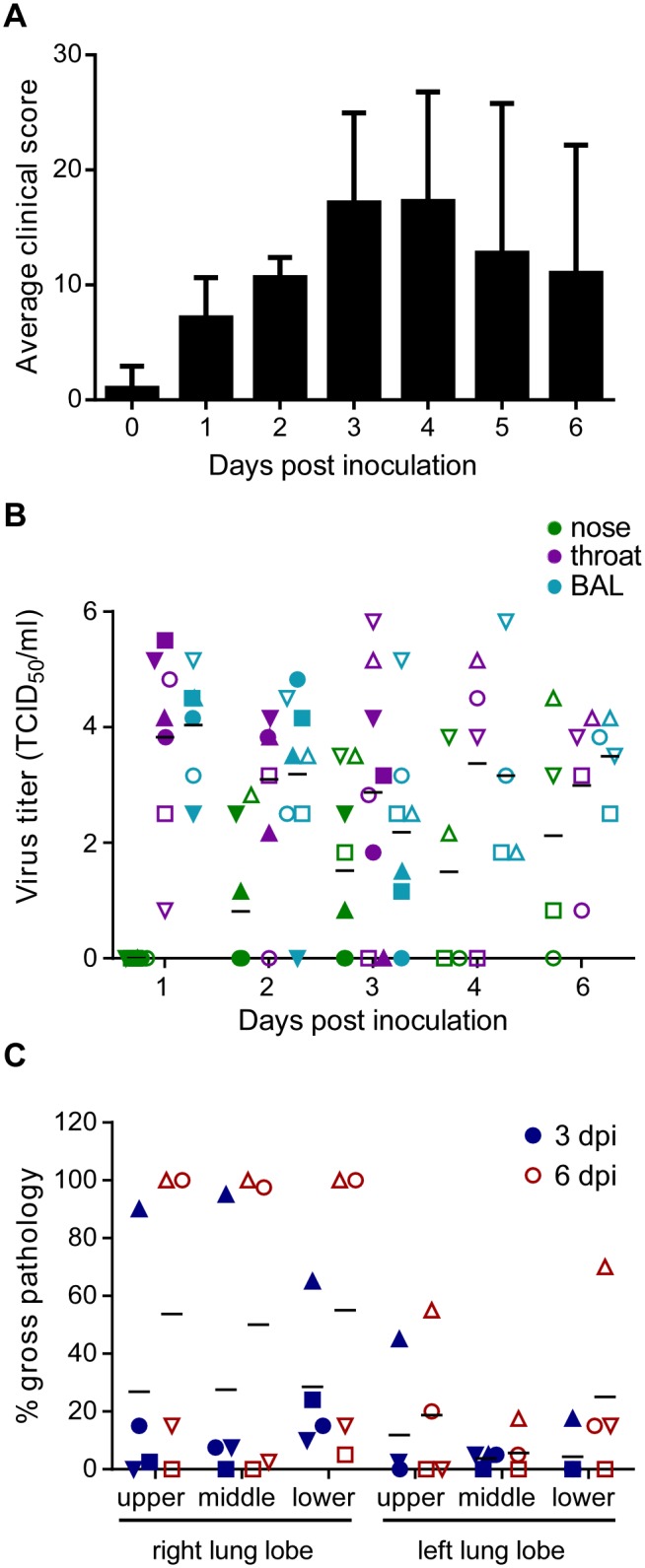
Clinical scores, virus shedding and lung pathology in cynomolgus macaques after infection with influenza virus A/Anhui/1/2013. (A) Eight cynomolgus macaques were inoculated with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013 and were observed twice daily for clinical signs of disease and scored using a previously described clinical scoring system (58). The average clinical score for all animals per time point is given. Clinical scores for individual animals can be found in Table S1 in the supplemental material. (B) Nose (green symbols) and throat (purple symbols) swabs and bronchoalveolar lavage fluid (blue symbols) were collected at 1, 2, 3, 4, and 6 days after inoculation. Virus titers in these samples were determined by titration in MDCK cells. (C) The percentage of area of each lung lobe affected by lesions was determined ventrally and dorsally in animals euthanized at 3 (blue symbols) and 6 (red symbols) dpi. The average percent gross pathology of dorsal and ventral measurements was calculated for each lung lobe of each animal. Of note, 4 animals were euthanized at 3 days postinoculation, and thus data in panels A and B from 4 dpi onwards are the averages for 4 animals. Symbols in panels B and C represent individual animals. Closed circles, H7N9-1; closed squares, H7N9-2; closed triangles, H7N9-3; closed inverted triangles, H7N9-4; open circles, H7N9-5; open squares, H7N9-6; open triangles, H7N9-7; open inverted triangles, H7N9-8.
Radiographic changes in lungs of animals inoculated with A/Anhui/1/2013.
At 1, 2, 3, 4, and 6 dpi, ventral-dorsal and lateral chest X-rays were taken to monitor the development of pneumonia. Radiographic changes in the lungs of inoculated animals were observed starting 2 and 3 dpi and were observed in all inoculated animals to various degrees (see Table S2 in the supplemental material). The interstitial infiltration was observed first in the lower right lung lobe and, in individual animals, spread over time to the right middle, left lower, left middle, and right upper lung lobes and developed into severe diffuse interstitial infiltration (Fig. 2; also, see Table S2 in the supplemental material).
FIG 2 .
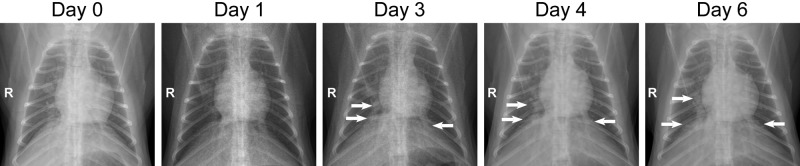
Radiographic changes in lungs of cynomolgus macaques inoculated with influenza virus A/Anhui/1/2013. Ventral-dorsal X rays were obtained from macaque H7N9-5 before inoculation (day 0; baseline) and at 1, 3, 4 and 6 days after inoculation with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013. A detailed analysis of radiographs taken from all eight infected macaques during infection is given in Table S2 in the supplemental material.
Virus shedding in cynomolgus macaques inoculated with A/Anhui/1/2013.
Clinical exams were performed at 0, 1, 2, 3, 4, and 6 dpi, and nasal, oropharyngeal, ocular, and rectal swabs were collected. Swabs were initially analyzed for the presence of viral RNA by real-time reverse transcription-PCR (RT-PCR). Because of the large number of swabs in which viral RNA was detected, all nasal, oropharyngeal, and ocular swabs were titrated on MDCK cells; rectal swabs were not titrated, as only 3 rectal swabs were positive by PCR. Oropharyngeal swabs were positive in virus titration by 1 dpi and remained positive in all animals until the end of the experiment at 6 dpi (Fig. 1B). Not all nasal swabs were positive by virus titration; most of the virus shedding via the nose occurred in animals H7N9-7 and H7N9-8 (Fig. 1B). Despite the ability of influenza A viruses of the H7 subtype to cause conjunctivitis, ocular swabs were only sporadically positive by virus titration: only the ocular swabs collected from H7N9-1 at 1, 2, and 3 dpi, the ocular swab collected from H7N9-4 at 1 dpi, and the ocular swab collected from H7N9-8 at 3 dpi were positive, with virus titers between 100.8 and 102.8 TCID50/ml (data not shown).
During clinical exams, bronchalveolar lavages (BAL) were performed, and the samples were analyzed for the presence of infectious virus. Virus could be isolated from the BAL fluid of all animals at 1 dpi; BAL fluid remained positive throughout the experiment, with significant amounts of virus still detected at 6 dpi, ranging from 3.2 × 102 TCID50/ml to 1.4 × 104 TCID50/ml (Fig. 1B).
Gross lung pathology in cynomolgus macaques inoculated with A/Anhui/1/2013.
Upon necropsy of 4 animals at 3 dpi, gross lesions were observed in the lungs of all animals. There was variation in the area of the lung affected between animals, but at least two lobes showed gross lesions in all animals, varying from 5% to 100% of tissue affected (Fig. 1C). In line with our observation on X-rays, the right lung lobes were more severely affected than the left lobes; this is likely a result of the intratracheal inoculation and the anatomy of the lung (17). By 6 dpi, the area of the lung displaying gross lesions had increased, with 100% of all three right lung lobes being affected in 2 of 4 animals (Fig. 1C).
Virus titers in tissues of cynomolgus macaques inoculated with A/Anhui/1/2013.
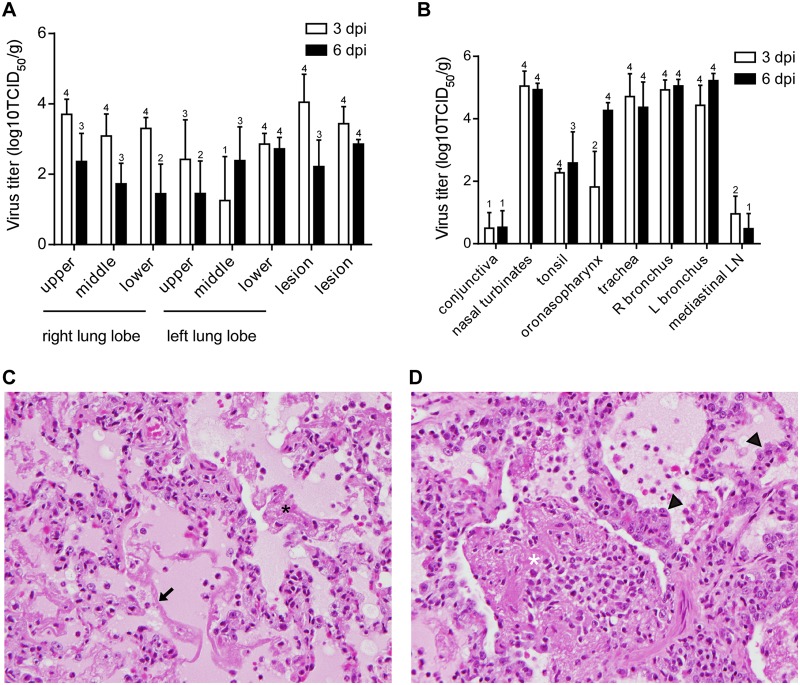
For each animal, virus titers were determined in tissue samples collected from all 6 lung lobes and 2 lung lesions. In line with our X-ray and gross pathology observations, at 3 dpi virus could be detected in all three lobes of the right lung but not in all three lung lobes of the left lung in all animals (Fig. 3A). Virus was present in the collected lung lesions, but unexpectedly, virus titers were not higher in lung lesions than in the collected lung lobe samples, indicating widespread virus replication throughout the lower respiratory tract. By 6 dpi, the virus titers in the lung lobes and lung lesions had decreased compared to those at 3 dpi, although this decrease was not statistically significant; the number of animals with positive virus titration also decreased (Fig. 3A).
FIG 3 .
Virus replication and histopathological changes in cynomolgus macaques inoculated with influenza virus A/Anhui/1/2013. Cynomolgus macaques were inoculated with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013; 4 animals were euthanized at 3 and 6 days postinoculation, and tissue samples were collected. (A and B) Virus titers in the indicated tissues collected at 3 and 6 days postinoculation were determined by titration in MDCK cells. Geometric mean titers were calculated; error bars represent standard deviations. Numbers above bars indicate the number of animals in which virus titration was positive (out of 4 animals per time point). R, right; L, left; LN, lymph node. (C and D) Histopathological changes in lungs of cynomolgus macaques inoculated with influenza virus A/Anhui/1/2013 at 3 (C) and 6 (D) days postinoculation. Lung tissue was collected and stained with hematoxylin and eosin. In severe cases of infection at 3 dpi, lesions were characterized by edema, alveolar fibrin (black asterisk), and hyaline membrane formation (arrow). At 6 dpi, edema, organizing fibrin (white asterisk), and type II pneumocyte hyperplasia (arrowheads) were observed. Magnification (C and D), ×400.
In other tissues of the respiratory tract, i.e., nasal turbinates, oropharynx, trachea, and right and left bronchus, virus titers were generally higher than in the lungs at 3 dpi and 6 dpi (Fig. 3B). Interestingly, although virus titers in the lung lobes and lung lesions decreased between 3 and 6 dpi, virus titers in the other respiratory tract tissues did not. Virus could also be detected in the tonsils and mediastinal lymph nodes of infected animals at 3 and 6 dpi (Fig. 3B). Virus could be detected in the conjunctiva of only one animal at 3 and 6 dpi.
The remaining tissues that were collected at 3 and 6 dpi (i.e., heart, liver, spleen, kidney, stomach, jejunum, ileum, transverse colon, and brain) were analyzed for the presence of viral RNA by real-time RT-PCR. Except for the liver, where vRNA was detected in 6 out of 8 animals, vRNA was detected sporadically in all tissues except in heart (see Table S3 in the supplemental material). Since cycle threshold values (CT) in almost all of these samples were higher than the level at which virus titration is usually successful (CT < 32), virus titration was not attempted on these samples.
Histopathology of respiratory tissues of cynomolgus macaques inoculated with A/Anhui/1/2013.
At 3 dpi, histopathology of the lungs was similar for all 4 animals necropsied and was characterized as mild to marked, acute, bronchointerstitial pneumonia. The pneumonia was characterized microscopically as mild to marked thickening of alveolar septa by fibrin, edema, neutrophils, and macrophages, and the alveoli contained small to large amounts of these same inflammatory components (Fig. 3C). Multifocally, hyaline membranes were observed. The lumens of terminal bronchioles frequently contained fibrin, edema, hyaline membranes, neutrophils, and macrophages, with multifocal necrosis and loss of lining epithelium. Inflammation and necrosis of bronchial submucosal glands were frequently noted, with mild, subacute periglandular inflammation or more severe changes that ranged from neutrophils and macrophages within ductular lumens to necrosis of acinar and ductular epithelium, occasionally affecting the entire gland. Larger bronchioles and bronchi were generally much less severely affected than terminal airways, with intact, viable lining epithelium and only occasional mild, neutrophilic luminal exudate.
At 6 dpi, histopathology of the lungs was characterized as mild to marked, subacute to chronic, bronchointerstitial pneumonia. Microscopic changes were again characterized by the presence of neutrophils, macrophages, fibrin, and edema within alveolar septa, alveoli, and terminal bronchioles (Fig. 3D). Additionally, type II pneumocyte hyperplasia was observed in extensive portions of each lung lobe, and large clumps of alveolar fibrin frequently engulfed neutrophils and macrophages and were lined and infiltrated by fibroblasts.
Immunohistochemistry (IHC) of sections of lung demonstrated low to moderate numbers of antigen-positive alveolar type I and type II pneumocytes, macrophages, and epithelium lining bronchioles, bronchi, and bronchial submucosal glands (Fig. 4). The numbers of positively stained alveolar pneumocytes and bronchial and bronchiolar submucosal gland epithelial cells were similar at 3 and 6 dpi. The number of positively stained pulmonary macrophages was increased at 6 dpi compared with 3 dpi. Positive staining in alveolar and submucosal macrophages was cytoplasmic, most likely indicating active phagocytosis of virus rather than replication of virus in these cells (Fig. 4H). Antigen-positive macrophages were also consistently noted within mediastinal lymph nodes, with increased numbers of these cells being noted at day 6 dpi. Low numbers of positively stained macrophages were noted in pharyngeal tonsils, oropharynges, tracheas, and extrapulmonary bronchi.
FIG 4 .
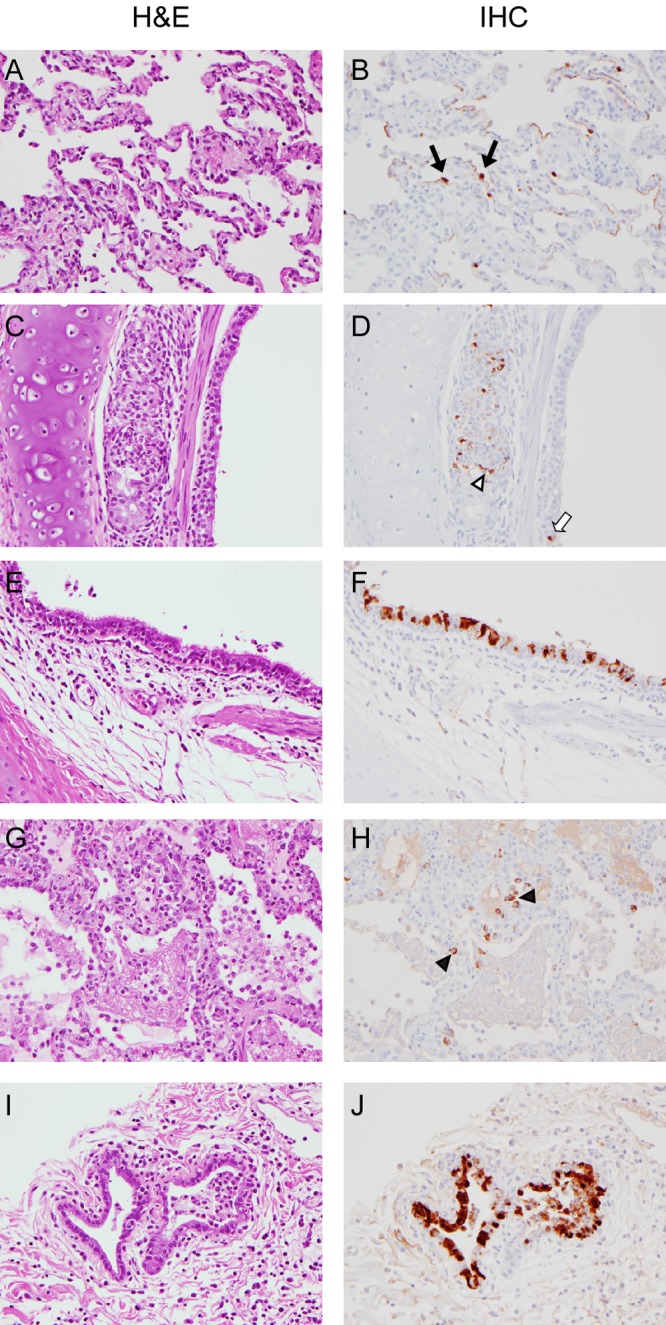
Immunohistochemical analysis of lungs of cynomolgus macaques inoculated with influenza virus A/Anhui/1/2013. Cynomolgus macaques were inoculated with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013; 4 animals were euthanized at 3 (A to D) and 6 (E to J) days postinoculation, and lung samples were collected. Matching tissue sections were stained with hematoxylin and eosin (H&E) or studied by immunohistochemistry (IHC) using an anti-NP monoclonal antibody (IHC; visible as red-brown staining). At 3 dpi, mild to marked thickening of alveolar septa by fibrin, edema, neutrophils, and macrophages was observed (A), and virus antigen was present in type I and type II (black arrows) pneumocytes (B). Inflammation and necrosis of bronchial submucosal glands were frequently noted (C), with virus antigen being present in submucosal gland (open arrowhead) and bronchial epithelium (white arrow). Infection of the bronchial lining epithelium was more pronounced by 6 dpi (E and F). Cytoplasmic staining of alveolar macrophages indicates active phagocytosis on 6 dpi (H; black arrowheads). (J) Virus replication in bronchial submucosal gland epithelium on 6 dpi. Magnification, ×400.
There was more variation between individual animals in histopathological lesions in the remaining tissues. Mild, subacute conjunctivitis was noted histologically in one of eight animals at 6 dpi, and low numbers of macrophages positive for influenza A virus antigen were present in the submucosa from that animal; the viral load in the eye swab collected from this animal (H7N9-7) at 6 dpi was still high (104 TCID50 equivalents/ml), likely indicating active virus replication. Influenza A virus-positive cells were noted in the epithelium of the nasal turbinates with minimal inflammation in 3 of 4 animals at 3 dpi (Fig. 5A and B). Although no microscopic abnormalities of the tonsil were noted, there were low to moderate numbers of positively stained tonsillar macrophages in all eight animals, with an increase in the amount of antigen detected between 3 and 6 dpi. Focal, subacute pharyngitis with ulceration was noted in one animal at 3 dpi; rare pharyngeal epithelial cells were positive according to IHC but not at the site of the ulcer (Fig. 5C and D). In one animal at 3 dpi and one at 6 dpi, influenza A virus antigen could be detected in respiratory epithelial cells from the nasopharynx. In other tissue sections of the pharynx, there were few macrophages that were positive by IHC. Tracheal inflammation affecting the mucosal lining and/or submucosal glands was noted in all macaques, with ulceration of lining epithelium occurring in one animal on 3 dpi and one on 6 dpi. Few epithelial cells and macrophages stained positive in IHC (Fig. 5E and F). Minimal to moderate subacute inflammation was noted in at least one of the extrapulmonary bronchi from each animal, and in one animal there was an extensive area of marked inflammation with ulceration. Upon IHC staining of the same sections of bronchi, few positively stained epithelial cells and/or macrophages were noted (Fig. 5G and H).
FIG 5 .
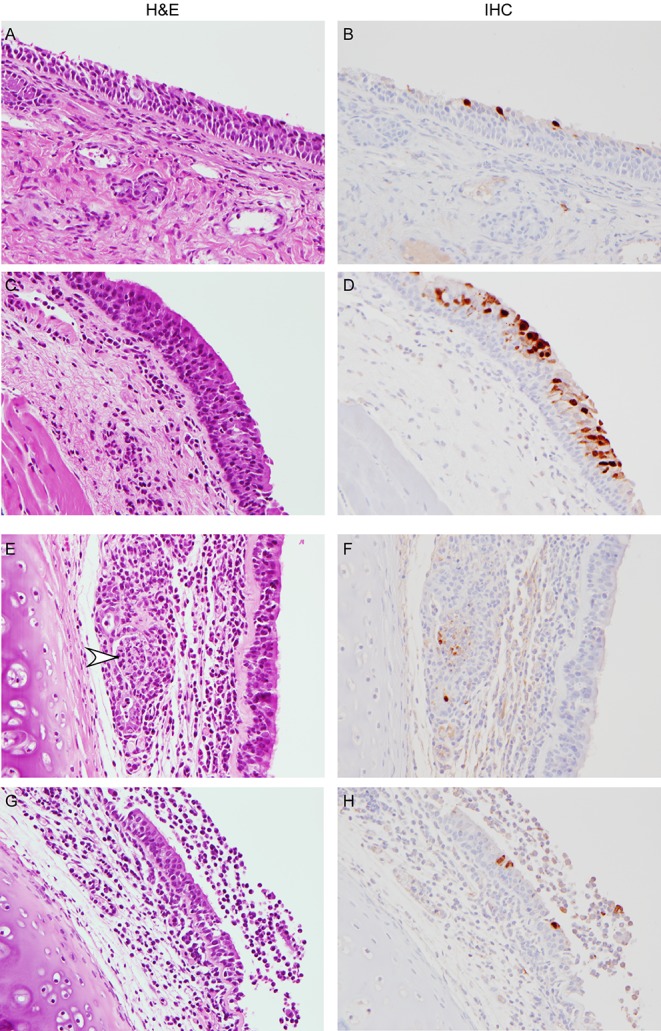
Extrapulmonary histopathological changes in cynomolgus macaques inoculated with influenza virus A/Anhui/1/2013. Cynomolgus macaques were euthanized at 3 (A and B) and 6 (C to H) days postinoculation, and tissue was collected and stained with hematoxylin and eosin (H&E) or immunohistochemistry (IHC) using an anti-NP monoclonal antibody (IHC; visible as red-brown staining). In the rostral nasal cavity (A and B) and nasopharynx (C and D), influenza virus A/Anhui/1/2013 mainly replicated in the lining epithelium. In the trachea (E and F), cell debris (arrowhead) with immunopositive staining is visible in a submucosal gland. In the bronchus (G and H), bronchial lumen exudate is visible, with virus replication in the epithelium lining the bronchus. Magnification, ×400.
Systemic cytokine and chemokine profiles in infected macaques.
Serum levels of 23 cytokines and chemokines were determined in samples obtained at 0, 1, 2, 3, 4, and 6 dpi. Transient differences were observed in granulocyte colony-stimulating factor (G-CSF), gamma interferon (IFN-γ), interleukin 1 receptor antagonist (IL-1RA), IL-2, IL-6, IL-8, IL-10, IL-15, monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), MIP-1β, tumor necrosis factor alpha (TNF-α), and vascular endothelial growth factor (VEGF) levels. At 1 dpi, there was a statistically significant increase in levels of G-CSF, IL-1RA, IL-6, IL-10, IL-15, MCP-1, and TNF-α (Fig. 6); levels of IFN-γ, IL-2, IL-8, MIP-1α, MIP-1β, and VEGF increased at that time point, but increases were not statistically significant (Fig. 6; also data not shown). By 6 dpi, all levels of cytokines and chemokines had returned to baseline.
FIG 6 .
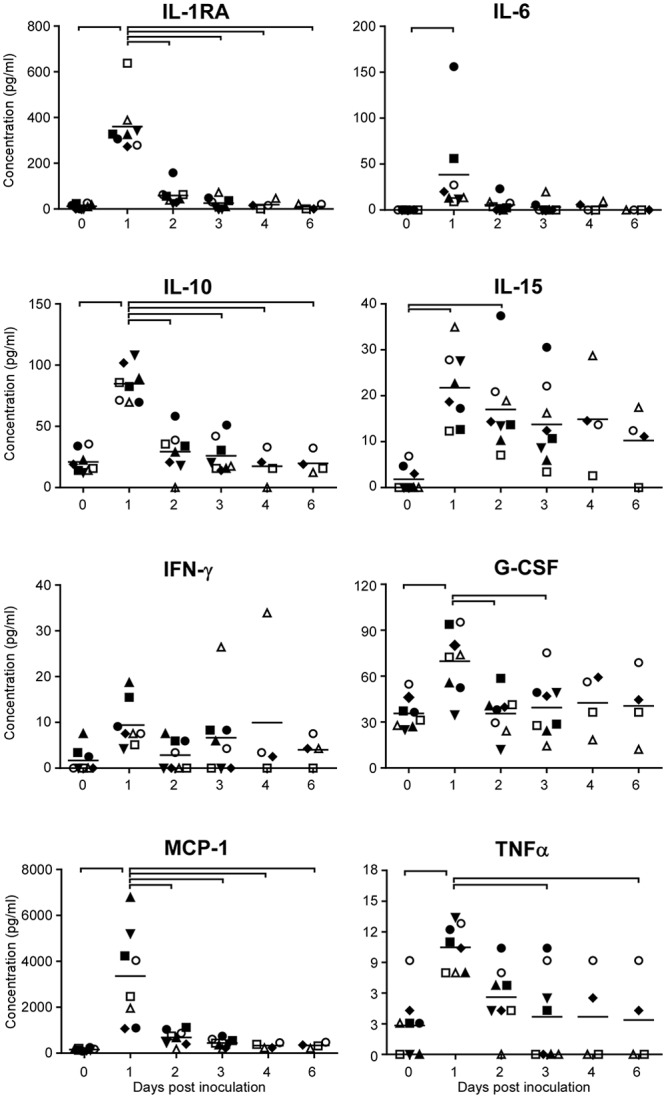
Cytokine and chemokine levels in serum samples of cynomolgus macaques infected with influenza virus A/Anhui/1/2013. Serum samples were collected 0, 1, 2, 3, 4, and 6 days postinoculation, and chemokine and cytokine levels were analyzed using the nonhuman primate cytokine Milliplex 23-plex map kit (Millipore). Only cytokines and chemokines with detectable levels that fluctuated during the course of the experiment are shown. Each symbol indicates one animal. ●, H7N9-1; ■, H7N9-2; ▲, H7N9-3; ▼, H7N9-4; ♦, H7N9-5; ○, H7N9-6; □, H7N9-7; Δ, H7N9-8. Bars at the tops of the graphs indicate statistically significant differences between time points (P < 0.05; one-way ANOVA with Bonferroni’s posttest).
Alterations in host gene expression upon infection with A/Anhui/1/2013.
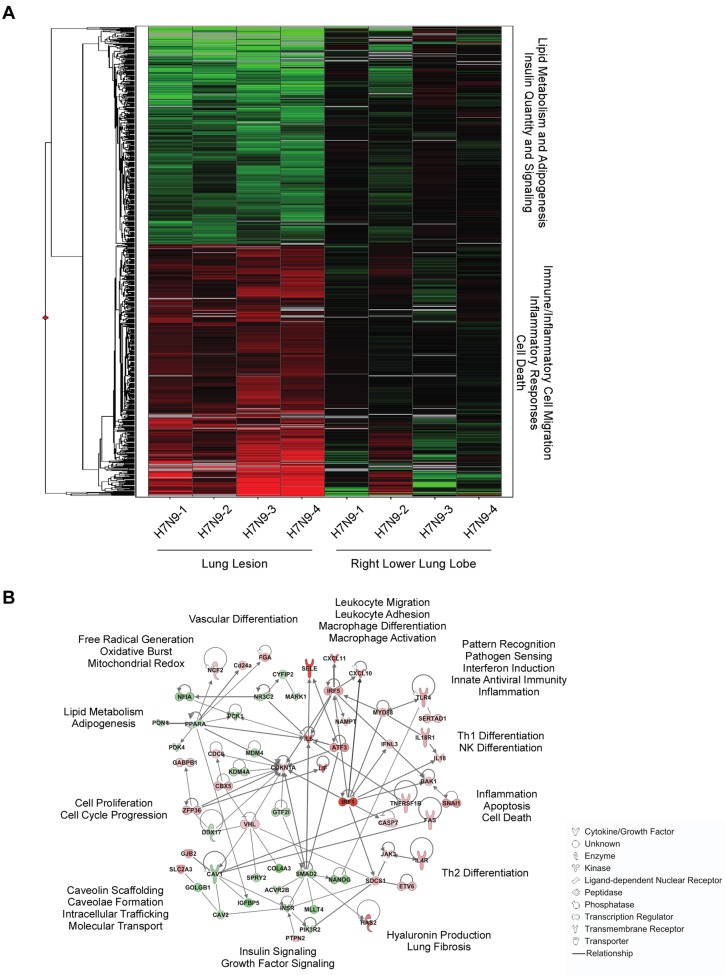
To elucidate global host responses specifically associated with sites of virus-induced airway injury in influenza virus A/Anhui/1/2013-infected macaques, we used microarrays to assess transcriptional profiles induced in lung lesions compared to the adjacent lung tissue. Welch’s t test (P < 0.05; change > 1.5-fold) identified 802 differentially expressed genes (DEG) in the lesions compared to lung samples from the same animals at 3 dpi (Fig. 7A; also, see Table S4 in the supplemental material); these 802 genes were differentially expressed in all animals. Of those, 429 were upregulated and 372 were downregulated in lesions compared to samples from the right lower lung lobe. Using Ingenuity pathway analysis (IPA), we assessed the functional significance of this molecular signature and constructed a network of critical molecules based on direct interactions in the IPA knowledge base (IPKB) (Fig. 7B). As expected, there are numerous transcripts upregulated from functional categories previously observed in cynomolgus macaque models of influenza infection (18–21), including known mediators of antiviral immunity, such as pattern recognition receptors and downstream signaling molecules (TLR4 and MYD88), interferons and interferon-stimulated genes (IFNL3), interferon-regulatory factors (IRF1, IRF5, and JAK3), and proinflammatory cytokines and inflammatory mediators (IL-6, NLR family, NLRP3, TNFRSF1B, TNFRSF6B, and TNFSF8). We also identified a number of upregulated genes associated with leukocyte migration and differentiation (CXCL10, CXCL11, SELE, IL4R, IL-18, and CSF2RA). This suggests that molecules that recruit infiltrating effector leukocytes are increased at the site of lesions and is in line with the observed influx of neutrophils and macrophages observed microscopically in the lungs. Moreover, hyaluronic acid synthase 2 (HAS2), a molecule known to be associated with lung injury, was strongly upregulated in the lung lesion samples.
FIG 7 .
Transcriptional signatures associated with influenza virus A/Anhui/1/2013 infection in cynomolgus macaques. Cynomolgus macaques were inoculated with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013; 4 animals were euthanized 3 days postinoculation, and tissue samples were collected from the right lower lung lobes and from lung lesions. (A) Heat map showing log2 expression ratios of 802 DEG in lung lesions relative to the right lower lung lobe in individual animals, as determined by Welch’s t test (P < 0.05, fold change ≥1.5), and grouped by hierarchical clustering. (B) Molecular interaction network built using DEG shown in panel A. Solid lines show direct molecular interactions. Red molecules are those upregulated in lesions relative to lung, while green molecules are downregulated. Text on the outside of the network indicates functional categories in which molecules in this part of the network cluster.
We also observed several functional categories of molecules that were downregulated in lesions relative to the adjacent lung tissue (Fig. 7B). These were associated predominantly with lipid metabolism and adipogenesis regulated by peroxisome proliferator-activated receptor alpha (PPARα). We also observed downregulation of growth factors, particularly those related to insulin signaling and regulation of glucose levels (INSR and IGFBP5).
By 6 dpi, 154 DEG were identified in lung tissue versus lung lesions of animals euthanized at 6 dpi (see Fig. S1 and Table S4 in the supplemental material). Due to substantial variability between animals, we were not able to determine any significantly enriched functional categories at this time point.
Next, the Upstream Analysis function in IPA was used to identify drugs predicted to act as significant upstream regulators of the key DEG identified at 3 dpi. Drugs that are predicted to inhibit pathological host responses were selected based on the activation z score. Negative activation z scores are predicted to cause opposite or inhibitory effects on significant genes associated with pathology in the lesions. These findings were confirmed using Connectivity Map (CMap), a resource allowing comparison with transcriptional signatures from multiple cell types treated with a library of chemical and genetic perturbagens (22), by determining if the connectivity score was also negative, indicating that the compound would have inhibitory effects on transcriptional signatures associated with lesion formation. We identified ten compounds in IPA (Table 1), four of which were perturbagens listed in CMap. We identified two compounds that met our criteria in IPA and CMap, rosiglitazone and simvastatin, predicted to have inhibitory effects on pathological host responses associated with lesions in influenza virus A/Anhui/1/2013-infected animals (Table 1). These molecules are both involved in regulating lipid biosynthesis and metabolism. Rosiglitazone is an FDA-approved diabetes drug that modulates PPAR activity and insulin sensitivity and has been shown to affect RNA virus replication (23–25), virus-induced inflammation (26, 27), and lung inflammation (28–30). Simvastatin is a FDA-approved statin which lowers blood cholesterol by inhibiting 3-hydroxy-3-methyl-glutaryl coenzyme A (HMG-CoA) reductase. It has been shown to reduce lung inflammation in mouse models of airway injury (31–34) and bacterial infections (35–37). However, simvastatin was not shown to have a significant effect on influenza A virus pathogenesis in mice (38–40). Besides rosiglitazone and simvastatin, mainly immunomodulatory drugs were predicted to have inhibitory effects on pathological host responses to A/Anhui/1/2013 infection.
Table 1 .
Top ten drugs and chemicals predicted by Ingenuity pathway analysis to inhibit lung lesion-specific host responses after infection with influenza virus A/Anhui/1/2013
| Drug | Drug class | IPA activation z score | IPA P value of overlap | CMap enrichment score | FDA-approved formulation |
|---|---|---|---|---|---|
| Rosiglitazone | Thiazolidinedione (PPAR antagonist; insulin sensitizer) | −2.358 | 8.41 × 10−6 | −0.146 | Yes (Avandia) |
| Dexamethasone | Glucocorticoid steroid (anti-inflammatory) | −2.234 | 0.000125 | 0.321 | Yes (generic) |
| Tyrphostin AG-1478 | Tyrosine kinase inhibitor (EGFR signaling inhibitor) | −2.425 | 0.00125 | 0.754 | No |
| Simvastatin | Statin (HMG-CoA reductase inhibitor) | −2.096 | 0.00226 | −0.535 | Yes (generic) |
| PD98059 | Kinase inhibitor (MEK inhibitor) | −2.838 | 0.00228 | NA | No |
| Infliximab | Monoclonal antibody (anti-TNF-α) | −2.236 | 0.00424 | NA | Yes (Remicade) |
| U0126 | Kinase inhibitor (MEK inhibitor) | −2.837 | 0.0045 | NA | No |
| Oleic acid | Monounsaturated fatty acid | −2.4 | 0.00556 | NA | No |
| Pyrrolidine dithiocarbamate | Cell cycle inhibitor, nitric oxide synthase inhibitor, metal chelator | −2.169 | 0.013 | NA | No |
| Curcumin | Antioxidant (anti-inflammatory) | −2.123 | 0.0703 | NA | No |
The ability of rosiglitazone to inhibit A/Anhui/1/2013 replication was tested in MDCK cells treated with different concentrations of this drug after infection with A/Anhui/1/2013 at a multiplicity of infection of 0.001. Twenty-four hours after addition of 100 µM rosiglitazone, but not at lower concentrations, a statistically significant (P = 0.0314; two-tailed unpaired t test) 14-fold reduction in virus titers in the supernatant of treated, infected cells compared to mock-treated cells was observed (see Fig. S2A in the supplemental material); a small cytotoxic effect of the drug was also noticed at this time point in an MTS [3-(4,5-dimethyl-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] assay (see Fig. S2B in the supplemental material). At 72 h after treatment, the inhibitory effect of rosiglitazone on A/Anhui/1/2013 replication could no longer be detected (data not shown).
DISCUSSION
The recent emergence of H7N9 influenza A virus in humans in China has shifted the focus away from HPAI H5N1 to H7N9 as one of the main pandemic threats. Although much attention has been drawn to the ability of this virus to be transmitted between ferrets, the pathogenicity of H7N9 influenza A virus in ferrets does not match the severity of disease observed in human cases (10–13). The physiology of the cynomolgus macaque lung is more similar to that of humans, and cytokine and chemokine responses to infection in macaques are similar to those in humans (14). Moreover, it was shown recently that the pattern of attachment of H7N9 influenza A virus to respiratory tissues of cynomolgus macaques is more similar to the attachment pattern in humans than that in other animal models frequently used for influenza A virus pathogenesis studies, including the ferret (15). Therefore, we studied the H7N9 infection of cynomolgus macaques in detail.
In agreement with the observed abundant attachment of the H7N9 influenza A virus to the human upper and lower respiratory tracts (41), and the replication of the H7N9 virus in ex vivo cultures of the human upper as well as lower respiratory tract (42), H7N9 virus replicated well in the upper and lower respiratory tracts of cynomolgus macaques, as indicated by virus titers in nasal turbinates, oronasopharynges, tracheas, bronchi, and lung tissue samples, reflecting the previously described receptor distribution of influenza virus H7N9 in the macaque respiratory tract (15). Surprisingly, the virus titers in nasal turbinates, oronasopharynges, tracheas, and bronchi did not decrease between 3 and 6 dpi, unlike the virus titers in the lung samples, which did decrease. This extended virus replication in the upper respiratory tract could result in prolonged virus shedding and thus an increased risk of virus transmission.
The histopathology of the lungs in fatal human H7N9 cases was similar to, albeit more severe than, that observed in cynomolgus macaques, with diffuse alveolar damage, infiltration of polymorphonuclear cells, formation of hyaline membranes, and, later after onset of symptoms, pneumocyte hyperplasia and fibroproliferative changes (4). However, clinically and histopathologically, H7N9 infection was not as severe in cynomolgus macaques as it has been described in humans, where H7N9 has a high case fatality rate. Although viral characteristics could cause differences in disease severity between humans and macaques, another possible explanation for the discrepancy in disease severity could be the high percentage of human cases with underlying medical complications (3).
Since some of the H7N9 influenza virus strains isolated from human cases have an R292K substitution in NA that renders them partially resistant to treatment with neuraminidase inhibitors (12, 43–45), it is important to identify drugs that either directly inhibit virus replication in the host or reduce the severity of disease and the level of lung injury after infection. By analyzing the gene expression profiles in lung lesions compared to adjacent infected, nonlesional lung tissue, we were able to predict drugs reported to act as upstream regulators of some of these genes that may thus play a role in the development of lung lesions during H7N9 infection. One of the predicted drugs, rosiglitazone, modestly reduced replication of influenza virus A/Anhui/1/2013 in vitro. A similar data analysis resulted in the identification of a compound that reduced replication of the Middle East respiratory syndrome coronavirus in vitro (46); however, the validity of this approach has not yet been tested in vivo. Thus, this analysis of the gene expression profile hints at new avenues of treatment to explore in in vivo models, rather than revealing novel treatments directly applicable in the clinic.
Although it is difficult to compare studies investigating the pathogenicity of different influenza A viruses in the macaque model, because of different inoculation routes and doses used and because sampling schemes do not overlap between studies, the H7N9 infection in macaques in our study was similar to that described previously (12). In both studies, animals developed transient clinical signs, with virus replication in the upper as well as lower respiratory tract and similar histopathological lesions in the lower respiratory tract. More importantly, H7N9 infection seemed to be clinically more severe than most infections with isolates of pandemic H1N1 (16); virus shedding from the throat was higher in H7N9-infected animals, and a larger area of the lung was affected with gross lesions (47). Inoculation of cynomolgus macaques with seasonal H3N2 influenza A virus resulted in infection with mild or even asymptomatic disease (48, 49). Cynomolgus macaques inoculated via the same route and with the same dose of 1918 Spanish influenza virus developed lethal disease (50), and HPAI H5N1 influenza A virus inoculated into macaques at a 40-fold-lower dose caused more severe disease than H7N9 influenza A virus in this study (51). Thus, the emerging H7N9 influenza virus is more pathogenic than seasonal influenza A virus and most isolates of the pandemic H1N1 virus but not as pathogenic as the 1918 Spanish influenza virus and HPAI H5N1 virus in cynomolgus macaques. However, the pathogenicity of the H7N9 virus may decrease if the virus adapts further to solely using α2,6-linked sialic acids as the receptor for entry, as pandemic influenza viruses to date have done (52–55). Exclusive attachment to α2,6-linked sialic acids would most likely result in a shift to replication mainly in the upper respiratory tract of humans, likely resulting in less severe disease, as has been described for the 2009 pandemic H1N1 virus (56) and upon adaptation of HPAI H5N1 virus to efficient transmission via respiratory droplets or aerosols (57).
MATERIALS AND METHODS
Ethics statement.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Rocky Mountain Laboratories and performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC), by certified staff in an AAALAC-approved facility.
Cells.
Madin-Darby canine kidney (MDCK) cells were cultured in Eagle’s modified essential medium (EMEM) (Gibco) supplemented with 10% fetal calf serum (FCS), 50 IU/ml penicillin, 50 µg/ml streptomycin, 2 mM glutamine, 0.75 mg/ml sodium bicarbonate, and nonessential amino acids.
Virus.
A/Anhui/1/2013 (passage E2/E1) was obtained from the Centers for Disease Control in Atlanta, GA, and passaged once in MDCK cells.
Animal study and sample collection.
Eight cynomolgus macaques (4 males, 4 females; age, 5 years; 4 to 6 kg) were inoculated with 7 × 106 TCID50 of A/Anhui/1/2013 via a combination of intratracheal (4 × 106 TCID50; 4 ml), intranasal (1 × 106 TCID50; 500 µl/nostril), oral (1 × 106 TCID50; 1 ml), and ocular (1 × 106 TCID50; 20 µl/eye) routes. The animals were observed twice daily for clinical signs of disease and scored using a previously described clinical scoring system (58). At 1, 2, 3, 4, and 6 days postinoculation, clinical exams were performed on anesthetized animals, and lateral X rays were taken and analyzed by a veterinarian. Nasal, oral, urogenital, and rectal swabs were collected in 1 ml Dulbecco’s modified essential medium (DMEM) with 50 U/ml penicillin and 50 µg/ml streptomycin; bronchoalveolar lavages (BAL) were performed using 10 ml sterile saline solution; blood was collected for hematology, blood chemistry analysis, and peripheral blood mononuclear cell (PBMC) isolation. The total white blood cell count, lymphocyte, platelet, reticulocyte, and red blood cell counts, hemoglobin and hematocrit values, mean cell volume, mean corpuscular volume, and mean corpuscular hemoglobin concentrations were determined from EDTA-containing blood with the Hemavet 950FS+ hemoglobin analyzer (Drew Scientific). PBMC were isolated by centrifugation over a Histopaque gradient (Sigma) as per the manufacturer’s recommendation. At 3 and 6 dpi, 4 macaques were euthanized, and samples of the conjunctivas, right and left eyes, nasal turbinates, tonsils, oronasopharynges, tracheas, right and left bronchi, all six lung lobes, lung lesions, mediastinal lymph nodes, hearts, livers, spleens, kidneys, stomachs, jejuna, ilea, transverse colons, olfactory bulbs, cerebella, brain stems, and bone marrow were collected. The percentage of the lung surface area affected by gross lung lesions was quantitated for each lung lobe, ventrally and dorsally, by a board-certified veterinary pathologist at the time of necropsy.
Histopathology and immunohistochemistry.
Histopathology and immunohistochemistry were performed on macaque tissues. After fixation for 7 days in 10% neutral buffered formalin and embedding in paraffin, tissue sections were stained with hematoxylin and eosin (H&E). To detect influenza A virus antigen, immunohistochemistry was performed using an anti-NP monoclonal HB-65 antibody (EVL, the Netherlands) as a primary antibody.
RNA extraction.
RNA was extracted from swabs, BAL fluid, and whole-blood samples using the QIAamp viral RNA kit (Qiagen) according to the manufacturer’s instructions. Tissues were stored at −80°C until further processing; tissue samples (30 mg) were homogenized in RLT buffer, and RNA was extracted using the RNeasy kit (Qiagen).
Quantitative real-time RT-PCR.
A one-step real-time RT-PCR targeted at the matrix gene of influenza A virus was performed using the Quantifast probe kit (Qiagen) according to instructions of the manufacturer using the primers and probe described in reference 59.
Virus titrations.
Viruses were titrated by endpoint dilution in MDCK cells. MDCK cells were inoculated with tenfold serial dilutions of culture supernatants. One hour after inoculation, cells were washed with phosphate-buffered saline (PBS) and supplemented with infection medium (EMEM supplemented with 50 IU/ml penicillin, 50 µg/ml streptomycin, 2 mM glutamine, 0.75 mg/ml sodium bicarbonate, nonessential amino acids, and 5 µg/ml trypsin). Three days after inoculation, the supernatants of infected cell cultures were tested for agglutination activity using turkey red blood cells as an indicator of infection of the cells. Infectious titers were calculated from 5 replicates by the Spearman-Karber method (60).
Serum cytokine and chemokine analysis.
Serum samples for analysis of cytokine/chemokine levels were inactivated with gamma radiation (5 megarads) according to standard operating procedures. Concentrations of granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, IFN-γ, IL-1β, IL-1 receptor antagonist, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12/23 (p40), IL-13, IL-15, IL-17, MCP-1, MIP-1α, MIP-1β, soluble CD40 ligand (sCD40L), transforming growth factor α, TNF-α, vascular endothelial growth factor (VEGF), and IL-18 were measured on a Bio-Plex 200 instrument (Bio-Rad) using the nonhuman primate cytokine Milliplex 23-plex map kit (Millipore) according to the manufacturer’s instructions.
Microarray data and functional analysis.
RNA was extracted using Qiagen micro-RNeasy spin columns per the manufacturer’s protocol. Low-yield samples were concentrated using the RNA Clean and Concentrator kit (Zymo Research, Irvine, CA). As multiple lesions were collected from each animal, equal masses of RNA from these samples (3 or 4 samples per animal) were pooled to investigate comprehensive signatures from multiple pathological sites within the same individual. Probe labeling was carried out using the Agilent low-input processing protocol and hybridized to Agilent rhesus macaque 8×60K microarrays (Agilent Technologies, Santa Clara, CA) using the manufacturer’s one-color analysis protocol. For comparisons of differentially expressed genes (DEG) in infected lungs and lung lesions, raw array data were uploaded to Genedata Analyst 7.6 (Genedata Inc., San Francisco, CA). Data were normalized using the quantile normalization method, and the log2 ratio expression was calculated relative to the mean probe values of the 4 right lower lung lobe samples per time point (lesion versus lung comparisons). Statistically significant DEG were identified using Welch’s t test (P < 0.05; fold change, ≥1.5). Hierarchical clustering of DEG was performed by the unweighted average method (unweighted pair group with arithmetic mean [UPGMA]) using Spotfire DecisionSite 9.1.1 (Tibco, Somerville, MA). Analysis of functional enrichment was performed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA), and upstream drug efficacy predictions were made using both the Upstream Analysis function of IPA and Connectivity Map 02 (Broad Institute, Cambridge, MA). Drugs predicted to inhibit pathological host responses were selected based on the activation z score. Negative activation z scores are predicted to cause opposite or inhibitory effects on significant genes associated with pathology in the lesions, and in this study we sought compounds with activation z scores less than −2 and a P value less than 0.05. These findings were confirmed using Connectivity Map (CMap) (22). CMap assigns enrichment scores ranging between −1 and 1, and we accepted compounds with CMap enrichment scores less than −0.1. These are compounds inducing transcriptional profiles that are negatively connected with the gene expression signature associated with lung pathology, confirming that these drugs may induce a transcriptional profile that is inhibitory to tissue damage and lesion formation.
Antiviral assay.
Confluent MDCK cells in 24-well culture plates were infected in triplicate with influenza virus A/Anhui/1/2013 at a multiplicity of infection of 0.001. After 1 h at 37°C, cells were washed once with PBS, and infection medium containing rosiglitazone (0 to 100 µM) was added to the cells. Cells were incubated for 24 h; supernatant was then harvested, stored at −80°C for subsequent virus titration, and replaced with fresh infection medium containing rosiglitazone. Supernatant was again collected at 72 h after infection and stored at −80°C for subsequent virus titration. To determine a potential cytotoxic effect of rosiglitazone, MDCK cells were simultaneously plated in 96-well culture plates and treated with rosiglitazone (0 to 100 µM). After 24 h incubation, cytotoxicity was tested using the CellTiter 96 AQueous one-solution cell proliferation assay (MTS) (Promega) according to the manufacturer’s instructions.
Microarray data accession number.
Raw microarray data have been deposited in NCBI’s Gene Expression Omnibus database (GSE48976) and are also available to the public at http://viromics.washington.edu.
SUPPLEMENTAL MATERIAL
Transcriptional signatures associated with influenza virus A/Anhui/1/2013 infection in cynomolgus macaques. Cynomolgus macaques were inoculated with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013; 4 animals were euthanized 6 days postinoculation, and tissue samples were collected from the right lower lung lobes and from lung lesions. The heat map shows log2 expression ratios of 154 DEG in lung lesions relative to the right lower lobe of the lung from individual animals, as determined by Welch’s t test (P < 0.05, fold change ≥1.5), and grouped by hierarchical clustering. Download
Effect of rosiglitazone on replication of influenza virus A/Anhui/1/2013 in MDCK cells. (A) MDCK cells were infected with influenza virus A/Anhui/1/2013 at a MOI of 0.001 and subsequently treated with different concentrations of rosiglitazone. At 24 h postinfection, supernatants were harvested, and virus titers in the supernatants were determined by endpoint titration on MDCK cells. Geometric mean virus titers are indicated; error bars indicate standard deviations, and asterisks indicate a statistically significant effect (P < 0.05; two-tailed unpaired t test). (B) The cytotoxic effect of the different rosiglitazone concentrations on MDCK cells was tested using an MTS assay. The average OD, measured at 405 nm, calculated from three individual wells is indicated; error bars indicate standard deviations. Download
Clinical scores of A/Anhui/1/2013-inoculated cynomolgus macaques. Animals were observed twice daily, and clinical parameters were recorded.
Summary of radiographic findings for A/Anhui/1/2013-inoculated cynomolgus macaques. The quality and location of interstitial infiltration observed from ventral-dorsal and lateral X rays are indicated.
Viral RNA in tissues from cynomolgus macaques infected with influenza virus A/Anhui/1/2013. Numbers are CT values obtained in real-time RT-PCR.
Overview of the differentially expressed genes shown in Fig. 7 and in Fig. S1 in the supplemental material, comparing samples from the right lower lung lobe or lung lesions collected at 3 and 6 days postinoculation to those from uninfected controls.
ACKNOWLEDGMENTS
We thank Dana Scott, Tina Thomas, Rebecca Rosenke, Dan Long, Doug Brining, Forrest Hoyt, Elizabeth Fischer, Darryl Falzarano, Anita Mora, Austin Athman, and animal caretakers of the Rocky Mountain Laboratories, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and Nicolas Tchitchek and Juliet Morrison of the University of Washington for assistance.
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health; A.L.R., A.O., S.C.P., J.C., and M.G.K. were funded through NIH/NIAID contract number HHSN272200800060C, grant P51 OD010425 51, to the Washington Primate National Research Center, Division of Nonhuman Primate Systems Biology, and grant 8R24 OD011157, Development of a Primate Genomics Resource.
Footnotes
Citation de Wit E, Rasmussen AL, Feldmann F, Bushmaker T, Martellaro C, Haddock E, Okumura A, Proll SC, Chang J, Gardner D, Katze MG, Munster VJ, Feldmann H. 2014. Influenza virus A/Anhui/1/2013 (H7N9) replicates efficiently in the upper and lower respiratory tracts of cynomolgus macaques. mBio 5(4)e01331-14. doi:10.1128/mBio.01331-14.
REFERENCES
- 1. Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 2. WHO 2014. Confirmed human cases of avian influenza A(H7N9) reported to WHO. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/13_ReportWebH7N9Number_20140221.pdf [Google Scholar]
- 3. Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N. Engl. J. Med. 368:2277–2285. 10.1056/NEJMoa1305584 [DOI] [PubMed] [Google Scholar]
- 4. Yu L, Wang Z, Chen Y, Ding W, Jia H, Chan JF, To KK, Chen H, Yang Y, Liang W, Zheng S, Yao H, Yang S, Cao H, Dai X, Zhao H, Li J, Bao Q, Chen P, Hou X, Li L, Yuen KY. 2013. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin. Infect. Dis. 57:1449–1457. 10.1093/cid/cit541 [DOI] [PubMed] [Google Scholar]
- 5. Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg. Infect. Dis. 15:859–865. 10.3201/eid1506.090072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361. 10.1073/pnas.0308352100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 18:20453. [PMC free article] [PubMed] [Google Scholar]
- 8. Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. 10.1038/nature12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. 2013. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect. 15:432–439. 10.1016/j.micinf.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 10. Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2013. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501:560–563. 10.1038/nature12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]
- 14. O’Donnell CD, Subbarao K. 2011. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect. 13:502–515. 10.1016/j.micinf.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siegers JY, Short KR, Leijten LM, de Graaf M, Spronken MI, Schrauwen EJ, Marshall N, Lowen AC, Gabriel G, Osterhaus AD, Kuiken T, van Riel D. 2014. Novel avian-origin influenza A (H7N9) virus attachment to the respiratory tract of five animal models. J. Virol. 88:4595–4599. 10.1128/JVI.03190-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Safronetz D, Rockx B, Feldmann F, Belisle SE, Palermo RE, Brining D, Gardner D, Proll SC, Marzi A, Tsuda Y, Lacasse RA, Kercher L, York A, Korth MJ, Long D, Rosenke R, Shupert WL, Aranda CA, Mattoon JS, Kobasa D, Kobinger G, Li Y, Taubenberger JK, Richt JA, Parnell M, Ebihara H, Kawaoka Y, Katze MG, Feldmann H. 2011. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J. Virol. 85:1214–1223. 10.1128/JVI.01848-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richt JA, Rockx B, Ma W, Feldmann F, Safronetz D, Marzi A, Kobasa D, Strong JE, Kercher L, Long D, Gardner D, Brining D, Feldmann H. 2012. Recently emerged swine influenza A virus (H2N3) causes severe pneumonia in cynomolgus macaques. PLoS One 7:e39990. 10.1371/journal.pone.0039990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, García-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, Aicher LD, Rosenzweig ER, Murali-Krishna K, Clark EA, Kotur MS, Fornek JL, Proll S, Palermo RE, Sabourin CL, Katze MG. 2009. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 106:3455–3460. 10.1073/pnas.0813234106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Go JT, Belisle SE, Tchitchek N, Tumpey TM, Ma W, Richt JA, Safronetz D, Feldmann H, Katze MG. 2012. 2009 pandemic H1N1 influenza virus elicits similar clinical course but differential host transcriptional response in mouse, macaque, and swine infection models. BMC Genomics 13:627. 10.1186/1471-2164-13-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Josset L, Engelmann F, Haberthur K, Kelly S, Park B, Kawoaka Y, García-Sastre A, Katze MG, Messaoudi I. 2012. Increased viral loads and exacerbated innate host responses in aged macaques infected with the 2009 pandemic H1N1 influenza A virus. J. Virol. 86:11115–11127. 10.1128/JVI.01571-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shoemaker JE, Fukuyama S, Eisfeld AJ, Muramoto Y, Watanabe S, Watanabe T, Matsuoka Y, Kitano H, Kawaoka Y. 2012. Integrated network analysis reveals a novel role for the cell cycle in 2009 pandemic influenza virus-induced inflammation in macaque lungs. BMC Syst. Biol. 6:117. 10.1186/1752-0509-6-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. 2006. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313:1929–1935. 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- 23. Guerrero CA, Murillo A, Acosta O, Acosta O. 2012. Inhibition of rotavirus infection in cultured cells by N-acetyl-cysteine, PPARgamma agonists and NSAIDs. Antiviral Res. 96:1–12. 10.1016/j.antiviral.2012.06.011 [DOI] [PubMed] [Google Scholar]
- 24. Wakui Y, Inoue J, Ueno Y, Fukushima K, Kondo Y, Kakazu E, Obara N, Kimura O, Shimosegawa T. 2010. Inhibitory effect on hepatitis B virus in vitro by a peroxisome proliferator-activated receptor-gamma ligand, rosiglitazone. Biochem. Biophys. Res. Commun. 396:508–514. 10.1016/j.bbrc.2010.04.128 [DOI] [PubMed] [Google Scholar]
- 25. Kim K, Kim KH, Ha E, Park JY, Sakamoto N, Cheong J. 2009. Hepatitis C virus NS5A protein increases hepatic lipid accumulation via induction of activation and expression of PPARgamma. FEBS Lett. 583:2720–2726. 10.1016/j.febslet.2009.07.034 [DOI] [PubMed] [Google Scholar]
- 26. Huang W, Rha GB, Han MJ, Eum SY, András IE, Zhong Y, Hennig B, Toborek M. 2008. PPARalpha and PPARgamma effectively protect against HIV-induced inflammatory responses in brain endothelial cells. J. Neurochem. 107:497–509. 10.1111/j.1471-4159.2008.05626.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramirez SH, Heilman D, Morsey B, Potula R, Haorah J, Persidsky Y. 2008. Activation of peroxisome proliferator-activated receptor gamma (PPARgamma) suppresses Rho GTPases in human brain microvascular endothelial cells and inhibits adhesion and transendothelial migration of HIV-1 infected monocytes. J. Immunol. 180:1854–1865. 10.4049/jimmunol.180.3.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu DS, Liu WJ, Chen L, Ou XM, Wang T, Feng YL, Zhang SF, Xu D, Chen YJ, Wen FQ. 2009. Rosiglitazone, a peroxisome proliferator-activated receptor-gamma agonist, attenuates acrolein-induced airway mucus hypersecretion in rats. Toxicology 260:112–119. 10.1016/j.tox.2009.03.016 [DOI] [PubMed] [Google Scholar]
- 29. Zhu M, Flynt L, Ghosh S, Mellema M, Banerjee A, Williams E, Panettieri RA, Jr, Shore SA. 2011. Anti-inflammatory effects of thiazolidinediones in human airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 45:111–119. 10.1165/rcmb.2009-0445OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee KS, Park SJ, Kim SR, Min KH, Jin SM, Lee HK, Lee YC. 2006. Modulation of airway remodeling and airway inflammation by peroxisome proliferator-activated receptor gamma in a murine model of toluene diisocyanate-induced asthma. J. Immunol. 177:5248–5257. 10.4049/jimmunol.177.8.5248 [DOI] [PubMed] [Google Scholar]
- 31. Chen W, Sharma R, Rizzo AN, Siegler JH, Garcia JG, Jacobson JR. 2014. Role of claudin-5 in the attenuation of murine acute lung injury by simvastatin. Am. J. Respir. Cell Mol. Biol. 50:328–336. 10.1165/rcmb.2013-0058OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin Y, Tachibana I, Takeda Y, He P, Kang S, Suzuki M, Kuhara H, Tetsumoto S, Tsujino K, Minami T, Iwasaki T, Nakanishi K, Kohmo S, Hirata H, Takahashi R, Inoue K, Nagatomo I, Kida H, Kijima T, Ito M, Saya H, Kumanogoh A. 2013. Statins decrease lung inflammation in mice by upregulating tetraspanin CD9 in macrophages. PLoS One 8:e73706. 10.1371/journal.pone.0073706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davis BB, Zeki AA, Bratt JM, Wang L, Filosto S, Walby WF, Kenyon NJ, Goldkorn T, Schelegle ES, Pinkerton KE. 2013. Simvastatin inhibits smoke-induced airway epithelial injury: implications for COPD therapy. Eur. Respir. J. 42:350–361. 10.1183/09031936.00042512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeki AA, Thai P, Kenyon NJ, Wu R. 2012. Differential effects of simvastatin on IL-13-induced cytokine gene expression in primary mouse tracheal epithelial cells. Respir. Res. 13:38. 10.1186/1465-9921-13-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu BQ, Luo JM, Wang YH, Shi YF, Liu H, Ba JH, Zhang TT. 2014. Inhibitory effects of simvastatin on Staphylococcus aureus lipoteichoic acid-induced inflammation in human alveolar macrophages. Clin. Exp. Med. 14:151–160. 10.1007/s10238-013-0231-z [DOI] [PubMed] [Google Scholar]
- 36. Boyd AR, Hinojosa CA, Rodriguez PJ, Orihuela CJ. 2012. Impact of oral simvastatin therapy on acute lung injury in mice during pneumococcal pneumonia. BMC Microbiol. 12:73. 10.1186/1471-2180-12-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDowell SA, Ma Y, Kusano R, Akinbi HT. 2011. Simvastatin is protective during Staphylococcus aureus pneumonia. Curr. Pharm. Biotechnol. 12:1455–1462. 10.2174/138920111798281027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Belser JA, Szretter KJ, Katz JM, Tumpey TM. 2013. Simvastatin and oseltamivir combination therapy does not improve the effectiveness of oseltamivir alone following highly pathogenic avian H5N1 influenza virus infection in mice. Virology 439:42–46. 10.1016/j.virol.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 39. Glück B, Schmidtke M, Walther M, Meerbach A, Wutzler P. 2013. Simvastatin treatment showed no prophylactic effect in influenza virus-infected mice. J. Med. Virol. 85:1978–1982. 10.1002/jmv.23682 [DOI] [PubMed] [Google Scholar]
- 40. Kumaki Y, Morrey JD, Barnard DL. 2012. Effect of statin treatments on highly pathogenic avian influenza H5N1, seasonal and H1N1pdm09 virus infections in BALB/c mice. Future Virol. 7:801–818. 10.2217/fvl.12.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Riel D, Leijten LM, de Graaf M, Siegers JY, Short KR, Spronken MI, Schrauwen EJ, Fouchier RA, Osterhaus AD, Kuiken T. 2013. Novel avian-origin influenza A (H7N9) virus attaches to epithelium in both upper and lower respiratory tract of humans. Am. J. Pathol. 183:1137–1143. 10.1016/j.ajpath.2013.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan MCW, Chan RWY, Chan LLY, Mok CKP, Hui KPY, Fong JHM, Tao KP, Poon LL, Nicholls JM, Peiris JS. 2013. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory tract. Lancet Respir. Med. 1:534–542. 10.1016/S2213-2600(13)70138-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z, Yuan Z. 2013. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 381:2273–2279. 10.1016/S0140-6736(13)61125-3 [DOI] [PubMed] [Google Scholar]
- 44. Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. 2013. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg. Infect. Dis. 19:1521–1524. 10.3201/eid1909.130724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yen HL, McKimm-Breschkin JL, Choy KT, Wong DD, Cheung PP, Zhou J, Ng IH, Zhu H, Webby RJ, Guan Y, Webster RG, Peiris JS. 2013. Resistance to neuraminidase inhibitors conferred by an R292K mutation in a human influenza virus H7N9 isolate can be masked by a mixed R/K viral population. mBio 4:e00396-13. 10.1128/mBio.00396-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. 2013. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 4:e00165-13. 10.1128/mBio.00165-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herfst S, van den Brand JM, Schrauwen EJ, de Wit E, Munster VJ, van Amerongen G, Linster M, Zaaraoui F, van Ijcken WF, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Andeweg AC, Kuiken T. 2010. Pandemic 2009 H1N1 influenza virus causes diffuse alveolar damage in cynomolgus macaques. Vet. Pathol. 47:1040–1047. 10.1177/0300985810374836 [DOI] [PubMed] [Google Scholar]
- 48. Rimmelzwaan GF, Baars M, van Beek R, van Amerongen G, Lovgren-Bengtsson K, Claas EC, Osterhaus AD. 1997. Induction of protective immunity against influenza virus in a macaque model: comparison of conventional and iscom vaccines. J. Gen. Virol. 78:757–765 [DOI] [PubMed] [Google Scholar]
- 49. Kobayashi SD, Olsen RJ, Lacasse RA, Safronetz D, Ashraf M, Porter AR, Braughton KR, Feldmann F, Clifton DR, Kash JC, Bailey JR, Gardner DJ, Otto M, Brining DL, Kreiswirth BN, Taubenberger JK, Parnell MJ, Feldmann H, Musser JM, Deleo FR. 2013. Seasonal H3N2 influenza A virus fails to enhance Staphylococcus aureus co-infection in a non-human primate respiratory tract infection model. Virulence 4:707–715. 10.4161/viru.26572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319–323. 10.1038/nature05495 [DOI] [PubMed] [Google Scholar]
- 51. Rimmelzwaan GF, Kuiken T, van Amerongen G, Bestebroer TM, Fouchier RA, Osterhaus AD. 2001. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 75:6687–6691. 10.1128/JVI.75.14.6687-6691.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23. 10.1006/viro.1994.1615 [DOI] [PubMed] [Google Scholar]
- 53. Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502–8512. 10.1128/JVI.74.18.8502-8512.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solórzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, García-Sastre A. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655–659. 10.1126/science.1136212 [DOI] [PubMed] [Google Scholar]
- 55. Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. 10.1126/science.1177238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Watanabe T, Shinya K, Watanabe S, Imai M, Hatta M, Li C, Wolter BF, Neumann G, Hanson A, Ozawa M, Yamada S, Imai H, Sakabe S, Takano R, Iwatsuki-Horimoto K, Kiso M, Ito M, Fukuyama S, Kawakami E, Gorai T, Simmons HA, Schenkman D, Brunner K, Capuano SV, III, Weinfurter JT, Nishio W, Maniwa Y, Igarashi T, Makino A, Travanty EA, Wang J, Kilander A, Dudman SG, Suresh M, Mason RJ, Hungnes O, Friedrich TC, Kawaoka Y. 2011. Avian-type receptor-binding ability can increase influenza virus pathogenicity in macaques. J. Virol. 85:13195–13203. 10.1128/JVI.00859-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brining DL, Mattoon JS, Kercher L, LaCasse RA, Safronetz D, Feldmann H, Parnell MJ. 2010. Thoracic radiography as a refinement methodology for the study of H1N1 influenza in cynomologus macaques (Macaca fascicularis). Comp. Med. 60:389–395 [PMC free article] [PubMed] [Google Scholar]
- 59. Munster VJ, Baas C, Lexmond P, Bestebroer TM, Guldemeester J, Beyer WE, de Wit E, Schutten M, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2009. Practical considerations for high-throughput influenza A virus surveillance studies of wild birds by use of molecular diagnostic tests. J. Clin. Microbiol. 47:666–673. 10.1128/JCM.01625-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuch [A contribution to the collective treatment of pharmacological experimental series]. Arch. Exp. Pathol. Pharmakologie 162:480–483. 10.1007/BF01863914 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptional signatures associated with influenza virus A/Anhui/1/2013 infection in cynomolgus macaques. Cynomolgus macaques were inoculated with 7 × 106 TCID50 of influenza virus A/Anhui/1/2013; 4 animals were euthanized 6 days postinoculation, and tissue samples were collected from the right lower lung lobes and from lung lesions. The heat map shows log2 expression ratios of 154 DEG in lung lesions relative to the right lower lobe of the lung from individual animals, as determined by Welch’s t test (P < 0.05, fold change ≥1.5), and grouped by hierarchical clustering. Download
Effect of rosiglitazone on replication of influenza virus A/Anhui/1/2013 in MDCK cells. (A) MDCK cells were infected with influenza virus A/Anhui/1/2013 at a MOI of 0.001 and subsequently treated with different concentrations of rosiglitazone. At 24 h postinfection, supernatants were harvested, and virus titers in the supernatants were determined by endpoint titration on MDCK cells. Geometric mean virus titers are indicated; error bars indicate standard deviations, and asterisks indicate a statistically significant effect (P < 0.05; two-tailed unpaired t test). (B) The cytotoxic effect of the different rosiglitazone concentrations on MDCK cells was tested using an MTS assay. The average OD, measured at 405 nm, calculated from three individual wells is indicated; error bars indicate standard deviations. Download
Clinical scores of A/Anhui/1/2013-inoculated cynomolgus macaques. Animals were observed twice daily, and clinical parameters were recorded.
Summary of radiographic findings for A/Anhui/1/2013-inoculated cynomolgus macaques. The quality and location of interstitial infiltration observed from ventral-dorsal and lateral X rays are indicated.
Viral RNA in tissues from cynomolgus macaques infected with influenza virus A/Anhui/1/2013. Numbers are CT values obtained in real-time RT-PCR.
Overview of the differentially expressed genes shown in Fig. 7 and in Fig. S1 in the supplemental material, comparing samples from the right lower lung lobe or lung lesions collected at 3 and 6 days postinoculation to those from uninfected controls.