ABSTRACT
The gut microbiota is essential for numerous aspects of human health. However, the underlying mechanisms of many host-microbiota interactions remain unclear. The aim of this study was to characterize effects of the microbiota on host epithelium using a novel ex vivo model based on mouse ileal organoids. We have explored the transcriptional response of organoids upon exposure to short-chain fatty acids (SCFAs) and products generated by two abundant microbiota constituents, Akkermansia muciniphila and Faecalibacterium prausnitzii. We observed that A. muciniphila metabolites affect various transcription factors and genes involved in cellular lipid metabolism and growth, supporting previous in vivo findings. Contrastingly, F. prausnitzii products exerted only weak effects on host transcription. Additionally, A. muciniphila and its metabolite propionate modulated expression of Fiaf, Gpr43, histone deacetylases (HDACs), and peroxisome proliferator-activated receptor gamma (Pparγ), important regulators of transcription factor regulation, cell cycle control, lipolysis, and satiety. This work illustrates that specific bacteria and their metabolites differentially modulate epithelial transcription in mouse organoids. We demonstrate that intestinal organoids provide a novel and powerful ex vivo model for host-microbiome interaction studies.
IMPORTANCE
We investigated the influence of the gut microbiota and microbially produced short-chain fatty acids (SCFAs) on gut functioning. Many commensal bacteria in the gut produce SCFAs, particularly butyrate, acetate, and propionate, which have been demonstrated to reduce the risk of gastrointestinal disorders. Organoids—small crypt-villus structures grown from ileal intestinal stem cells—were exposed to SCFAs and two specific gut bacteria. Akkermansia muciniphila, found in the intestinal mucus, was recently shown to have a favorable effect on the disrupted metabolism associated with obesity. Faecalibacterium prausnitzii is a commensal gut bacterium, the absence of which may be associated with Crohn’s disease. We showed that in our model, A. muciniphila induces stronger effects on the host than F. prausnitzii. We observed that A. muciniphila and propionate affect the expression of genes involved in host lipid metabolism and epigenetic activation or silencing of gene expression. We demonstrated that organoids provide a powerful tool for host-microbe interaction studies.
INTRODUCTION
The gut microbiota plays an important role in the regulation of human health and pathogenesis of disease, such as obesity, type 2 diabetes, and inflammatory bowel disease (IBD) (1–4).
Common microbial metabolites, such as butyrate, acetate, propionate, and other short-chain fatty acids (SCFAs), have been shown to affect multiple intestinal cell signaling pathways. Besides serving as an energy source, SCFAs can signal through several G protein-coupled receptors to elicit a wide range of cellular responses (5). Butyrate has been shown to inhibit histone deacetylases (HDACs), thereby inducing histone hypermethylation, leading to changes in gene transcription (6). In addition, butyrate was found to induce a specific transcriptional response in the human colon, affecting genes and pathways involved in fatty acid oxidation, epithelial integrity, and apoptosis (7). Elucidation of the functional and mechanistic in vivo implications of these host-microbiota interactions holds great potential for the development of therapeutic targets for many metabolic (and possibly other) diseases (8).
The gut microbiota has been shown to be involved in regulation of intestinal barrier function and nutrient absorption, as well as fat metabolism and storage in mice (9–11). However, effects of the microbiota on the host metabolism are highly diet, species, and strain dependent, making it difficult to draw definitive conclusions regarding underlying mechanisms (12, 13). To circumvent issues of diet and species discrepancy and to study direct effects of specific bacteria on intestinal epithelial function at the molecular and cellular levels, several in vitro studies with both human and rodent intestinal epithelial cell lines exposed to bacterial monocultures or SCFAs have been performed. Grootaert et al. (14) studied the effects of bacterial monocultures (Clostridium perfringens, Enterococcus faecalis, Bacteroides thetaiotaomicron, and Escherichia coli) and SCFAs on human intestinal epithelial cell lines, where both bacterial monocultures and SCFAs were shown to modulate fasting-induced adipose factor/angiopoietin-like protein (Fiaf/Angptl4) expression. The thoroughly investigated Fiaf protein is a multifunctional signal protein expressed in several tissues (15) and is upregulated during fasting, hypoxia, and adipocyte differentiation (16, 17) and has been found to link the gut microbiota and obesity (18). In addition, Fiaf has been suggested to modulate lipid metabolism in different tissues (19). These studies extended our understanding of the role of microbial communities in regulation of host cellular function, but the effects analyzed were only representative for one epithelial cell type. Moreover, the cell lines used, which have been maintained for decades in the laboratory, may not show the dynamic responses found in primary and differentiated intestinal cells. Hence, the effect of the gut microbiota on other epithelial cell types and mucosal function remains to be elucidated.
To study and understand the dynamic interactions between the intestinal epithelium and the microbiota, a need for robust, physiologically representative, and reproducible model systems has emerged (20). Organoids, cultured from crypts or single Lgr5+ stem cells, hold great potential as a sophisticated intestinal model (21). These stem-cell-based gut organoids make an attractive ex vivo model system in two ways. First, organoids show self-renewing capacity. Second, they recapitulate the in vivo tissue architecture, consisting of both stem cells and differentiated functional epithelial cells, namely, enterocytes, goblet cells, enteroendocrine cells, and Paneth cells, the latter previously impossible to culture (21, 22).
The aim of this study was to determine the effects of important intestinal bacteria and their products on host ileal epithelium gene expression.
For this purpose, we have selected two symbiotic and numerically abundant members of the human gut microbiota, Akkermansia muciniphila and Faecalibacterium prausnitzii. A. muciniphila is a mucus-colonizing member of the microbiota and may constitute up to 3% of the gut microbiota (23). Its mucin degradation activity leads to the production of propionate and acetate (24). An extensive study regarding the effects of A. muciniphila on host intestinal function revealed modulation of host intestinal epithelial genes involved in basal metabolism (25). F. prausnitzii is also an abundant intestinal anaerobe that can make up approximately 4% of the mainly luminal microbiota (26). It induces an anti-inflammatory immune response in a mouse model of inflammation, while butyrate, the main SCFA produced by F. prausnitzii, was indicated to be unresponsive in the disease suppression (27). Both bacteria are considered promising next generation probiotics (28).
The effects of bacterial species and SCFA on host gene expression were previously assessed in mouse ileal intestinal tissue and Caco-2 cells (25, 29, 30) The latter phenotypically resemble differentiated ileal intestinal enterocytes, both morphologically and physiologically, rather than colonic epithelial cells (31).
In addition to the colon, the microbiota of the distal ileum, which consists of up to 108 bacterial cells/g, including A. municiphila and F. prausnitzii (32), is another source of SCFA production. For these reasons, we applied distal ileal organoids to elucidate the effect of these microbiota members and their products on intestinal epithelial cells.
We exposed mature ileal organoids to supernatant collected from A. muciniphila and F. prausnitzii cultures, as well as to individual butyrate, propionate, and acetate solutions.
Subsequently, an extensive analysis of gene transcription modulations and pathway analysis was performed by means of a microarray analysis.
We focused on 5 genes coding for the following products that have been shown to be involved in cell cycle control, adipocyte function, and peripheral lipid metabolism: (i) Fiaf, involved in the deposition of triglycerides in adipocytes (19); (ii) G protein-coupled receptor 43 (Gpr43), which binds SCFAs and is involved in several pathological conditions, such as obesity and inflammatory diseases (33); (iii) histone deacetylase 3 (Hdac3) and (iv) Hdac5, epigenome-modifying enzymes responsible primarily for deacetylation of lysine residues within histones (6); and (v) peroxisome proliferator-activated receptor gamma (Pparγ), previously shown to be involved in microbiota-induced expression of Fiaf (34).
The results revealed distinct transcriptional responses elicited by the intestinal symbionts and indicate that A. muciniphila has the strongest impact, notably on the lipid metabolism, as may be expected from its location within the intestinal mucus layer.
RESULTS
Growth of A. muciniphila and F. prausnitzii and SCFA analysis of the conditioned media.
A. muciniphila and F. prausnitzii were grown anaerobically, harvested at the end of their exponential phase, and used for later incubation studies. To further understand the role of the conditioned medium, its SCFA and residual sugar compositions were determined. The medium conditioned by A. muciniphila contained residual concentrations of 0.71 mM glucose and 0.77 mM l-fucose, while the SCFAs included 3.65 mM acetate and 7.14 mM propionate. The medium conditioned by F. prausnitzii contained a residual amount of 0.16 mM glucose, while the SCFAs included 1.51 mM acetate, 5.51 mM formate, 7.06 mM propionate, and 8.03 mM butyrate. It should be noted that the small amount of propionate in the conditioned growth medium of F. prausnitzii is due to the addition of this SCFA to stimulate growth, mimicking the rumen conditions (35). At 7 days after splitting (Fig. 1), mature intestinal organoids were exposed to either the cultured cells or the conditioned medium from both strains for 3 h, followed by the gene expression analysis as described below.
FIG 1 .
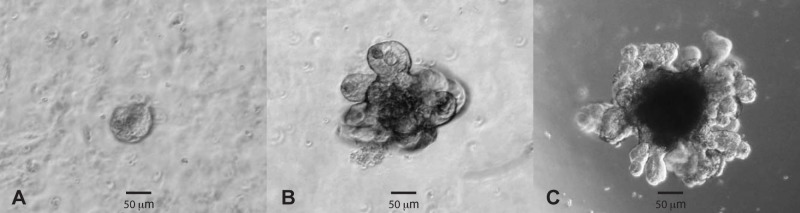
Bright-field images of mouse small-intestinal organoids cultured in Matrigel for 1 (A), 3 (B), and 7 (C) days after splitting.
Exposure to A. muciniphila and F. prausnitzii results in strain-specific effects on gene expression of intestinal organoids.
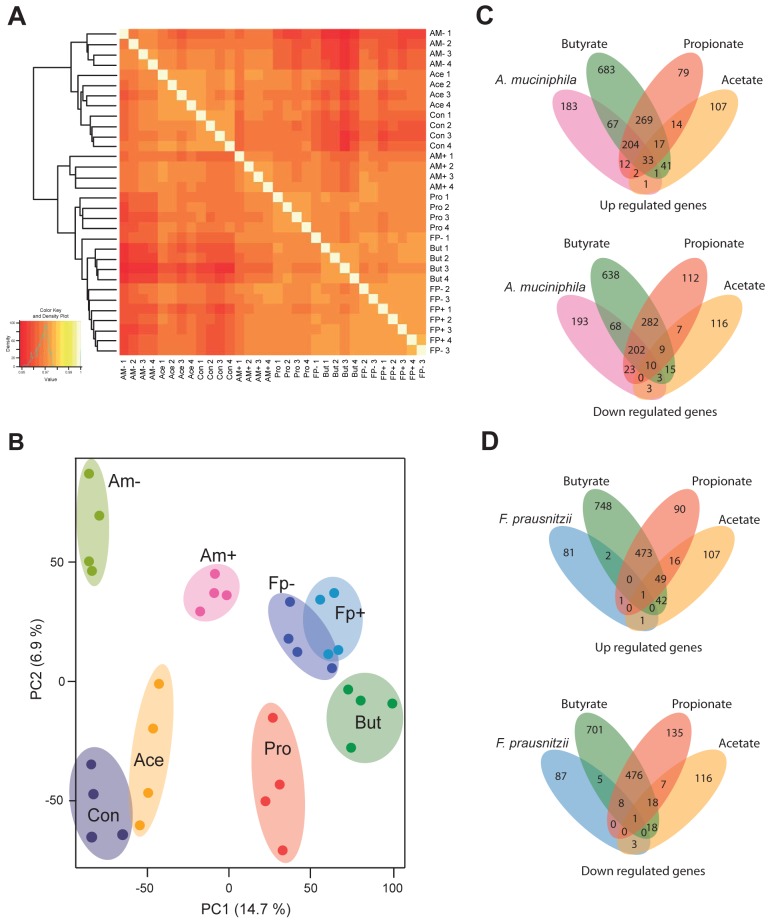
Organoids exposed to either of the bacterial monoculture-conditioned media showed distinct transcriptomic responses, as indicated by the hierarchical clustering of genes of replicates exposed to same conditions (Fig. 2A). Analysis of the data through an unsupervised clustering method (principal component analysis [PCA]) further supported the specific effects of the conditioned media on the clustering of expressed genes of the intestinal organoid genes (Fig. 2B). On top of this, organoids exposed to A. muciniphila-conditioned medium (Am+ in Fig. 2B) showed a distinct pattern of gene expression compared to its control consisting of only growth medium (Am− in Fig. 2B). Hence, it is evident that A. muciniphila compounds induce a different cluster of genes than just its culture medium as these are well separated in the PCA plot. In contrast, an overlapping set of clusters of genes are found in organoids that have been exposed to F. prausnitzii-conditioned medium (Fp+ in Fig. 2B) and its control medium (Fp− in Fig. 2B), suggesting that metabolites from F. prausnitzii do not have a major effect on the gene expression profile. Interestingly, the number of overlapping genes affected by both A. muciniphila- and F. prausnitzii-conditioned media was very low, namely 7 in total (see Fig. S1 in the supplemental material).
FIG 2 .
Distinct transcriptome signatures in intestinal organoids upon stimulation with bacterial monocultures and SCFAs. (A) Heat map and hierarchical clustering representing the array correlation plot of microarray data from all replicates. (B) PCA plot of the microarray data showing the distribution of different experimental treatment groups. Clusters represent samples exposed to A. muciniphila culturing medium (Am−), A. muciniphila-conditioned medium (Am+), F. prausnitzii culturing medium (Fp−), F. prausnitzii-conditioned medium (Fp+), acetate (Ace), butyrate (But), propionate (Pro), and standard organoid culturing medium (Con). (C) Venn diagrams representing the number of genes up- and downregulated by exposure of organoids to A. muciniphila-conditioned medium relative to their respective controls (unconditioned microbial culture medium) compared to exposure to the SCFAs butyrate, propionate, and acetate. (D) Venn diagrams representing the number of genes up- and downregulated by exposure to F. prausnitzii-conditioned medium relative to the respective controls compared to exposure to butyrate, propionate, and acetate.
Regarding the effects of exposure to SCFA, our results demonstrate that each SCFA elicits a specific response on gene expression in the intestinal epithelium (Fig. 2C and D; see Fig. S1C and S1D). However, propionate and butyrate share the largest number of host epithelial genes that were affected by exposure. The greatest effect on gene expression was observed after exposure to butyrate (Fig. 2B). Acetate did not induce major gene expression changes relative to the control treatment, and the cluster of genes affected by acetate was relatively close to genes exposed to the control condition (Fig. 2B). The intrinsic biological variation within groups was relatively small, amounting to <5% (calculated as Pearson’s coefficient), and no extreme outliers were detected.
A. muciniphila and its metabolite propionate regulate expression of transcription factors and genes involved in host metabolic pathways.
A. muciniphila-conditioned medium affected the expression of 1,005 genes in intestinal organoids by fold changes of 1.5 and higher (P = 0.01), of which 503 were upregulated, and 502 were downregulated (Fig. 2C). The number of genes affected by exposure to F. prausnitzii-conditioned medium was only 190, of which 86 genes were upregulated and 104 genes were downregulated (Fig. 2D). The differences and similarity in genes affected by the SCFA were also compared (see Fig. S1C and S1D in the supplemental material); however, the main focus was the analysis of genes affected by the two intestinal bacteria, and the comparison of the effects of individual SCFAs produced (butyrate, propionate, and acetate). Genes affected by A. muciniphila-conditioned medium showed the greatest overlap with organoid genes affected by exposure to propionate or butyrate (204 genes upregulated and 202 genes downregulated by all three conditions) (Fig. 2C). Genes with a specific response to individual SCFAs were analyzed and compared to genes affected by A. muciniphila. The A. muciniphila-conditioned medium induced the expression of genes with the greatest overlap with those induced upon exposure to butyrate (67 genes upregulated and 68 genes downregulated) (Fig. 2C). Compared to propionate, the A. muciniphila-conditioned medium showed overlap of 12 upregulated genes and 23 downregulated genes specifically affected by propionate (Fig. 2C). Very little overlap was observed between genes affected in organoids exposed to A. muciniphila-conditioned medium and acetate (with 1 gene upregulated and 0 downregulated under both conditions) (Fig. 2C). Overall, 33 genes were upregulated in organoids treated under any of the four conditions of A. muciniphila, butyrate, propionate, and acetate (Fig. 2C), whereas 10 genes were affected in organoids exposed to any of these four conditions (Fig. 2C).
Organoids exposed to F. prausnitzii-conditioned medium showed a very limited number of genes that were similarly affected by exposure to SCFAs (Fig. 2D)—namely less than 10 genes were affected by exposure to any of the four conditions.
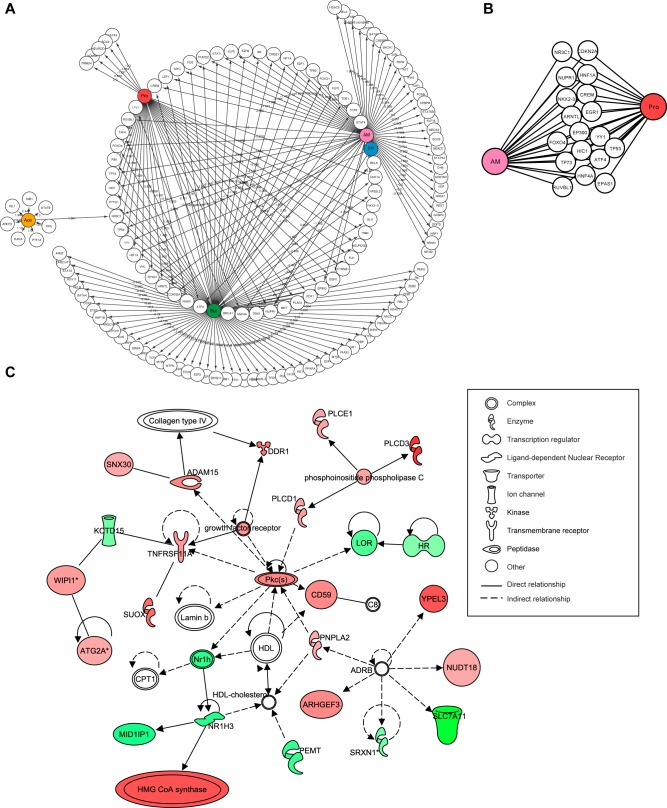
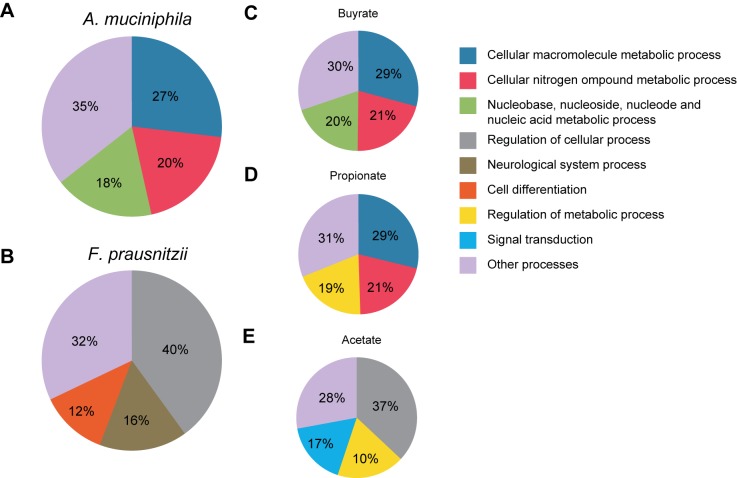
To analyze the effect of microbiota and SCFA on the main regulators of gene expression in the intestinal epithelium, we focused on the effects of these conditions on transcription factor expression (Fig. 3A). We observed that major overlap of effects on transcription factor expression was evident after treatment with A. muciniphila and propionate and butyrate. However, butyrate additionally affected expression of specific transcription factors in the intestinal epithelium of organoids not affected by other conditions (Fig. 3A). The expression of a very small number of transcription factors was affected specifically by exposure of organoids to acetate or propionate (Fig. 3A). Subsequently, the overlapping transcription factors affected by both A. muciniphila and its main metabolite, propionate, were analyzed. Many transcription factors regulating lipid metabolism and proliferation were affected by both conditions, such as Hnf4α and members of p53 family of proteins (Tp53 and Tp73), respectively (Fig. 3B) (36). Supporting these effects of A. muciniphila and propionate on transcription factor expression, Ingenuity pathway analysis (IPA) demonstrated lipid metabolism as one of the top 10 altered associated networks affected by exposure to A. muciniphila-conditioned medium (Fig. 3C). The main regulators of lipid metabolism, such as nuclear receptor Lxr (Nr1h3), Cpt1, and 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) synthase (37), were all altered in their expression in response to A. muciniphila exposure, supporting previous findings in mice where cholesterol homeostasis and lipoprotein metabolic processes were affected after A. muciniphila colonization (25). More detailed information and the effects of the other three conditions on molecular, cellular, and biological pathways are shown in Table S1 and Table S2 in the supplemental material. Database for Annotation, Visualization and Integrated Discovery (DAVID) analyses of gene ontologies in intestinal organoids exposed to A. muciniphila, F. prausnitzii, propionate, butyrate, and acetate confirmed our findings by IPA and demonstrated large overlap between the effects of A. muciniphila with propionate and butyrate on genes specifically involved in metabolic processes (Fig. 4). Acetate and F. prausnitzii affected different biological pathways than the other three conditions, both mainly affecting general cellular processes, including cellular differentiation (F. prausnitzii) and signal transduction (acetate) (Fig. 4B and 3E).
FIG 3 .
A. muciniphila regulates the expression of many host transcription factors and genes involved in cellular metabolic function. (A) Network representation of transcription factors affected by A. muciniphila, F. prausnitzii, and SCFAs. (B) Transcription factors affected in organoids after exposure to both A. muciniphila and its metabolite propionate. (C) IPA network demonstrating the effect of A. muciniphila on expression of genes involved in lipid metabolism. Nodes in green and red correspond to down- and upregulated genes, respectively. Noncolored nodes are proposed by IPA and suggest potential targets functionally coordinated with the differential genes.
FIG 4 .
Many metabolic processes are affected in organoids after exposure to A. muciniphila and propionate. Pie charts show the distribution of affected genes in relation to A. muciniphila (A), F. prausnitzii (B), butyrate (C), propionate (D), and acetate (E) based on their annotations in biological functions (Gene Ontology).
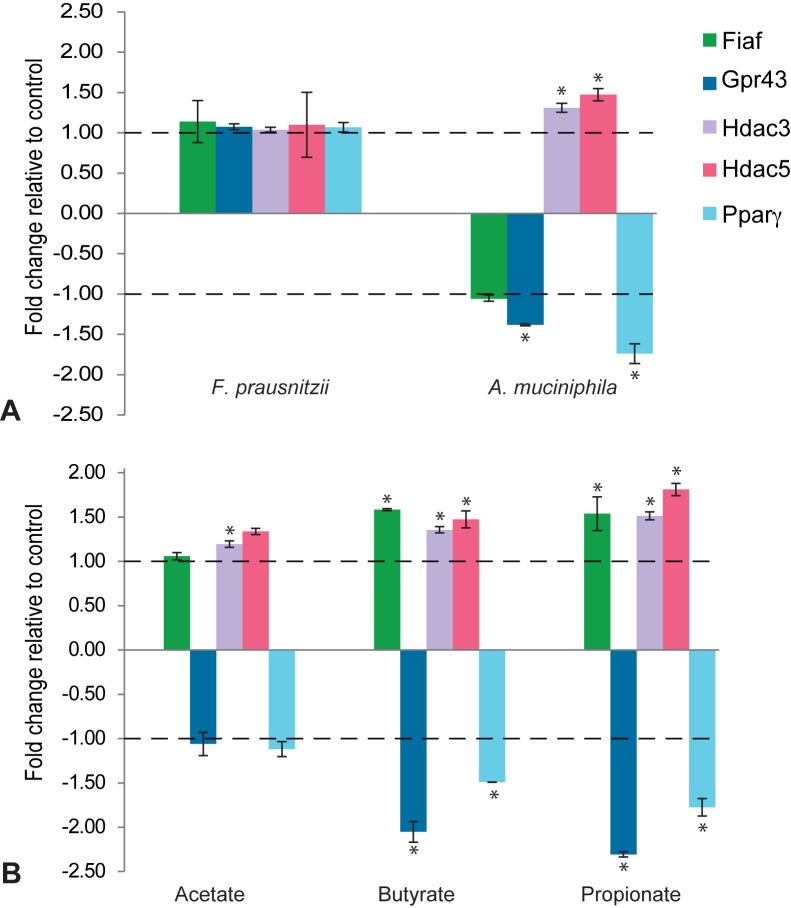
A. muciniphila, propionate, and butyrate, but not F. prausnitzii and acetate, regulate expression of Fiaf, Gpr43, and HDACs in intestinal epithelial organoids.
Previous studies suggested that SCFAs, mainly butyrate and propionate, modulate adiposity regulation via direct effect on Fiaf, Gpr, and Pparγ gene expression (34, 38). Furthermore, transcription of a broad range of genes through HDAC modulation was shown after exposure of intestinal cell lines to butyrate (6). Therefore, we decided to analyze the expression of these specific genes in organoids in response to exposure to SCFAs (Fig. 5). Our data show that, F. prausnitzii has no significant effect on expression of Fiaf, Gpr43, HDACs, or Pparγ (Fig. 5). A. muciniphila leads to significantly decreased expression of Gpr43 and Pparγ and increased expression of Hdac3 and Hdac5 (Fig. 5). It should be noted that the observed A. muciniphila-induced decrease of Fiaf is not significant. These results are partially supported by the effects of both butyrate and propionate stimulation, leading to significant increase in Fiaf expression, while the expression of Gpr43 and Pparγ is significantly decreased (Fig. 5). In addition, the expression of Hdac3 is significantly increased by all three SCFAs (Fig. 5). Acetate had no significant effect on the expression of Fiaf, Gpr43, Hdac5, or Pparγ, while the expression of Hdac3 was significantly increased by acetate (Fig. 5). Expression profiles of organoids exposed to F. prausnitzii did not show any overlap with the changes observed after exposure to individual SCFAs (Fig. 5).
FIG 5 .
Effects of A. muciniphila, propionate, and butyrate on expression of metabolic genes coding for Fiaf, Gpr43, Hdac3, and Hdac4 in intestinal organoids. (A) Log fold change of gene expression after exposure to F. prausnitzii and A. muciniphila relative to control conditions (A) and after exposure to butyrate, propionate, and acetate relative to control conditions (B) (mean fold change ± standard deviation [SD]; P < 0.01; n = 4).
DISCUSSION
The present study demonstrates the applicability of small intestinal epithelial organoids as a novel model system to study host-microbiota interactions, as well as high similarity to in vivo host epithelial response to microbiota. Interestingly, we observed that different members of the human gut microbiota elicit different gene responses in the organoids. We observed a strong regulation of host epithelial genes involved in lipid metabolism and lipolysis by A. muciniphila and the microbial metabolites propionate and butyrate. These results are well in agreement with previous in vitro and in vivo studies describing the effects of microbial monocultures and SCFAs on mouse host intestinal epithelium (9, 11, 25, 19).
Notably, we observed very little overlap between the genes up- or downregulated upon exposure to individual SCFAs and genes affected by exposure to conditioned media that also contained SCFAs (Fig. 2C and D). This may be caused by the fact that the conditioned media contained a complex mixture of compounds that might induce a different transcriptional response. Also, those genes similarly affected by culture media alone were subtracted and are therefore excluded from this comparison.
Although the functional relevance of some of the observed up- and downregulation of genes is difficult to reconcile with known cellular pathways, altogether, the observations with the mouse organoids align with those from other mouse studies, illustrating the usefulness of the organoid model.
Overall, modifications in gene expression in organoids upon exposure to A. muciniphila, F. prausnitzii, or SCFAs involved mostly changes in expression of genes that contribute to regulation of metabolic pathways and genes involved in cellular growth and survival networks. This implies that the microbiota strongly affects the molecular and physiological function of the intestinal epithelium and underscores the importance of studying the interaction of both systems in addressing relevant (patho)physiological research questions, rather than studying isolated host epithelium and intestinal microbial systems. As previously demonstrated for A. muciniphila and Lactobacillus plantarum, different microbial inhabitants of the host may induce expression-specific mouse host genes at different locations (25). We confirmed this finding using the organoid system, in which we demonstrate very strong gene expression modifications upon A. muciniphila product exposure, while F. prausnitzii products only alter the expression of a limited number of genes in organoids after hours of exposure. The specificities of A. muciniphila and F. prausnitzii could be explained by the different physiologies of the two bacterial species, the metabolites they produce, and the different segments of the intestine they colonize.
Genes affected by A. muciniphila were involved in cellular metabolism, cell growth, and survival. The effect of A. muciniphila on cell survival was previously demonstrated in vivo in germfree mice by (25). In agreement with our findings, it was also demonstrated in germfree mice colonized with A. muciniphilla that a large number of genes involved in cell survival, more specifically in cell death receptor signaling, were affected in ileal tissue (25). The same study demonstrated the effect of A. muciniphila on lipid homeostasis and metabolism in cecum samples from germfree mice colonized with A. muciniphila, confirming our results regarding transcriptomic changes in organoids exposed to A. muciniphila-conditioned medium. We observed several key transcription factors and genes involved in fatty acid, cholesterol, and bile acid metabolism, such as those coding for Lxr, Cpt1, and HMG-CoA synthase, which were affected by A. muciniphila. This reveals a possible molecular mechanism underlying the effects of A. muciniphila on lipid metabolism in intestinal epithelium of the host (25). Similar conclusions were evident from in vitro findings in the human enterocyte cell line Caco-2, where cholesterol biosynthesis was downregulated upon SCFA administration (39).
SCFAs, mostly butyrate, have previously been identified as potent regulators of HDACs (6), associated with various biological processes, including transcriptional regulation of interleukin-8 (IL-8) and monocyte chemoattractant protein 1 (MCP-1) expression (40). Alenghat et al. (41) recently showed that Hdac3 expression is reduced in tissues from mice and humans with inflammatory bowel disease and that mice with gut epithelial cell-specific deletion of Hdac3 show loss of Paneth cells and an altered gut microbiota composition. This suggests a critical role for Hdac3 in maintaining the symbiotic balance between host and gut microbiota. Here, both Hdac3 expression and Hdac5 expression were significantly increased after exposure to physiological concentrations of both butyrate and propionate, supporting the hypothesis that these SCFAs regulate transcriptional response of the host. Notably, we show that in addition to butyrate and propionate, A. muciniphila products also affected the expression of HDACs, suggesting regulation of host transcriptional response by A. muciniphila via histone acetylation modifications.
Fiaf is postulated to play a role in the protection from obesity in germfree mice by selectively being suppressed in the intestinal epithelium of normal mice by conventionalization and by being fasting responsive (18, 42). However, a direct role of Fiaf as a mediator in the relationship between gut colonization and adiposity has been disputed as well (12, 13). Hepatic Fiaf has been shown to directly modulate lipid biosynthesis in liver tissue (19). Here, we analyzed the expression of Fiaf in response to both bacteria and SCFAs. We demonstrate that physiologically relevant SCFA concentrations (5 mM) lead to increased intestinal Fiaf expression in organoids. Furthermore, we show that the increase in Fiaf is accompanied by increased expression of genes regulating cholesterol metabolism. This is in agreement with previous rat studies that show that SCFAs regulate cholesterol synthesis in both liver and intestinal tissue (43) and with human intestinal cell line studies, where SCFAs were shown to regulate cholesterol biosynthesis (39) and to induce Fiaf expression (34).
In conclusion, our data illustrate that different symbiotic microbial species trigger specific transcriptome responses in the intestinal epithelium, most likely depending on the metabolic products produced. Further studies are required in order to translate these transcription dynamics into physiologically relevant responses. We propose that organoids provide a novel, physiologically relevant, gut model system to further explore microbiota-epithelial interactions underlying nutrition- and microbiota-induced health effects.
MATERIALS AND METHODS
Mouse ileal crypt isolation.
Ileal tissue from an adult male wild-type (WT) C57BL/6 mouse (Charles River) was excised longitudinally and flushed with cold phosphate-buffered saline (PBS). The upper epithelium (villi) was scraped, and remaining tissue was washed again with cold PBS. Subsequently, the tissue was cut into 5-mm pieces and incubated in 5 mM EDTA for 30 min in order to release the crypts from the underlying muscle layer. After incubation, the released crypts were removed from the muscle layer and used for the organoid culture protocol, as described previously (21).
Organoid culture.
In short, murine (WT C57BL/6) small intestinal crypts were isolated and embedded in 250 µl Matrigel (BD Biosciences) and submerged in 500 µl basal culture medium supplemented with penicillin-streptomycin, HEPES, Glutamax, N2, B27, N-acetylcysteine, and the murine growth factors epidermal growth factor (EGF), noggin, and R-spondin-1 (ENR), as previously described (21). Organoids were passaged every 7 days with a 1:4 splitting ratio (Fig. 1).
Exposure to bacterial cultures and SCFAs.
A. muciniphila (ATTC BAA-835) (24) was grown anaerobically at 37°C in a basal liquid medium that contained the following (per liter of deionized water): 0.4 g KH2PO4, 0.669 g Na2HPO4 plus 2H2O, 0.3 g NH4Cl, 0.3 g NaCl, 0.1 g MgCl2 plus 6H2O, 10 g Casitone, 1 mM l-threonine, 1 ml trace mineral solution, 5 mM l-fucose, and 5 mM d-glucose.
F. prausnitzii strain A2-165 (DSM 17677) was grown anaerobically at 37°C in a liquid medium that contained the following (per liter of deionized water): 10 g Casitone, 2.5 g yeast extract, 4 g NaHCO3, 1 ml resazurin (0.1% [wt/vol]), 0.45 g K2HPO4, 0.45 g KH2PO4, 0.9 g (NH4)2SO4, 0.9 g NaCl, 0.09 g MgSO4, 0.09 g CaCl2, 2.5 mM acetate, 9 mM propionate, 1 mM n-valerate, 1 mM isovalerate, 1 mM isobutyrate, 0.28 g KOH, 2.5 ml ethanol (95%), 10 mg hemin, 10 µg biotin, 10 µg cobalamin, 30 µg para-aminobenzoic acid, 50 µg folic acid, 150 µg pyridoxamine, 1 g l-cysteine, and 25 mM glucose (35). The concentrations of the following organic acids and sugars were determined in the culture medium before and after growth using a high-performance liquid chromatography (HPLC) device equipped with a Shodex Sugar SH-G and Shodex SH1821 column as described previously (44): glucose, mannose, galactose, l-fucose, glucose-N-acetylglucosamine, lactate, formate, acetate, propionate, 1,2-propendiol, and butyrate. Overnight-grown cultures were pelleted by centrifugation at 20 000 × g for 10 min, and supernatant was collected. At 7 days after splitting, organoids were exposed to 250 µl A. muciniphila supernatant, 250 µl unconditioned A. muciniphila culture medium, 85 µl F. prausnitzii supernatant, or 85 µl F. prausnitzii culture medium for 3 h at 37°C, prior to RNA extraction. A 250-µl concentration of A. muciniphila supernatant and culture medium SCFAs sodium butyrate, sodium propionate, and sodium acetate (Sigma-Aldrich, Zwijndrecht, The Netherlands) was administered at a final concentration of 5 mM. At 7 days after splitting, mature organoids were exposed to individual SCFAs for 3 h at 37°C prior to RNA extraction.
RNA extraction.
Cultured organoids were pooled (approximately 300 organoids per sample), and RNA was isolated using Trizol (Invitrogen, Breda, The Netherlands), followed by purification using the RNeasy kit (Qiagen) and DNase treatment. Samples were assayed for quantity and quality with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and NanoDrop spectrophotometer (NanoDrop, Wilmington, DE).
Data are presented from n = 4 independent biological replicates from one line of organoids.
Microarray and statistical analysis.
Total mRNA from the organoids was amplified, biotinylated, and randomly hybridized to MouseRef-8 v2 Expression bead arrays (Illumina, Inc., San Diego, CA), followed by scanning and feature extraction. Gene expression data from the Illumina beadchip microarrays proved to be highly reproducible, as indicated by low intersample variation among technical replicates.
Quality control, RNA labeling, hybridization, and data extraction were performed at ServiceXS (Leiden, The Netherlands). RNA concentrations were measured using the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). RNA quality and integrity were determined using Lab-on-Chip analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) and on Shimadzu MultiNA RNA analysis chips (Shimadzu Corporation, Kyoto, Japan). Biotinylated cRNA was prepared using the Illumina TotalPrep RNA amplification kit (Ambion, Inc., Austin, TX) according to the manufacturer’s specifications with an input of 200 ng total RNA. Per sample, 750 ng of the obtained biotinylated cRNA samples was hybridized onto the Illumina MouseRef-8 v2 (Illumina, Inc., San Diego, CA). Each BeadChip contains eight arrays. Hybridization and washing were performed according to the instructions in the Illumina Direct Hybridization Assay Guide (Illumina, Inc. San Diego, CA). Scanning was performed on the Illumina iScan (Illumina, Inc., San Diego, CA). Image analysis and extraction of raw expression data were performed with the Illumina GenomeStudio v 2011.1 Gene Expression software with default settings (no background subtraction and no normalization).
Data were extracted using GenomeStudio. Quality control and normalization (quantile) of microarray data were performed using the lumi package of Bioconductor, available at www.bioconductor.org. Nonexpressed genes were removed by filtering on the detection value (P ≥ 0.99 in at least one sample). This resulted in gene expression values for 25,697 probes. For differential expression analysis, Limma software was used. Subsequently, expression data of the transcriptomics were log transformed (base 2). Probes were used for biological interpretation if the P value threshold of <0.01 was passed and if the fold change between two conditions was |1.5|. In order to characterize those genes specifically affected by microbial compounds we normalized transcription profiles by subtracting the genes similarly affected by the unconditioned culture medium from genes affected by microbially conditioned medium. Further information on the function and biological role of the probes was derived from Ingenuity pathway analysis (IPA) (Ingenuity Systems, Redwood City, CA), the Database for Annotation, Visualization and Integrated Discovery (DAVID), and Cytoscape.
Microarray data accession number.
Raw microarray data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE59644.
SUPPLEMENTAL MATERIAL
(A and B) Venn diagrams representing the number of genes up- and downregulated by exposure of organoids to F. prausnitzii- or A. muciniphila-conditioned medium relative to their respective controls (differentially regulated and overlapping between different exposure methods). (C and D) Venn diagrams representing the number of genes up- and downregulated by exposure of organoids to butyrate, propionate, and acetate relative to control medium without SCFAs. Download
Top five molecular and cellular pathways affected in organoids as analyzed by IPA software. The numbers of molecules and P values are extracted from IPA pathway analysis. Only significantly affected networks are indicated. In boldface, pathways involved in gene expression regulation and cell growth and survival are indicated.
Top three associated networks affected by different exposures to microbial species or SCFAs as analyzed by IPA software.
Top five fold changes of genes up- or downregulated in organoids in response to butyrate, along with their function as analyzed by IPA. Only significantly affected genes are indicated (P = 0.01).
ACKNOWLEDGMENTS
This work was supported through TNO’s Research Program on Healthy Nutrition and by grant 25017 (Microbes Inside) from the European Research Council (ERC) to W.M.d.V.
We thank Mehdi Hassan Zahde Nadjari and Steven Aalvink for technical assistance and our colleagues at TNO as well as the members of the Clevers lab (Hubrecht Laboratory) for helpful discussions.
Footnotes
Citation Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G. 2014. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio 5(4):e01438-14. doi:10.1128/mBio.01438-14.
REFERENCES
- 1. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- 5. Matsuki T, Pédron T, Regnault B, Mulet C, Hara T, Sansonetti PJ. 2013. Epithelial cell proliferation arrest induced by lactate and acetate from Lactobacillus casei and Bifidobacterium breve. PLoS One 8:e63053. 10.1371/journal.pone.0063053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. 2008. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 19:587–593. 10.1016/j.jnutbio.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 7. Vanhoutvin SA, Troost FJ, Hamer HM, Lindsey PJ, Koek GH, Jonkers DM, Kodde A, Venema K, Brummer RJ. 2009. Butyrate-induced transcriptional changes in human colonic mucosa. PLoS One 4:e6759. 10.1371/journal.pone.0006759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roeselers G, Bouwman J, Venema K, Montijn R. 2012. The human gastrointestinal microbiota—an unexplored frontier for pharmaceutical discovery. Pharmacol. Res. 66:443–447. 10.1016/j.phrs.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 9. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101:15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cani PD, Bibiloni R, Knauf C, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic diet—induced obesity and diabetes in mice. Diabetes 57:1470–1481. 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- 11. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 110:9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. 2010. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 104:919–929. 10.1017/S0007114510001303 [DOI] [PubMed] [Google Scholar]
- 13. Swartz TD, Sakar Y, Duca FA, Covasa M. 2013. Preserved adiposity in the Fischer 344 rat devoid of gut microbiota. FASEB J. 27:1701–1710. 10.1096/fj.12-221689 [DOI] [PubMed] [Google Scholar]
- 14. Grootaert C, Van de Wiele T, Van Roosbroeck I, Possemiers S, Vercoutter-Edouart AS, Verstraete W, Bracke M, Vanhoecke B. 2011. Bacterial monocultures, propionate, butyrate and H2O2 modulate the expression, secretion and structure of the fasting-induced adipose factor in gut epithelial cell lines. Environ. Microbiol. 13:1778–1789 [DOI] [PubMed] [Google Scholar]
- 15. Kersten S, Lichtenstein L, Steenbergen E, Mudde K, Hendriks HF, Hesselink MK, Schrauwen P, Müller M. 2009. Caloric restriction and exercise increase plasma ANGPTL4 levels in humans via elevated free fatty acids. Arterioscler. Thromb. Vasc. Biol. 29:969–974. 10.1161/ATVBAHA.108.182147 [DOI] [PubMed] [Google Scholar]
- 16. Dutton S, Trayhurn P. 2008. Regulation of angiopoietin-like protein 4/fasting-induced adipose factor (Angptl4/FIAF) expression in mouse white adipose tissue and 3T3-L1 adipocytes. Br. J. Nutr. 100:18–26. 10.1017/S0007114507882961 [DOI] [PubMed] [Google Scholar]
- 17. Murata M, Yudo K, Nakamura H, Chiba J, Okamoto K, Suematsu N, Nishioka K, Beppu M, Inoue K, Kato T, Masuko K. 2009. Hypoxia upregulates the expression of angiopoietin-like-4 in human articular chondrocytes: role of angiopoietin-like-4 in the expression of matrix metalloproteinases and cartilage degradation. J. Orthop. Res. 27:50–57. 10.1002/jor.20703 [DOI] [PubMed] [Google Scholar]
- 18. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101:15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lichtenstein L, Berbée JF, van Dijk SJ, van Dijk KW, Bensadoun A, Kema IP, Voshol PJ, Müller M, Rensen PC, Kersten S. 2007. Angptl4 upregulates cholesterol synthesis in liver via inhibition of LPL− and HL-dependent hepatic cholesterol uptake. Arterioscler. Thromb. Vasc. Biol. 27:2420–2427. 10.1161/ATVBAHA.107.151894 [DOI] [PubMed] [Google Scholar]
- 20. Roeselers G, Ponomarenko M, Lukovac S, Wortelboer HM. 2013. Ex vivo systems to study host–microbiota interactions in the gastrointestinal tract. Best Pract. Res. Clin. Gastroenterol. 27:101–113. 10.1016/j.bpg.2013.03.018 [DOI] [PubMed] [Google Scholar]
- 21. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 22. Yui S, Nakamura T, Toshiro S, Yasuhiro N, Tomohiro M, Zheng X, Ichinose S, Takashi N, Ryuichi O, Tsuchiya K, Clevers H, Watanabe M. 2012. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 18:618–623. 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
- 23. Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. 2008. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74:1646–1648. 10.1128/AEM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54:1469–1476. 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- 25. Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. 2011. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2:166. 10.3389/fmicb.2011.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neef A, Sanz Y. 2013. Future for probiotic science in functional food and dietary supplement development. Curr. Opin. Clin. Nutr. Metab. Care 16:679–687. 10.1097/MCO.0b013e328365c258 [DOI] [PubMed] [Google Scholar]
- 29. Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki SI, Kuwahara A. 2008. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J. Physiol. Pharmacol. 59(Suppl 2):251–262 [PubMed] [Google Scholar]
- 30. Peng L, He Z, Chen W, Holzman IR, Lin J. 2007. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr. Res. 61:37–41. 10.1203/01.pdr.0000250014.92242.f3 [DOI] [PubMed] [Google Scholar]
- 31. Pinto M, Robine-Leon S, Appay M-D, Kedinger M, Triadou N, Dussaulx E, Lacroix B, Simon-Assmann P, Haffen K, Fogh J, Zweibaum A. 1983. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 47:323–330 [Google Scholar]
- 32. Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. 2012. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6:1415–1426. 10.1038/ismej.2011.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, Koppen A, Mudde K, Haenen D, Al-Lahham S, Roelofsen H, Houtman R, van der Burg B, Mandrup S, Bonvin AM, Kalkhoven E, Müller M, Hooiveld GJ, Kersten S. 2013. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol. Cell. Biol. 33:1303–1316. 10.1128/MCB.00858-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141–2146. 10.1099/ijs.0.02241-0 [DOI] [PubMed] [Google Scholar]
- 36. Stroup D, Chiang JY. 2000. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7alpha-hydroxylase gene (CYP7A1). J. Lipid Res. 41:1–11 [PubMed] [Google Scholar]
- 37. Colin S, Bourguignon E, Boullay AB, Tousaint JJ, Huet S, Caira F, Staels B, Lestavel S, Lobaccaro JM, Delerive P. 2008. Intestine-specific regulation of PPARalpha gene transcription by liver x receptors. Endocrinology 149:5128–5135. 10.1210/en.2008-0637 [DOI] [PubMed] [Google Scholar]
- 38. Grootaert C, Van de Wiele T, Van Roosbroeck I, Possemiers S, Vercoutter-Edouart AS, Verstraete W, Bracke M, Vanhoecke B. 2011. Bacterial monocultures, propionate, butyrate and H2O2 modulate the expression, secretion and structure of the fasting-induced adipose factor in gut epithelial cell lines. Environ. Microbiol. 13:1778–1789. 10.1111/j.1462-2920.2011.02482.x [DOI] [PubMed] [Google Scholar]
- 39. Alvaro A, Solà R, Rosales R, Ribalta J, Anguera A, Masana L, Vallvé JC. 2008. Gene expression analysis of a human enterocyte cell line reveals downregulation of cholesterol biosynthesis in response to short-chain fatty acids. IUBMB Life 60:757–764. 10.1002/iub.110 [DOI] [PubMed] [Google Scholar]
- 40. Fusunyan RD, Quinn JJ, Fujimoto M, MacDermott RP, Sanderson IR. 1999. Butyrate switches the pattern of chemokine secretion by intestinal epithelial cells through histone acetylation. Mol. Med. 5:631–640 [PMC free article] [PubMed] [Google Scholar]
- 41. Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CG, Mullican SE, Choi I, Grunberg S, Sinha R, Wynosky-Dolfi M, Snyder A, Giacomin PR, Joyce KL, Hoang TB, Bewtra M, Brodsky IE, Sonnenberg GF, Bushman FD, Won KJ, Lazar MA, Artis D. 2013. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 504:153–157. 10.1038/nature12687 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bäckhed F, Crawford PA, O’Donnell D, Gordon JI. 2007. Postnatal lymphatic partitioning from the blood vasculature in the small intestine requires fasting-induced adipose factor. Proc. Natl. Acad. Sci. U. S. A. 104:606–611. 10.1073/pnas.0605957104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hara H, Haga S, Aoyama Y, Kiriyama S. 1999. Short-chain fatty acids suppress cholesterol synthesis in rat liver and intestine. J. Nutr. 129:942–948 [DOI] [PubMed] [Google Scholar]
- 44. Stams AJ, Van Dijk JB, Dijkema C, Plugge CM. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl. Environ. Microbiol. 59:1114–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A and B) Venn diagrams representing the number of genes up- and downregulated by exposure of organoids to F. prausnitzii- or A. muciniphila-conditioned medium relative to their respective controls (differentially regulated and overlapping between different exposure methods). (C and D) Venn diagrams representing the number of genes up- and downregulated by exposure of organoids to butyrate, propionate, and acetate relative to control medium without SCFAs. Download
Top five molecular and cellular pathways affected in organoids as analyzed by IPA software. The numbers of molecules and P values are extracted from IPA pathway analysis. Only significantly affected networks are indicated. In boldface, pathways involved in gene expression regulation and cell growth and survival are indicated.
Top three associated networks affected by different exposures to microbial species or SCFAs as analyzed by IPA software.
Top five fold changes of genes up- or downregulated in organoids in response to butyrate, along with their function as analyzed by IPA. Only significantly affected genes are indicated (P = 0.01).