ABSTRACT
Glycans form the primary nutritional source for microbes in the human gut, and understanding their metabolism is a critical yet understudied aspect of microbiome research. Here, we present a novel computational pipeline for modeling glycan degradation (GlyDeR) which predicts the glycan degradation potency of 10,000 reference glycans based on either genomic or metagenomic data. We first validated GlyDeR by comparing degradation profiles for genomes in the Human Microbiome Project against KEGG reaction annotations. Next, we applied GlyDeR to the analysis of human and mammalian gut microbial communities, which revealed that the glycan degradation potential of a community is strongly linked to host diet and can be used to predict diet with higher accuracy than sequence data alone. Finally, we show that a microbe’s glycan degradation potential is significantly correlated (R = 0.46) with its abundance, with even higher correlations for potential pathogens such as the class Clostridia (R = 0.76). GlyDeR therefore represents an important tool for advancing our understanding of bacterial metabolism in the gut and for the future development of more effective prebiotics for microbial community manipulation.
IMPORTANCE
The increased availability of high-throughput sequencing data has positioned the gut microbiota as a major new focal point for biomedical research. However, despite the expenditure of huge efforts and resources, sequencing-based analysis of the microbiome has uncovered mostly associative relationships between human health and diet, rather than a causal, mechanistic one. In order to utilize the full potential of systems biology approaches, one must first characterize the metabolic requirements of gut bacteria, specifically, the degradation of glycans, which are their primary nutritional source. We developed a computational framework called GlyDeR for integrating expert knowledge along with high-throughput data to uncover important new relationships within glycan metabolism. GlyDeR analyzes particular bacterial (meta)genomes and predicts the potency by which they degrade a variety of different glycans. Based on GlyDeR, we found a clear connection between microbial glycan degradation and human diet, and we suggest a method for the rational design of novel prebiotics.
INTRODUCTION
The human gastrointestinal tract harbors an extensive array of commensal microorganisms. Species composition is highly diverse both within and between individuals (1), and the activities of these organisms affect the host through many pathways, including the production of short-chain fatty acids (SCFA) that regulate epithelial cell growth and immune system development, displacement of potential pathogens, detoxification of protein fermentation products, and gas production (2–6). The beneficial or detrimental outcomes of these effects depend largely on the community structure, environmental factors, diet, and the genetic background of the host (7, 8). In order to gain a deeper understanding of the gut ecosystem as a whole, a systems biology approach integrating genomic, metabolic, and enzymatic information is an invaluable tool (9, 10).
Large-scale sequencing analysis of the human gut microbiome has led to many tantalizing and highly important gut microbiome-human health associations, but typically little or no mechanistic insight is provided (11). Simplified in vitro models aim to bridge this gap, but the reliance of these models on a few strains and on defined culture media makes them difficult to relate to the complexities of the human gut (12, 13). A primary issue is the fact that very few simple metabolites escape digestion in the small intestine (14), which makes complex carbohydrates and their derivatives, collectively termed glycans, the predominant nutrients for microbes in the colon (15, 16). These glycans are poorly accounted for in any current systems biology or pathway-related frameworks or databases, and thus they represent a major hurdle in leveraging the full weight of systems biology methods in the gut microbiome field. Incorporating mechanisms into microbiome research will therefore first require a large-scale accounting and analysis of glycan degradation by the gut microbiota.
While some human colonic bacteria simply require acetate or branched-chain fatty acids (17), the detailed growth requirements for the majority of gut bacteria remain unknown (18). Characterizing these requirements will shed light on the different metabolic niches organisms fill and may enable the design of dietary interventions that promote growth of particular beneficial microbes, an approach collectively termed “prebiotics” (19). While several glycans are currently marketed around the world as prebiotics, few have been validated through high-quality human trials (19, 20). Furthermore, dietary enrichment of a specific prebiotic compound may permit preferential expansion of a microbial group that is well adapted to its use, but the outcomes for the gut community as a whole can be unpredictable (21).
In this study, we investigated the connections between diet and glycan metabolism of the human gut microbiota. Whereas the study of the metabolic activity conducted by gut microbiota has been the focal point of a wide range of computational studies (9, 11), current approaches have been highly limited in their ability to analyze glycan degradation. We present a novel algorithm (termed GlyDeR) for predicting the glycan degradation patterns of any bacterium with a sequenced genome. The algorithm is based on manual curation of nearly 150 carbohydrate-active enzymes (CAZymes) and is applied to a set of 10,000 glycan structures and 203 microbial genomes. Given a particular bacterial (meta)genome, GlyDeR can be used to reverse engineer the predicted potency by which a bacterium degrades a variety of different glycans. These predictions correlate with known KEGG reactions and expand upon the limited, previously available glycan degradation data 100-fold. We determined that the microbiota of herbivores and carnivores have stronger degradation affinities for plant-derived and animal-derived glycans, respectively, and that a Western diet in humans correlates more strongly with meat-derived glycans than a non-Western diet. Finally, we show that species-specific glycan degradation profiles are associated with and can be used to predict bacterial species abundance, making GlyDeR a valuable tool for the future rational design of novel prebiotics, by deliberately manipulating the microbiome based on nutrient availability.
RESULTS
Construction of the glycan degradation (GlyDeR) pipeline.
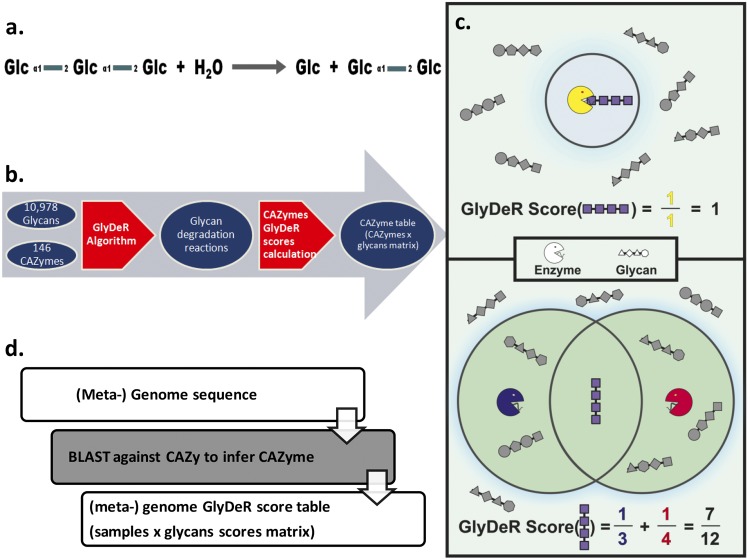
Although the exact biochemistry of glycan degradation is missing from all currently available databases, considerable knowledge is embedded in the descriptions of the CAZymes that catalyze these degradation reactions and is typically represented by enzymatic commission (EC) numbers. We leveraged this knowledge to develop a new computational pipeline that uses enzymatic and structural data sources to predict the degradation of every glycan in the KEGG database (22) that has a sequenced (meta)genome. That is, given (meta)genomic data as input, GlyDeR yields phenotypic (glycan degradation) data as output. The construction of the pipeline comprises two steps. (i) The first step relies on a novel algorithm that we developed, which we term glycan degradation (GlyDeR). The algorithm takes as input a manually curated annotation of all the reactions that each known CAZyme is capable of performing (see Table S1 in the supplemental material) and a network representation of all the glycans in KEGG, in which the nodes are the monosaccharides and the edges are the glycosidic linkages (Fig. 1a; see also Table S6 in the supplemental material). We converted the CAZyme annotations to the computer-based rules that dictate their mechanism for breaking a given glycan into two subcomponents. The manual curation of this critical step was done using the help of experts with knowledge of the biochemistry of glycan metabolism. GlyDeR then executes these rules recursively on all the glycans, to generate 141,561 GlyDeR reactions, each linking a specific enzyme to a glycan substrate and its products (an example reaction is given in Fig. 1a, and a more detailed explanation is provided in Materials and Methods). (ii) In the second step, GlyDeR reactions are mapped back to CAZymes in order to produce a table where the rows are CAZymes, the columns are glycans, and each entry contains a CAZyme score for CAZyme i and glycan j. If CAZyme i is unable to break glycan j, then the score is 0, otherwise the score is calculated as follows:
where gi is the number of glycans that are broken by CAZyme i. The entire construction process is summarized in Fig. 1b. A CAZymes table that contains all of the CAZyme scores can be found in Table S5 in the supplemental material.
FIG 1 .
The GlyDeR platform. (a) A visual representation of the glycan degradation reaction performed for EC 3.2.1.115, breaking down Kojitriose into Kojibiose and glucose. (b) A schematic representation of the construction of the computational pipeline. Information is taken from multiple databases and analyzed as follows. Step 1 (lred arrow on left): by using CAZyme information and the GlyDeR algorithm, glycan degradation reactions are reconstructed. Step 2 (red arrow on right): a CAZyme table is constructed that represents the potency with which different CAZymes break different glycans. (c) GlyDeR score calculation. (Top) The organism has one enzyme (yellow PacMan) dedicated to the degradation of one glycan (purple); therefore, the GlyDeR score for the purple glycan equals 1. (Bottom) The organism has two enzymes capable of degrading 3 and 4 glycans, respectively, and therefore the GlyDeR score for the purple glycan equals 7/12. (d) GlyDeR utilization. (Meta)genomes are annotated for CAZymes by using CAZy, SEED, and KEGG databases, and with the CAZyme table a GlyDeR score can be calculated, reflecting the capacity of a (meta)genome to degrade a specific glycan.
Use of the GlyDeR pipeline.
Microbial (meta)genomes were annotated for CAZymes by using BLAST analysis (23) against three reference databases: the Carbohydrate-Active Enzymes (CAZy) database (24), the Seed-RAST annotation (25), and KEGG (26) (see Materials and Methods). Then, CAZyme scores were assigned to genes, and a GlyDeR score was calculated for each glycan i and (meta-) genome j as follows:
where nji is the number of genes in (meta)genome j which translate to a CAZyme that can break glycan i.
The GlyDeR score represents the predicted potency with which the glycan can be degraded by that (meta)genome, taking into account how many CAZymes can degrade the glycan and with decrements for the score of a promiscuous enzymes with low specificities (see Materials and Methods). For example, an organism that contains three enzymes that degrade maltotetraose, each of which also degrades four other glycans, would have a GlyDeR score of 3/5 (two examples are provided in Fig. 1c). The use of GlyDeR is captured in Fig. 1d.
Validating the GlyDeR pipeline.
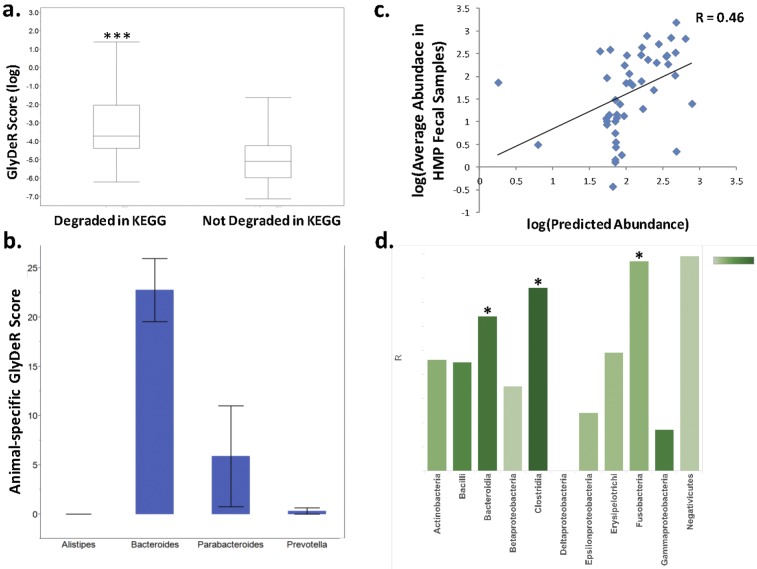
To assess the biological relevance of GlyDeR, we performed a cross-validation procedure that examined its consistency in capturing known degradation reactions in KEGG (see Materials and Methods). We found that the products of GlyDeR reactions were highly enriched with known rather than hypothetical glycans (P = 10−19, hypergeometric test; see also Fig. S2a in the supplemental material). As further validation, we compared the predicted genome-specific GlyDeR scores of each bacterial strain with the glycans that, according to KEGG, the strain is able to break (KEGG glycans). Since the above information from KEGG was not used to construct the set of GlyDeR reactions, a circular argument was avoided. We found a significantly higher mean GlyDeR score for KEGG glycans across all strains compared to non-KEGG glycans (see Fig. S2a). Notably, our analysis produced GlyDeR scores for over 100 times the number of unique glycan degradation reactions that are currently reported in KEGG (116,388 versus 1,374), highlighting the limited scope of glycan metabolism information captured in the KEGG database.
Characterization of glycan degradation patterns across the major gut bacterial phyla.
We first applied GlyDeR to a cohort of 203 reference gut microbial genomes retrieved from the Human Microbiome Project (HMP) (27). All of the available information on these strains is listed in Table S2 of the supplemental material. We used GlyDeR to study the extent to which different microbial phyla metabolize different glycans. Initially, we examined whether phylogenetic clusters are reflected in glycan degradation patterns. We therefore computed for each of the HMP genomes a GlyDeR profile, i.e., a vector of its GlyDeR scores for all glycans. The species-specific GlyDeR “signature” describes the potency by which a given species can catabolize each of the ~10,000 reference glycans in our database and hence provides an overall view of its glycan utilization capabilities. We then mapped each species to its respective phylum and performed principal coordinates analysis (PCoA) on the Bray-Curtis dissimilarities between the species GlyDeR profiles. This yielded clusters of phyla that were statistically distinct (multivariate analysis of variance [ANOVA] test, Wilke’s lambda < 0.001) (see Fig. S2b in the supplemental material). Still, there were apparent significant differences in the average glycan degradation capacities of genera belonging to a given phylum (see Fig. S2c).
Bacteroides species are highly effective degraders of animal-derived glycoproteins.
Recently, it has been shown that human diets high in animal protein are associated with high levels of Bacteroides, whereas diets rich in plant-derived carbohydrates and very low in animal protein display enrichment for Prevotella (28–30). Among the phyla in the HMP data set, we found Bacteroidetes to be the most efficient degraders of animal-derived glycans (see Fig. S2e in the supplemental material). Notably, this trend was apparent for the Bacteroides genus but absent for Prevotella, which also belongs to that phylum (Fig. 2b). Furthermore, all 19 of the highest-scoring species with GlyDeR belonged to the Bacteroides genus (see Table S3 in the supplemental material), consistent with their known roles as primary glycan degraders in the gut (31–33).
FIG 2 .
Glycan degradation of gut microbiota reference genomes. (a) Distribution of species-specific GlyDeR scores (y axis) for all the glycans in KEGG. GlyDeR scores with corresponding reactions in KEGG appear on the left, while those with no KEGG reaction appear on the right (Student’s t = 6.14, P < 0.0001). (b) Bar plot comparing the animal-specific glycan degradation potential of different bacterial genera within the Bacteroidetes phylum. Each bar depicts the sum of GlyDeR scores of organisms belonging to their respected phylum. The height of the bar represents the mean, while the error bars reflect standard errors. (c) The log-log scatterplot shows the average abundance of 48 HMP strains within 325 human fecal samples (y axis) and the linear regression-predicted abundance of each individual strain (x axis) (linear regression correlation coefficient = 0.46, P = 0.0016). (d) Bar chart denoting the correlation value (height of the bar) between actual and predicted abundance from linear regression models built for each class of bacteria (x axis) based on the taxon’s GlyDeR features. The color of the bar reflects the number of species in the class. The feature extraction is explained in Materials and Methods.
Recent papers have also shown that glycans found in human milk, such as human milk oligosaccharides (HMOs), are utilized mainly by several Bifidobacterium and Bacteroides species (31, 34). According to GlyDeR, 21 out of the 23 HMP genomes that are capable of degrading HMOs belong to Bacteroides species (the other degraders were Parabacteroides sp. D13 and Bifidobacterium bifidum) (see Table S4 in the supplemental material). To further investigate this point, we examined two of the most abundant animal-derived glycoproteins in the human diet: ovalbumin (35) and casein (36). Indeed, we confirmed that members of Bacteroides degrade these glycoproteins more efficiently than any other genus (see Fig. S2f in the supplemental material). Given that many dietary animal glycans are derived from proteins (e.g., glycoproteins and proteoglycans), we propose that the high capability of Bacteroides to degrade animal glycans might explain why their abundance is increased in Westerners (30, 37).
Glycan degradation patterns can be used to predict bacterial abundance.
We studied the relationship between the glycan degradation scores of a given bacterial taxon and its abundance in the gut. We matched the abundance of 16S rRNA marker gene sequences from 325 human individual gut samples found in the HMP database with the aforementioned 203 microbial reference genomes (see Materials and Methods). For each taxon, we extracted 6 features that characterized its glycan degradation capacity, including plant-specific glycans, animal-specific glycans, disaccharides, oligosaccharides, short polysaccharides, and long polysaccharides (see Table S3 in the supplemental material). Each feature represents the sum of GlyDeR scores for the glycans that belong in the class. Based on these features, we built a linear regression model for the abundance of these taxa in the samples. In order to apply the linear regression model, we filtered out taxa that were not detected in any sample and taxa that were highly varied (see Materials and Methods for criteria), resulting in 48 predictable taxa for the analysis. This regression yielded a correlation coefficient of 0.46 (Fig. 2c), a score markedly higher than the correlation achieved using a model based on CAZyme abundances in a genome (R = 0.11). We next built similar regression models independently for each class of bacteria. Remarkably, the Clostridia class had the highest combination of R (0.76) and P (0.0001) values; other classes with significantly predictive models were Bacteroidia and Fusobacteria (Fig. 2d; see also Table S8 in the supplemental material). These results suggest that glycan supplements can be tailored to control certain species abundances, especially those of potentially pathogenic Clostridia.
In an effort to include taxa that were initially omitted in the analysis above due to their high levels of variation, we first clustered the HMP samples according to their 16S rRNA data into 2 main groups by using KMeans (see Materials and Methods), and we recalculated the average taxon abundance separately for each cluster. The same procedure for building predictors of bacterial taxon abundance based on their genome-specific GlyDeR features was then used, and this yielded 53 predictable taxa in the first cluster and 71 predictable taxa in the second cluster, with concomitant increases in the correlation coefficients (0.51 and 0.57, respectively). Based on the two clusters, we assembled a list of 25 strains with highly predictable abundances (see Table S9 in the supplemental material), and with only one exception, all the strains belonged either to the Bacteroidia or Clostridia classes (see Discussion). Notably, based on the regression formulas of all three models, the degradation capacity of long polysaccharides had the highest effect on bacterial abundance. It is therefore likely that because most long polysaccharides are not digested by the human host prior to reaching the colon, an ability to degrade them provides a significant selective advantage for gut microbes.
Glycan degradation profiles of mammalian species are associated with their diet.
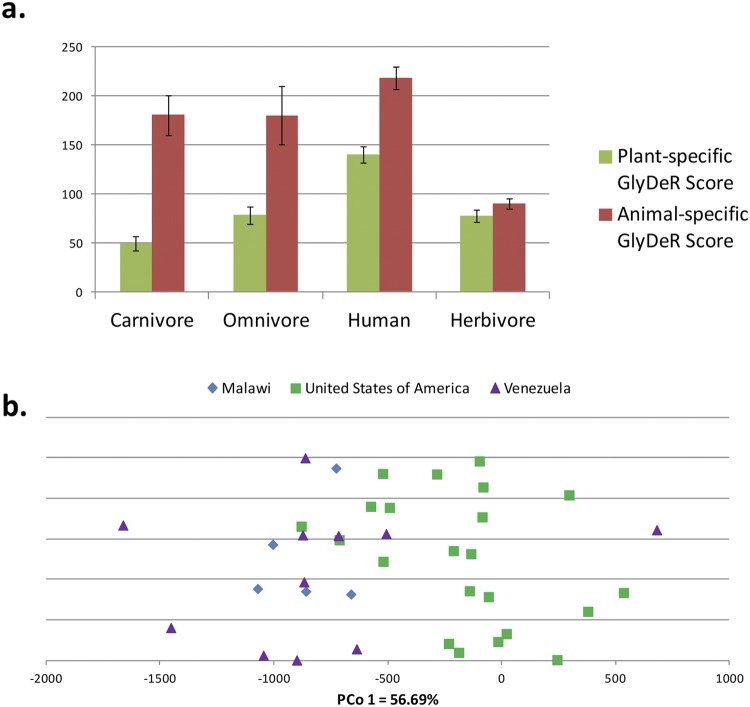
Because diet is the prime determinant of colonic glycan composition and gut microbiota vary according to general dietary patterns (38), we expected that glycan degradation would systematically vary between the microbiota of different mammalian hosts based on their diet. To test this, we analyzed variation in diet and glycan degradation profiles across different mammalian species, using metagenomic sequencing data from 57 fecal samples across 34 different species, including 18 human samples (38). According to the host’s diet, each sample was characterized as being either herbivorous, carnivorous, or omnivorous. To correct for research biases arising from uneven annotations of CAZymes between species, we normalized each sample by the total number of CAZymes in it before running PCoA on the GlyDeR profiles. PCoA revealed a clear spectrum of samples over the first principal coordinate, from herbivores, through omnivores, to carnivores (see Fig. S3a in the supplemental material). With a subset of glycans that were categorized into either plant-derived or animal-derived glycans, we discovered a striking relationship between the diet of a host organism and the glycans predicted to be degraded by its gut microbiota: microbiota from herbivores tend to degrade plant-derived glycans (P = 0.04 compared to carnivores, Wilcoxon test) (Fig. 3a), while microbiota from carnivores prefer animal-derived glycans (P = 0.0001 compared to herbivores, Wilcoxon test) (Fig. 3a). Interestingly, the degradation efficiencies of omnivores and human gut microbiota place them as intermediates between herbivores and carnivores (Fig. 3a). Notably, the higher overall degradation efficiencies for animal-based glycans is probably due to the higher number of animal-derived glycans (1,898) compared to plant-derived glycans (594) in the database.
FIG 3 .
The connection between glycan degradation and diet. (a) GlyDeR profiling analysis of the Muegge data set. Bars showing the average sum of plant-specific and animal-specific GlyDeR scores of the samples grouped according to their host diet and normalized by the number of CAZymes in each sample. A fourth group was created to segregate humans from all other omnivores. The plant- and animal-specific GlyDeR scores of herbivores and carnivores are significantly different (P = 0.04 and P = 0.0001, respectively). (b) The Yatsunenko data set. A scatterplot showing the sample projections on the first principal coordinate and colored according to the country of origin. Samples from individuals younger than 2 years old were omitted (see text).
To further explore where humans stand with respect to the dietary spectrum, we trained a support vector machine (SVM) classifier to distinguish between herbivore and carnivore samples based on their inferred glycan degradation profiles (see Materials and Methods). The classifier predicted all but one sample correctly in a leave-one-out cross-validation (area under the curve [AUC] = 0.93, F = 0.96) and notably outperformed a classifier based only on the abundance of CAZymes found in each sample (which had three misclassifications; AUC = 0.71, F = 0.84), confirming the added predictive value of GlyDeR. We next applied this classifier to the 11 available nonhuman omnivore samples and classified 6 and 5 of the samples as herbivores and carnivores, respectively. These samples were missing direct dietary labeling; however, a comparison of these classifications versus the fiber index for these mammals (39) showed a nice correspondence with the predicted dietary regimens of the animals (Table 1). We next applied the classifier to predict the dietary habits for the human samples, which were unknown, resulting in 15 out of the 18 samples being labeled as carnivores. Thus, at least in the small population sample analyzed here, humans may be closer to carnivores in some functional aspects of their gut microbiota.
TABLE 1 .
Mammalian host diet predictions based on GlyDeR profilesa
| Sample no. | Mammalian species | SVM diet predicted | Fiber index | Correspondence? |
|---|---|---|---|---|
| 4461343 | Hamadryas baboon | Herbivore | 50–500 | Yes |
| 4461344 | Hamadryas baboon | Herbivore | 50–500 | Yes |
| 4461347 | North American black bear | Carnivore | 0-50 | Yes |
| 4461348 | Black lemur | Carnivore | NAb | NA |
| 4461351 | Goeldi’s marmoset | Carnivore | 0–50 | No |
| 4461353 | Chimpanzee | Herbivore | 50–500 | No |
| 4461354 | Chimpanzee | Herbivore | 50–500 | No |
| 4461374 | Ring-tailed lemur | Herbivore | 50–500 | No |
| 4461375 | White-faced saki | Herbivore | 0–50 | No |
| 4461376 | Spectacled bear | Carnivore | 50–500 | No |
| 4461378 | Prevost’s squirrel | Carnivore | 0–50 | Yes |
An SVM classifier was trained based on the GlyDeR profiles of herbivores and carnivores. A diet fiber index for these species was obtained from Ley et al. (40), with the percentages in each diet of acid-detergent fiber (ADF) and neutral-detergent fiber defined. A higher index suggests a diet that is more plant-based. The last column displays the correspondence between the GlyDeR-predicted diet of the animal and its fiber index, where values of 0 to 50 correspond to carnivores and values of 50 to 500 correspond to herbivores.
NA, not available.
We next explored whether humans who live in different geographical areas with markedly different diets exhibit different GlyDeR profiles. We analyzed the Yatsunenko et al. data set (29), which contains metagenomic sequences from fecal samples of 110 humans who live in Venezuela, Malawi, and the United States. Malawian and Venezuelan diets are dominated by plant-derived polysaccharides, while typical U.S. diets contain large quantities of meat (29). As before, we ran GlyDeR and performed PCoA on all the GlyDeR profiles. Because infants display large variabilities over the first coordinate (see Fig. S3b in the supplemental material) and have an unusual diet relative to adults, we filtered out all samples from individuals younger than 2 years old. This led to a clear separation over the first coordinate between low meat consumers (Malawians and Venezuelans) and high meat consumers (Americans) (Fig. 3b). The ratio of animal- to plant-specific GlyDeR scores revealed significant differences between samples from different countries of origin (ANOVA; F = 6.56, P < 0.005), with a higher animal/plant ratio in the United Staets (P < 0.003) and Venezuela (P < 0.03) than in Malawi. The ratio for the United States was slightly but not significantly higher than for Venezuela (Tukey-Kramer honestly signficant difference test; see Fig. S3c).
DISCUSSION
In this analysis, we aimed to determine the association between human diet and microbial metabolism in the gut. We maintain that in order to properly study this relationship, one must incorporate the degradation of glycans into the equation.
We detected diet-driven adaptations at both the level of single species (Fig. 2b) and of communities (Fig. 3a). We found species of the Bacteroidetes phylum to be the most efficient degraders of animal-derived glycans and human milk oligosaccharides. While this trend was apparent for Bacteroides, it was absent for Prevotella, another key member of that phylum. Diets that are high in animal protein have been associated with high levels of Bacteroides, whereas enrichment of Prevotella has been associated with diets rich in plant-derived carbohydrates and very low in animal protein (28–30). Given that many dietary animal glycans are derived from proteins (i.e., glycoproteins and proteoglycans), we propose that the high capabilities of Bacteroides and Parabacteroides to degrade animal glycans explains why their abundance is increased in Westerners (30, 37).
The plethora of novel glycans and their predicted glycan degradation efficiencies supplied by our method may prove to be highly important for designing prebiotic interventions. For example, the inability of some prebiotics to result in significant changes to the gut microbiota may be due to the use of glycans that are utilized too universally by the communities and thus do not provide a competitive advantage to individual, beneficial microbiota community members. GlyDeR provides researchers and clinicians with the ability to predict exactly what glycans are best metabolized by a given desirable taxon and excluded by others in a very specific way. As a striking example, a linear regression model based on GlyDeR-related features was capable of accurately predicting the abundance of bacterial strains that displayed low intersample variance. Degradation of long polysaccharides was the most predictive feature in the model, an unsurprising result considering the importance of these glycans as the main carbon and energy sources for colonic bacteria. Finally, our results were improved significantly by dividing the HMP samples into two clusters and reanalyzing each cluster individually. This supports the notion that microbiome analysis should not be general, but should rather be based carefully on the background community structure.
Our GlyDeR profiling revealed that the relative abundances of many taxa, especially those of Clostridia, are significantly correlated with their ability to degrade glycans. It was recently shown that Clostridium difficile and other pathogenic gut bacteria rely on microbiota-liberated mucosal glycans during their expansion in the gut following antibiotic treatment (40). Thus, it may be possible to design prebiotics that help increase the levels of beneficial Clostridia and prevent the expansion of pathogenic strains. More generally, since the breakdown of a given substrate can be highly species specific (18), the prediction of bacterial glycan degradation efficiencies may prove to be an important tool for designing nutritional interventions to help alter microbial communities.
In analyzing the mammalian fecal samples data reported by Muegge et al. (38), we demonstrated that differences in microbial community compositions carry functional importance—that is, the microbiota of herbivores and carnivores have stronger affinities to plant- and animal-derived glycans, respectively. To the best of our knowledge, this is the first time that a computational framework has been able to provide such observations. The lack of large-scale in vitro glycan utilization assays makes straightforward validation of many of our predictions difficult at present. Nevertheless, our ability to train an accurate classifier to predict the diet of a host based on its microbiota glycan degradation profile, and the correspondence between the classifier’s predictions and animal nutrition (Table 1), both provide a strong operative testimony to the veracity and utility of GlyDeR.
Although humans are generally thought of as omnivores, there is an ongoing debate on the subject of our dietary history and adaptations. Tackling this question through the lens of our microbiota, we used the aforementioned binary herbivore-carnivore classifier in order to classify humans. Remarkably, the classifier predicted 15 out of 18 human subjects to be carnivores. This result is less surprising considering that all of the human subjects were U.S. residents and that the United States is the highest meat-consuming country per capita in the world (41). In contrast to the U.S. population, the diets of individuals from Malawi and Venezuela mainly include plant-derived polysaccharides (these populations consume, on average, 8.3 and 76.8 kg of meat per year, as opposed to 120.2 kg per year in the United States [41]). We consequently found a lower ratio for animal versus plant degradation potency in the microbiota of individuals from these countries (see Fig. S4c in the supplemental material). Notably, GlyDeR does not predict a reduced potency of plant degradation within the U.S. population. Therefore, it seems that the capacity of Western individuals to degrade glycans has not diminished over the course of evolution, but merely shifted toward the direction of carnivores.
Taken together, these results further advance our understanding of human diet-specific adaptations, but conclusions must be drawn with caution. First, the data upon which GlyDeR relies are often incomplete. For instance, only 74 out of the 146 CAZymes mapped to at least one HMP genome (see Fig. S2c in the supplemental material). Furthermore, 31 CAZymes were not capable of breaking any glycan, either because some glycan structures are missing from the database or because of inaccurate enzymatic annotation (see Fig. S2c). Second, several biases may arise from the existence of nonproportional representation of different glycan categories in the KEGG Glycan database. For example, the animal-derived GlyDeR scores are always higher than plant-derived scores because there are three times more animal-derived glycans in the database. We therefore suggest looking only at relative scores and conducting comparisons within a given glycan category (e.g., comparing the degradation of animal-derived glycans in Malawi and the United States) and not between different glycan categories. Finally, the GlyDeR platform does not take into account many important factors, such as enzyme transcription levels, kinetic parameters, and downstream biochemical pathways for glycan utilization. Nevertheless, GlyDeR is the first computational analysis framework that successfully enables one to directly model how the microbiota can respond to dietary glycans from a mechanistic point of view.
The current study was focused on developing a novel method for the study of glycan degradation processes and establishing this method’s value in assessing a wide spectrum of diet-related trends. Future studies will examine whether GlyDeR could capture subtler differences in microbiomes that are derived from individuals with more homogeneous backgrounds (e.g., only meat-eaters). Another open question is the identification of strain differences within a given bacterial species. Since the current study involved only a selected set of 203 microbial genomes, we did not have sufficient coverage of any single species in order to establish significant strain differences in glycan degradation. Since the number of sequenced genomes is constantly increasing, we expect that a larger analysis would be able to provide insight into the glycan degradation capabilities of different strains within species. Finally, an important issue to tackle is the aspect of microbial cross-feeding. GlyDeR is fully capable of analyzing metabolic interactions in the degradation of complex substrates that demand more than one strain for breakdown, and it therefore provides a golden opportunity to examine glycan degradation processes that occur in synthetic communities (and ultimately in natural ones).
While metagenomics are still the gold standard for high-throughput functional analysis of microbial communities, 16S rRNA sequencing is a strong alternative in the many circumstances where metagenomic data are not available, or prohibitively expensive. To this end, we plan to extend and integrate GlyDeR into routine 16S rRNA analyses (e.g., with the help of PICRUSt [42]), as well as incorporate GlyDeR within the larger framework of genome-scale metabolic modeling (9, 43–46). Within this integrated framework, glycan analysis will hopefully become a standard tool in the arsenal of microbial researchers.
MATERIALS AND METHODS
We developed various tools for glycan degradation data integration, manipulation, and analysis, and these can be divided into 3 categories: (i) data retrieval, for which we describe here the sources of information for this study and how they were used; (ii) reconstruction of glycan degradation reactions and cross-validation, by which we reconstructed novel glycan degradation reactions by implementing our GlyDeR algorithm; (iii) data analysis, that is, the steps we performed in order to analyze single taxa and 16S rRNA and metagenomics sequence data and generated microbial (meta)genome glycan degradation potency predictions.
Data retrieval.
Information about glycans and the enzymes that might break them down is spread across many databases and tools. The types of data sources used to infer genome-based glycan degradation capacities included bacterial taxa, genome annotations, and glycans.
(i) Bacterial taxa.
A catalog of 281 taxa was downloaded on 8 October 2011 from The Human Microbiome Project website (http://www.hmpdacc.org/) using the following filters: NCBI superkingdom Bacteria; HMP isolation body site: gastrointestinal tract; project status complete; NCBI submission status annotation (and sequence) public on NCBI site. The catalog contains the following annotation fields: HMP ID, GOLD ID, organism name, domain, NCBI taxon ID, NCBI superkingdom, NCBI phylum, NCBI class, NCBI order, NCBI family, NCBI genus, NCBI species, all body sites, all body subsites, current finishing level, NCBI project ID, Genbank ID, Gene count, size (kb), GC content, Greengenes ID, NCBI 16S accession, strain repository ID, oxygen requirement, cell shape, motility, sporulation, temperature range, optimum temperature, Gram stain, type strain.
(ii) Genome annotations.
All of the HMP taxa were searched against the Seed database (http://pubseed.theseed.org/) using the key of NCBI Taxon Id number as a cross-reference. A total of 204 matches were detected, and their RAST genome annotations were extracted by using the web services of API.
(iii) Glycans.
The entire KEGG Glycan database (http://www.genome.jp/kegg/glycan/) was downloaded on 7 January 2011. The database contains information on 10,978 glycans. We used the following annotation fields from the annotations: G number, name, KCF file, and class. An additional biological origin field was retrieved from an external source, as described below.
The KCF file for each glycan describes a graphical representation of its two-dimensional structure. This representation takes into account the monomeric building blocks (nodes) and the glycosidic linkages (edges) of the glycan. Textual and visual representations of the KCF graph for glycan G00010 are given in Fig. S1a and b in the supplemental material.
The glycans database was subsequently filtered according to the following criteria related to nodes, edges connecting two nodes, and synonym glycans (i.e., glycans with identical structures but different ID numbers). Since the database contains more than 800 nodes denoting glycan-related building blocks, many of which are extremely rare, we chose to focus on a subset of 35 nodes that corresponded to the most prominent sugar monosaccharides, prevalent modifications, amino acids found in glycoproteins, and ceramide found in glycolipids. Therefore, we removed from the analysis all of the glycans which contained nodes not part of this subset. The full list of known nodes can be found in Table S7 in the supplemental material.
Similar to the nodes, an edge connecting two nodes in KEGG Glycan mostly has a standard form denoting whether the sugar at the nonreducing side is in an alpha- or beta-conformation, as well as the numbers of the carbons participating in the glycosidic linkage, e.g., “Glc a1-3 Glc” denotes glucose(α-1,3)glucose. However, there are some rare edges that have a different form. In order to maintain consistent reaction rules, we defined an edge to be legal if it had the common form of “R z$-$ R,” where R is any node except for Thr (threonine), Ser (serine), Ser/Thr (serine or threonine), Asn (asparagine), S (sulfate), or P (phosphate); z is either a or b; $ is any number. Since threonine, serine, asparagine, sulfate and phosphate are not monosaccharides, the glycosidic linkages they are involved in are not created via a carbon atom and therefore the edge description is different. In this case, the rule we used is “R z$-R*,” where R is a regular node and R* is a nonmonosaccharide node. All other edges were marked as illegal, and their glycans were omitted from the analysis.
Some glycans in the database have different IDs but identical structures; therefore, we denote these as “synonym glycans.” Synonym glycans were grouped together, and one glycan from each group was chosen to represent the entire group for further analyses.
We developed glycan structure definitions in order to process information in our database to conform to our subsequent glycan degradation (GlyDeR) reactions. We identified several types of glycans: regular glycans, linear repeating glycans, nonlinear repeating glycans, and polysaccharides. Regular glycans are those glycans with a fixed and known length (see Fig. S1c in the supplemental material). Linear repeating glycans are built completely from a repeating sugar segment (repeating parts are marked with asterisks in Fig. S1d). Nonlinear repeating glycans have a repeating linear segment but also contain modifications on some of the sugars, which make them nonlinear (see Fig. S1e). Polysaccharides are glycans that meet one of the following conditions: it is a repeating glycan, it has the value “polysaccharide” in its class field in the KEGG Glycan database, or it has more than 10 nodes.
(iv) EC numbers and glycan degradation rules.
We obtained a list of 146 CAZymes with a textual description of their enzymatic function within the CAZy database (http://www.cazy.org/). The CAZy database describes families of structurally related catalytic and carbohydrate-binding modules (or functional domains) of enzymes that degrade, modify, or create glycosidic bonds. We retrieved from the database all the EC numbers that belong to following families: EC 2.3.1, acyltransferases, transferring groups other than amino-acyl groups; EC 2.4.1, glycosyl transferases; EC 3.1.1, carboxylic ester hydrolases; EC 3.2.1, glycoside hydrolases; EC 3.5.1, hydrolases acting on carbon-nitrogen bonds, other than peptide bonds, in linear amides; EC 4.2.2, polysaccharide lyases.
Based on the information available for these EC numbers in ExPASy (http://expasy.org/) and KEGG (http://www.genome.jp/kegg/), we manually generated a table that linked each EC number with the following fields: enzyme name, linkages broken, contained subglycan (linkage must be part of the subglycan), contains only (nodes), glycan released, Endo versus Exo, DP preference (number of nodes), terminal side preference, enzymatic reaction, and comments. These fields were later used to generate glycan degradation reactions by defining and implementing a set of rules for analysis of the KCF file of all the glycans (see Fig. S1d in the supplemental material for further informaton on the reconstruction of glycan degradation reactions). These fields can be further described as follows:
Enzyme Name—The accepted name of the enzyme. KEGG Reactions—The reactions from KEGG that map to the EC number. Linkages Broken—In the case of glycosidic linkages the value is a string representing two nodes and an edge that connects them based on the KCF graph representation of the glycan structure. For example, in the case of deacetylation reactions, this value is “Ac-R,” denoting the removal of an acetyl group from node R. Contained Subglycan—A G# identifier of a glycan structure contained within the structure of a larger glycan, e.g., “Glc b1-4 Glc” is a subglycan of Man a1-3 Glc b1-4 Glc. Contains Only (nodes)—A G# identifier for one or more nodes that the glycan must contain and only contain. Glycan Released—A G# identifier that defines a glycan that must be one of the products after the reaction with the enzyme takes place. Endo vs. Exo—“exo” enzymes, which remove only terminal sugars (the edges of the terminal nodes), “endo” enzymes, which break all glycosidic bonds except for terminal ones (remove all edges except the ones of the terminal nodes), and “both” enzymes, which can break any bond (remove any edge). DP preference—This field reflects the degree of polymerization of the glycans upon which the enzyme works. This number is also the exact/minimal (+)/maximal (−) number of nodes that the KCF graph for the glycan structure must contain. Terminal Side Preference—This field is unique for exo-acting enzymes and describes their specificity toward the reducing or nonreducing end. The KCF graph is directional, hence “reducing” means only the removal of the right-most node is allowed, “nonreducing” refers to the left-most, and “both” allows the removal of both edges. Notice that the terminal node of the reducing end is always at position 1 in the KCF graph, except for repeating glycans. Enzymatic Reaction—A textual description of the enzymatic reaction performed by a given enzyme (EC number) was obtained from http://enzyme.expasy.org/. Comments—Specific comments about an enzyme (EC number) were also taken from http://enzyme.expasy.org/. (Special characters used in the above descriptions included the following. $ signifies any number. R signifies any type of the following sugars [nodes; abbreviations are those used for the http://enzyme.expasy.org/ website]: ara, Araf, D/LAra, D/LAraf, LAra, LAraf, D/LAraf, D/LAra, Api, Apif, D/LApi, D/LApif, 3,6-Anhydro-LGal, L3,6-anhydro-Gal, 3,6-Anhydro-Gal, GalA, D/LGalA, GalNAc, GalfNAc, D/LGalNAc, GalN, GlcNAc, D/LGlcNAc, GlcA, D/LGlcA, GlcN, D/LGlcN, Glc, Glcf, D/LGlc, Fru, Fruf, D/LFru, D-Fruf, Man, Manf, D/LMan, ManA, Rha, D/LRha, LRha, D/LRha, Gal, Galf, D/LGal, D/LGalf, Fuc, D/LFuc, Fucf, LFuc, D/LFuc, Xyl, D/LXyl, Xylf, Neu, Neu5Ac, Neu5Gc, MurNAc. # indicates an “or” association, and & denotes an “and” association.) Overall, this workflow resulted in 141,561 glycan degradation reactions, of which 9,325 were reactions that degrade KEGG glycans and newly reconstructed glycans and 132,236 were intermediate glycan-degrading reactions.
(v) CAZyme annotation.
We used sequence similarity to match the genes which belong to the HMP taxa with specific CAZymes. We therefore performed BLAST analysis for all of the genomes of the HMP taxa against the bacterial protein sequences found in the CAZy database. Each enzyme family in CAZy contains a set of manually curated enzymes that have been determined to execute a specific catalytic function. We used the NCBI BLAST utility and filtered errors at the level of 10 e−10 and matches below 97% identity. At that point, we had a mapping between genes in the HMP taxa and CAZyme families. Because many families contain a one-to-many mapping between a family and its associated EC numbers, we had to refine this annotation. We therefore extracted the genes predicted for enzymatic annotations from the SEED and KEGG databases. While the CAZyme annotations are more comprehensive, they are sometimes not as accurate as the manually curated ones. Thus, we integrated the information obtained from all of these sources by using the following logic: for proteins that were mapped to families of 1 EC number in CAZy, we accepted this annotation. For proteins that were mapped to families of multiple EC numbers, we first checked if they had an available annotation in SEED or KEGG, and if that was the case, then we checked if this annotation belonged to one of the multiple annotations in CAZy. If it did, then we accepted the KEGG/SEED annotation.
(vi) Subcellular localization annotation.
To define the subcellular localization (SCL) of reactions, we used the RAST genome annotation as a first proxy. We mined the function and subsystem fields of the annotation for special keywords. For our purposes, we were only interested in whether the enzyme exerts its function inside the cell or outside. Enzymes were defined as intracellular if their associated genes contained the keywords cytoplasm, cytosol, or cytoplasmic. Enzymes were defined as cross-membrane if their associated genes contained one of the keywords periplasm, periplasmic, inner membrane, or cytoplasmic membrane. Finally, enzymes were defined as extracellular if their associated genes contained one of the keywords cellulosome, outer membrane, secreted, cell wall, or extracellular. For enzymes that were not associated with any meaningful keyword, we took advantage of the LOCtree localization prediction software (https://rostlab.org/owiki/index.php/LOCtree). LOCtree uses a protein amino acid sequence to predict the SCL. It supplies five possible SCLs: cytosol, inner membrane, periplasmatic, outer membrane, and secreted. Enzymes with the value cytosol were classified as intracellular, enzymes with the values secreted or outer membrane were classified as extracellular, and enzymes with the values periplasmatic or inner membrane were classified as cross-membrane, i.e., enzymes that exert their function on the cross-membrane between the cell and its environment. To fix possible erroneous annotations, we refined our localization selection based on specific knowledge of the glycan degradation biochemistry. A literature survey suggested that there are no polysaccharides within the bacterial cytoplasm (with glycogen being the only exception). Thus, enzymes that were predicted as intracellular or cross-membrane were filtered out if the glycan that they processed was either repeating, defined as a polysaccharide, or had more than 10 subcomponents.
(vii) Biological origin of glycans.
We accessed the CarbBank database (47) and mapped the KEGG glycans to it using the KEGG Glycan ID (G number) as a cross-reference. CarbBank contains detailed descriptions of where a specific glycan can be found in nature. We parsed these data in order to define certain glycans as either plant-derived or animal-derived.
(viii) Degree of polymerization of glycans.
Glycans are routinely categorized into one of four possible degrees or classes of polymerization. With respect to classes, glycans were defined as disaccharides if they contained 2 nodes, oligosaccharides if they contained 3 to 10 nodes, short polysaccharides if they contained >10 nodes, and long polysaccharides if they had a repeating structure.
Construction of the CAZyme table (a key step in the GlyDeR pipeline).
We manually curated all of the CAZymes (146 EC numbers) and mapped each one to a set of computer-based rules dictating the mechanism by which it can break a given glycan (i.e., split its graph into two separate components). These rules account for structural features such as the glycosidic linkages the enzyme can break, the cleavage mechanism, the chemical neighborhood, and the degree of polymerization of the glycan (see Table S3a and b for a list of the rules). We then executed these rules on all the glycans that appear in the KEGG Glycan database, which yielded 141,561 glycan degradation (GlyDeR) reactions. In the following section we describe the logic behind the reconstruction of glycan degradation reactions by identifying for each glycan which enzymes are able to break it and how the degradation reaction will look. GlyDeR reactions are then mapped back to CAZymes in order to produce a table where the rows are CAZymes, the columns are glycans, and each entry contains a CAZyme score, calculated as follows:
where ei is an enzyme that can degrade glycan j and gk is the number of glycans that it breaks down. The entire construction process is summarized in Fig. 1b. A CAZymes table which contains all of the CAZyme scores can be found in Table S5 in the supplemental material.
(i) Glycan degradation (GlyDeR) rules.
A reaction is represented by its substrate(s), product(s), the enzyme(s) responsible for the catalysis, and a subcellular localization. For the GlyDeR reaction generation process, we used all the computer-based glycan breaking rules described in Table S1 of the supplemental material. For an EC number-related enzyme (rule) to break a glycan, the glycan and the resultant reaction must comply with all the limitations defined in the fields of the given rule, namely, the glycan must contain at least one of the glycosidic linkages (or nodes containing an acetyl group in case of deacetylation reactions) described in the Linkages Broken field; the glycosidic linkage hydrolyzed must appear in the terminal edges of the glycan if the value in the Endo Vs. Exo is set to Endo, and vice versa; the number of nodes the glycan contains must conform to the value described in the DP Preference field; in case the Endo versus Exo field is set to Exo, the terminal side of the glycosidic linkage hydrolyzed must be located on the right side of the graph of the glycan if the value in the Terminal Side Preference field is set to Reducing and on the left side if this field is set to Nonreducing. If this field is set to “Both,” then location of this linkage on both sides is allowed. The glycan must contain the structure of a glycan (nodes and edges) described in the Contained Subglycan field, and the linkage being broken must also be part of this subglycan. The reaction must contain the glycan described in the Glycan Released field as one of its products. Figure 1a gives an example of an Exo-acting enzyme breaking a regular glycan.
(ii) Deacetylation rules.
Some of the EC numbers (enzymes) we analyzed have a deacetylation activity, i.e., they have the capability to remove acetyl groups. In the KEGG Glycan database, monosaccharides containing an acetyl group are described as a single unique node, e.g., the node GlcNAc corresponds to N-acetyl-glucosamine. Therefore, if an enzyme has the capability to remove an acetyl group, we simply remove the substring “Ac” from the label of the node and make it the product of the reaction, e.g., GlcNAc ↔ GlcN + Ac.
(iii) Reconstruction of new glycans.
We manually constructed a set of 107 glycans which we determined were important but that were missing from the KEGG Glycan database. To distinguish these glycans from the ones previously available in the database, we gave these new glycans the prefix TAU instead of G, which is assigned by KEGG. A list of all the TAU glycans can be found in the supplemental material. Furthermore, in most cases the products of the degradation reactions did not have a preexisting G number, meaning they currently do not exist in the KEGG Glycan database. Working under the assumption that most glycans in nature are still uncharacterized in databases, we decided to add these new glycans automatically. Thus, whenever a reaction produced a new glycan, we gave this glycan a unique ID beginning with “TAUS” (to distinguish it from original glycans, designations for which begin with G or TAU).
Data analysis. (i) Single-taxon data analysis.
To define microbial genome-specific GlyDeR scores, after building the CAZyme table we associated the CAZymes with the genomes of the HMP gut taxa. For every taxon-specific gene we calculated, based on its enzymatic annotation and the enzyme’s subcellular localization, a GlyDeR score. Given a bacterial taxon i and glycan j, the GlyDeR score was calculated as follows:
where ek is an enzyme that can degrade glycan j, nik is the number of genes in its genome which translate to enzyme ek, and gk is the number of glycans broken by enzyme ek. This metric decrements the contribution of CAZymes that are more promiscuous versus those specifically geared to degrade the glycan in question (Fig. 1b). For some categories of glycans such as long polysaccharides and plant-specific glycans, we defined a category-specific score, GSic, which is the sum of GlyDeR scores for glycans that belong in that group:
where GSij is the GlyDeR score for genome i and glycan j and c is the collection of glycans that belong to category C. This scoring system has the feature that summing the GlyDeR scores over all the glycans in a given genome gives the total number of CAZymes in the genome, according to the following equation:
where j ∈ j is the set of all glycans and i is the index of a specific taxon. Subsequently, we defined the GlyDeR profile of a bacterial taxon as follows:
For the GlyDeR reaction consistency check and cross-validation, when applied to all the glycans available in KEGG, the GlyDeR pipeline produced a list of 114,573 intermediate glycan products, most of which were novel and thus do not appear in the original KEGG database. We refer to these as hypothetical glycans in the text. To test the consistency of GlyDeR, we performed a cross-validation process where we picked a random subset of 1,000 glycans from KEGG and applied GlyDeR to degrade them. We then tested whether the products obtained from these 1,000 glycans were enriched with known versus novel intermediate glycans. A hypergeometric test indicated that the products were highly enriched for known glycans (P = 10−19) (see Fig. S2a in the supplemental material). A sensitivity analysis with subsets of different initial random sets and sizes still resulted in highly significant enrichments (data not shown). This result testifies that GlyDeR is capable of recapitulating the biochemical knowledge imprinted in the CAZymes that constitutes its computational foundation.
For principal coordinates analysis on GlyDeR profiles, we calculated the pairwise Bray-Curtis dissimilarities between all the GlyDeR profiles and performed PCoA on the resulting dissimilarity matrix to project the differences in degradation into two dimensions (see Fig. S2d in the supplemental material). The GlyDeR analysis file for the HMP taxa is given in Table S3 of the supplemental material and lists for each genome the unique CAZymes, total CAZymes, plant-specific GlyDeR score, animal-specific GlyDeR score, and bacterium-specific GlyDeR score.
For the GlyDeR-related features definition, we extracted 6 features that characterized the several dimensions of a (meta)genome glycan degradation potential. These features were the GlyDeR scores for plant-specific glycans, animal-specific glycans, disaccharides, oligosaccharides, short polysaccharides, and long polysaccharides. Each feature represented the sum of the GlyDeR scores for the glycans that belong in the class.
(ii) 16S rRNA sequence data analysis.
We retrieved the 16S rRNA sequence data and metadata from fecal samples belonging to 325 healthy human individuals from the HMP Data Analysis and Coordination Center (DACC) (48). Because we were not interested in time series data, we only used samples from the initial time point. 16S rRNA sequences were mapped to the HMP genomes based on sequence similarity and further used to build an operational taxonomic unit table that described the abundances of the HMP taxa in each sample. For this purpose, we used the QIIME software (49) with the following exact commands:pick_otus.py -i HMP_samples_seqs -r HMP_taxa_ref_seqs -m uclust_ref -C
make_otu_table.py -i pick_otus_output
(iii) Metagenomics sequence data analysis.
We retrieved the Muegge et al. (38) and Yatsunenko et al. (29) data sets from MG-RAST (50). For both data sets, we downloaded the FragGeneScan gene-calling output, which maps each original read to 0, 1, or more open reading frames (ORFs). This way, many reads could be assigned to a single ORF and so the abundance of each ORF was taken into account. Next, ORFs were assigned to CAZymes and to specific subcellular localizations, as described above. The percent identity used for the BLAST search was changed to 60%, a value commonly used in metagenomic annotation projects (51). We then constructed a CAZymes abundance table to describe the abundances of all the CAZymes in each sample. In order to calculate a sample-specific GlyDeR score, we used the sample CAZymes abundance table and precalculated CAZyme scores table. Thus, the GlyDeR score, GSkj, of glycan j in sample k was defined as follows:
where ek is an enzyme that can degrade glycan j, nik is the number of genes in sample k that map to enzyme ek, and gk is the number of glycans broken by enzyme ek. Dmax/Dk is a normalization factor that denotes the ratio between the depth (i.e., the total number of sequenced reads) of the sample with the maximum depth (Dmax) and the depth of the current sample, Dk. Next, we defined the GlyDeR profile of sample k as follows:
(iv) Multivariate regression between GlyDeR features and bacterial abundance.
We analyzed the 16S rRNA sequences from the HMP fecal samples in order to determine the abundance of our 203 bacterial taxa in each sample. To increase the signal-to-noise ratio (SNR), we excluded species with a high abundance variability based on the following criterion:
The 6 features described in the previous section were used to build a linear regression model with bacterial abundance as the dependent variable: bacterial abundance = (−13.248 × plant-specific GlyDeR score) + (28.1822 × disaccharides score) − (9.6206 × oligosaccharides score) + (32.701 × long polysaccharides score) + 42.7354. To eliminate the possibility of overfitting the data, we used a standard 10-fold cross-validation method. All calculations were performed using WEKA (51).
To assess the added value of using the GlyDeR features over genomic information alone, we defined for each HMP taxon a vector containing the genomic copy number of the CAZymes used in the analysis. We then built a similar linear regression model with these 82 CAZymes used as features and bacterial abundance in the HMP samples as the dependent variable.
Because of the high variability in bacterial taxa abundance across the samples, we used the KMeans algorithm to cluster the samples. We chose to use 3 clusters because this option resulted in the lowest cubic clustering criterion (CCC). However, one cluster was composed of only one outlier taxon, so we omitted it from further analysis. Indeed, after removing the outlier point, the lowest CCC was achieved for k = 2. We built a cluster-specific linear regression model, as described above: cluster 1 bacterial abundance = (−40.6116 × plant-specific GlyDeR score) + (33.283 × long polysaccharides score) + 31.6149; cluster 2 bacterial abundance = (−15.9773 × oligosaccharides score) + (69.8184 × long polysaccharides score) + 70.697.
We defined a list of 25 bacterial taxa with highly predictable accuracy. A taxon was included in the list if the standard error of its predicted abundance obeyed the following rule for either cluster 1 or cluster 2:
(v) Classification of dietary patterns based on GlyDeR scores.
The GlyDeR scores of all herbivore and carnivore mammals from the Muegge et al. data set were used to train a binary SVM classifier. The SMO implementation of this classification algorithm in WEKA (51) was used for the computation. To estimate the accuracy of the classifier, we used a standard leave-one-out cross-validation. To apply this classifier to the remaining human and nonhuman omnivore samples, we hid the labels of the samples and classified them as either carnivore or herbivore.
SUPPLEMENTAL MATERIAL
KEGG glycans. (a to e) Examples of different types of glycans found in the KEGG Glycans database. (a) A textual representation of the KCF graph for glycan G00010. (b) A visual representation of the KCF graph for glycan G00010. (c) A regular glycan. (d) A linear repeating 4 glycan. (e) A nonlinear repeating glycan. (f) Reconstruction of GlyDeR reactions. The GlyDeR algorithm receives a glycan graph structure and an EC number as input and generates the appropriate glycan products, while considering the following rules: glycosidic linkages hydrolyzed, endo- versus exo-acting enzyme, degree of polymerization preference, reducing versus nonreducing end preference, contained subglycan, glycan released. In the example depicted, these fields had the following values, respectively: Glc a1-2 Glc, exo, 10+, nonreducing, unknown, TAU00015 (the glycan ID of glucose). Download
Glycan degradation of the gut microbiota reference genomes. (a) A cross-validation process was performed to show that GlyDeR reaction products are enriched with existing glycans rather than hypothetical ones. The Venn diagram depicts a significant overlap between the products created by GlyDeR (green) and the original glycans in the KEGG Glycan database (purple). (b) PCoA of the glycan degradation profiles of the species, colored according to their respected phyla. (c) Bar chart depicting the median GlyDeR score of the species belonging to each genus and colored according to their respected phyla. (d) A heat map denoting the average relative glycan degradation potency of the different bacterial genera in the study. Each entry was calculated based on the average sum of GlyDeR scores per genus for a specific degree of polymerization (DP) category (e.g., disaccharides) and normalized by the overall sum of GlyDeR scores of all the genera for the same DP category. (e) Bar plot depicting the Total GlyDe scores (and error bars) of strains belonging to each phylum. Purple indicates all glycans, red indicates glycans derived from animals, blue indicates glycans derived from bacteria, and green indicates glycans derived from plants. (f) Bar plot comparing the glycan degradation potential of the different bacterial genera for ovalbumin (blue) and casein (red). Each bar depicts the median of the GlyDe scores of organisms belonging to their respected genus. (g) Nonmetric multidimensional scaling (NMDS) of the glycan degradation profiles of the species, colored according to their respected phyla. (h) Shepard plot of obtained versus observed (target) ranks, indicating the quality of the NMDS result. Download
The connection between glycan degradation and diet. (a) The Muegge et al. data set and PCoA of the GlyDeR profiles of all the samples. The first principal coordinate shows a gradient is formed, starting from herbivores (red), to omnivores (green) and carnivores (blue). (b and c) The Yatsunenko et al. data set. (b) The box plots represent the variations in GlyDeR profiles of the samples over the first principal coordinate and separated into bins according to the age of the host. (c) Box plots showing the differences between the animal- to plant-specific GlyDeR score ratios of adults in Malawi, Venezuela, and the United States. Download
Description of the manually curated GlyDeR rules that describe each CAZyme (EC number)
Summary of all 203 HMP bacterial strains used in the analysis
GlyDeR outputs for all the HMP taxa
GlyDeR scores of 8 human milk oligosaccharides available in KEGG
CAZyme table
Detailed account of the glycans used throughout this analysis
Monosaccharides and other basic chemical entities used as nodes in the graphs representing glycan structures in KEGG and incorporated into our system
Summary of linear regression coefficients and root mean square error scores between actual and predicted abundances of specific bacterial classes in the HMP samples
Taxa with highly predictable abundance levels
ACKNOWLEDGMENT
This work was supported by the U.S.-Israel Binational Science Foundation (BSF).
Footnotes
Citation Eilam O, Zarecki R, Oberhardt M, Ursell LK, Kupiec M, Knight R, Gophna U, Ruppin E. 2014. Glycan degradation (GlyDeR) analysis predicts mammalian gut microbiota abundance and host diet-specific adaptations. mBio 5(4):e01526-14. doi:10.1128/mBio.01526-14.
REFERENCES
- 1. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Keefe SJ. 2008. Nutrition and colonic health: the critical role of the microbiota. Curr. Opin. Gastroenterol. 24:51–58. 10.1097/MOG.0b013e3282f323f3 [DOI] [PubMed] [Google Scholar]
- 3. Goodman AL, Gordon JI. 2010. Our unindicted coconspirators: human metabolism from a microbial perspective. Cell Metab. 12:111–116. 10.1016/j.cmet.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holmes E, Li JV, Marchesi JR, Nicholson JK. 2012. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 16:559–564. 10.1016/j.cmet.2012.10.007 [DOI] [PubMed] [Google Scholar]
- 5. Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 6. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- 7. Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S, Nicholson JK. 2012. Therapeutic modulation of microbiota-host metabolic interactions. Sci. Transl. Med. 4:137rv6. 10.1126/scitranslmed.3004244 [DOI] [PubMed] [Google Scholar]
- 9. Borenstein E. 2012. Computational systems biology and in silico modeling of the human microbiome. Brief. Bioinform. 13:769–780. 10.1093/bib/bbs022 [DOI] [PubMed] [Google Scholar]
- 10. Zengler K, Palsson BO. 2012. A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 10:366–372. 10.1038/nrmicro2763 [DOI] [PubMed] [Google Scholar]
- 11. Dirk G, Mihai P, Patrick DS, Curtis H, Jonathan AE. 2012. Bioinformatics for the Human Microbiome Project. PLoS Comput. Biol. 8:e1002779. 10.1371/journal.pcbi.1002779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macfarlane GT, Macfarlane S. 2007. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr. Opin. Biotechnol. 18:156–162. 10.1016/j.copbio.2007.01.011 [DOI] [PubMed] [Google Scholar]
- 13. Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gérard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Zoetendal E, Kleerebezem M, Smidt H, Van de Wiele T. 2010. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 76:5237–5246. 10.1128/AEM.00759-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elia M, Cummings JH. 2007. Physiological aspects of energy metabolism and gastrointestinal effects of carbohydrates. Eur. J. Clin. Nutr. 61(Suppl 1):S40–S74. 10.1038/sj.ejcn.1602938 [DOI] [PubMed] [Google Scholar]
- 15. Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10:323–335. 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106:5859–5864. 10.1073/pnas.0901529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654–1661. 10.1128/AEM.66.4.1654-1661.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flint HJ, Duncan SH, Scott KP, Louis P. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9:1101–1111. 10.1111/j.1462-2920.2007.01281.x [DOI] [PubMed] [Google Scholar]
- 19. Macfarlane GT, Macfarlane S. 2011. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 45(Suppl):7. 10.1097/MCG.0b013e31822fecfe [DOI] [PubMed] [Google Scholar]
- 20. Willem FB, Christophe MC, Kristin V, Van T, Van de wiele W, Willy V, Jan AD. 2011. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit. Rev. Food Sci. Nutr. 51:178–191. 10.1080/10408390903044768 [DOI] [PubMed] [Google Scholar]
- 21. Sonnenburg JL, Fischbach MA. 2011. Community health care: therapeutic opportunities in the human microbiome. Sci. Transl. Med. 3:78ps12. 10.1126/scitranslmed.3001626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. 2006. KEGG as a glycome informatics resource. Glycobiology 16:63R–70R. 10.1093/glycob/cwj010 [DOI] [PubMed] [Google Scholar]
- 23. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 24. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:8. 10.1093/nar/gkn953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Stevens RL, Vonstein V, Xia F. 2012. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7:e48053. 10.1371/journal.pone.0048053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27:29–34. 10.1093/nar/27.20.e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Francesco VD, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res. 19:2317–2323. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Judith RK, Gary DW. 2012. The gut microbiota, environment and diseases of modern society. Gut Microbes 3:374–382. 10.4161/gmic.21333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- 33. Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. 10.1016/j.cell.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marcobal A, Sonnenburg JL. 2012. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 18(Suppl 4):12–15. 10.1111/j.1469-0691.2012.03863.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harvey DJ, Wing DR, Küster B, Wilson IB. 2000. Composition of N-linked carbohydrates from ovalbumin and co-purified glycoproteins. J. Am. Soc. Mass Spectrom. 11:564–571. 10.1016/S1044-0305(00)00122-7 [DOI] [PubMed] [Google Scholar]
- 36. Robitaille G, Ng-Kwai-Hang KF, Monardes HG. 1991. Association of kappa-casein glycosylation with milk production and composition in Holsteins. J. Dairy Sci. 74:3314–3317 [DOI] [PubMed] [Google Scholar]
- 37. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A. 107:14691–14696. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. 10.1126/science.1198719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut Microbes. Science 320:1647–1651. 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. FAO 2009. FAOSTAT databases. United Nations Food and Agriculture Organization, Rome, Italy [Google Scholar]
- 42. Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. 10.1038/nbt2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28:977–982. 10.1038/nbt.1672 [DOI] [PubMed] [Google Scholar]
- 44. Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. 2011. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2:589. 10.1038/ncomms1597 [DOI] [PubMed] [Google Scholar]
- 45. Thiele I, Heinken A, Fleming RM. 2013. A systems biology approach to studying the role of microbes in human health. Curr. Opin. Biotechnol. 24:4–12. 10.1016/j.copbio.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 46. Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BØ. 2011. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6:1290–1307. 10.1038/nprot.2011.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doubet S, Albersheim P. 1992. CarbBank. Glycobiology 2:505. 10.1093/glycob/2.6.505 [DOI] [PubMed] [Google Scholar]
- 48. Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Jacob B, Ratner A, Liolios K, Pagani I, Huntemann M, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG/M-HMP: a metagenome comparative analysis system for the Human Microbiome Project. PLoS One 7:e40151. 10.1371/journal.pone.0040151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caporaso J, Kuczynski J, Stombaugh J, Bittinger K. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. 2010. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb. Protoc. 2010:pdb.prot5368. 10.1101/pdb.prot5368 [DOI] [PubMed] [Google Scholar]
- 51. Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. 2009. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter 11:10–18 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
KEGG glycans. (a to e) Examples of different types of glycans found in the KEGG Glycans database. (a) A textual representation of the KCF graph for glycan G00010. (b) A visual representation of the KCF graph for glycan G00010. (c) A regular glycan. (d) A linear repeating 4 glycan. (e) A nonlinear repeating glycan. (f) Reconstruction of GlyDeR reactions. The GlyDeR algorithm receives a glycan graph structure and an EC number as input and generates the appropriate glycan products, while considering the following rules: glycosidic linkages hydrolyzed, endo- versus exo-acting enzyme, degree of polymerization preference, reducing versus nonreducing end preference, contained subglycan, glycan released. In the example depicted, these fields had the following values, respectively: Glc a1-2 Glc, exo, 10+, nonreducing, unknown, TAU00015 (the glycan ID of glucose). Download
Glycan degradation of the gut microbiota reference genomes. (a) A cross-validation process was performed to show that GlyDeR reaction products are enriched with existing glycans rather than hypothetical ones. The Venn diagram depicts a significant overlap between the products created by GlyDeR (green) and the original glycans in the KEGG Glycan database (purple). (b) PCoA of the glycan degradation profiles of the species, colored according to their respected phyla. (c) Bar chart depicting the median GlyDeR score of the species belonging to each genus and colored according to their respected phyla. (d) A heat map denoting the average relative glycan degradation potency of the different bacterial genera in the study. Each entry was calculated based on the average sum of GlyDeR scores per genus for a specific degree of polymerization (DP) category (e.g., disaccharides) and normalized by the overall sum of GlyDeR scores of all the genera for the same DP category. (e) Bar plot depicting the Total GlyDe scores (and error bars) of strains belonging to each phylum. Purple indicates all glycans, red indicates glycans derived from animals, blue indicates glycans derived from bacteria, and green indicates glycans derived from plants. (f) Bar plot comparing the glycan degradation potential of the different bacterial genera for ovalbumin (blue) and casein (red). Each bar depicts the median of the GlyDe scores of organisms belonging to their respected genus. (g) Nonmetric multidimensional scaling (NMDS) of the glycan degradation profiles of the species, colored according to their respected phyla. (h) Shepard plot of obtained versus observed (target) ranks, indicating the quality of the NMDS result. Download
The connection between glycan degradation and diet. (a) The Muegge et al. data set and PCoA of the GlyDeR profiles of all the samples. The first principal coordinate shows a gradient is formed, starting from herbivores (red), to omnivores (green) and carnivores (blue). (b and c) The Yatsunenko et al. data set. (b) The box plots represent the variations in GlyDeR profiles of the samples over the first principal coordinate and separated into bins according to the age of the host. (c) Box plots showing the differences between the animal- to plant-specific GlyDeR score ratios of adults in Malawi, Venezuela, and the United States. Download
Description of the manually curated GlyDeR rules that describe each CAZyme (EC number)
Summary of all 203 HMP bacterial strains used in the analysis
GlyDeR outputs for all the HMP taxa
GlyDeR scores of 8 human milk oligosaccharides available in KEGG
CAZyme table
Detailed account of the glycans used throughout this analysis
Monosaccharides and other basic chemical entities used as nodes in the graphs representing glycan structures in KEGG and incorporated into our system
Summary of linear regression coefficients and root mean square error scores between actual and predicted abundances of specific bacterial classes in the HMP samples
Taxa with highly predictable abundance levels