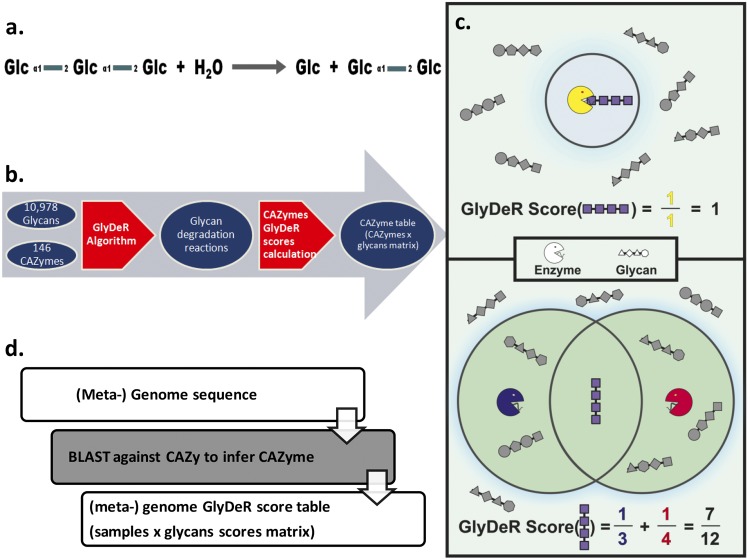
FIG 1 .
The GlyDeR platform. (a) A visual representation of the glycan degradation reaction performed for EC 3.2.1.115, breaking down Kojitriose into Kojibiose and glucose. (b) A schematic representation of the construction of the computational pipeline. Information is taken from multiple databases and analyzed as follows. Step 1 (lred arrow on left): by using CAZyme information and the GlyDeR algorithm, glycan degradation reactions are reconstructed. Step 2 (red arrow on right): a CAZyme table is constructed that represents the potency with which different CAZymes break different glycans. (c) GlyDeR score calculation. (Top) The organism has one enzyme (yellow PacMan) dedicated to the degradation of one glycan (purple); therefore, the GlyDeR score for the purple glycan equals 1. (Bottom) The organism has two enzymes capable of degrading 3 and 4 glycans, respectively, and therefore the GlyDeR score for the purple glycan equals 7/12. (d) GlyDeR utilization. (Meta)genomes are annotated for CAZymes by using CAZy, SEED, and KEGG databases, and with the CAZyme table a GlyDeR score can be calculated, reflecting the capacity of a (meta)genome to degrade a specific glycan.