Abstract
Oral squamous cell carcinoma (OSCC) is the most common malignant neoplasm of the oral cavity, representing ~90% of all oral carcinomas and accounting for 3–5% of all malignancies. The WWOX gene (WW-domain containing oxidoreductase) is a candidate tumor suppressor gene located at 16q23.3–24.1, spanning the second most common fragile site, FRA16D. In this report, the role of the WWOX gene was investigated in 20 tumors and 10 normal oral mucosas, and we demonstrated an altered WWOX gene in 50% (10/20) of OSCCs. Using nested RT-PCR, mRNA transcription was altered in 35% of the tumors, with the complete absence of transcripts in 2 samples as well as absence of exons 6–8 (2 tumors), exon 7 (1 tumor), exon 7 and exon 6–8 (1 tumor) and partial loss of exons 8 and 9 (1 tumor). To determine if the aberrant transcripts were translated, Western blots were performed in all samples; however, only the normal protein was detected. By immunohistochemistry, a reduction in Wwox protein expression was observed, affecting 40% of the tumors when compared with normal mucosa. In addition, a novel somatic mutation (S329F) was found. The presence of alterations in mRNA transcription correlated with the reduced expression of Wwox protein in the tumors. These results show that the WWOX gene is frequently altered in OSCC and may contribute to the carcinogenesis processes in oral cancer.
Keywords: WWOX, oral squamous cell carcinoma, tumor suppressor gene, fragile sites
Oral squamous cell carcinoma (OSCC) is the sixth most common type of cancer worldwide, accounting for 3–5% of all malignancies and characterized by a high degree of invasiveness and metastasis to cervical lymph nodes. It also has one of the lowest 5-year survival rates among all major tumor types including breast, prostate and colon cancers.1 It is generally accepted that OSCC results from a multistep process of accumulated genetic damage leading to cell deregulation, with disruption in cell signaling, DNA-repair and cell-cycle, which are fundamental to homeostasis. Although frequent genetic abnormalities have been demonstrated in OSCC and in oral dysplastic epithelium,2,3 the precise molecular mechanisms of development or progression of OSCC still remain unclear.
WWOX (WW-domain containing oxidoredutase) is a candidate tumor suppressor gene that was recently identified in chromosome 16q23.3–24.1. It was determined that the WWOX gene spans the common fragile site region FRA16D.4–6 Chromosomal fragile sites are often hot spots for translocations, deletions, gene amplification and the integration of oncogenic viruses. These are chromosomal regions that frequently exhibit DNA strand breaks, often following exposure to chemicals that delay DNA replication.7 Of the common chromosomal fragile site loci, FRA3B and FRA16D are the most frequently expressed.8 The tumor suppressor gene fragile histidine triad (FHIT) was found to span the FRA3B fragile site,9 and abnormal FHIT transcription and low FHIT expression were detected in various human cancers, including oral squamous cell carcinomas.10,11 Studies on those 2 most frequently affected common fragile site loci, FRA3B and FRA16D, have provided compelling evidence that these regions are indeed prone to DNA instability in cancer cells.12 It has been shown that exposure to environmental carcinogens such as smoking and alcohol consumption increases the potential for chromosome breakage at fragile sites FRA3B and FRA16D in esophageal and non-small-cell lung cancer.13–15 Therefore, as smoking and alcohol consumption are major risk factors for OSCC,16 given the fact that environmental carcinogens may preferentially induce alterations in fragile regions, and that alterations of the WWOX gene have been found in esophageal squamous carcinomas,17 in this report we investigated whether the WWOX gene at FRA16D plays a role in tumorigenesis in a series of primary human OSCCs.
Material and methods
Human tissue
Fresh tissue and blood samples were consecutively obtained from 20 smokers undergoing therapeutic surgical resection for OSCC at the Hospital Luxemburgo between August 2003 and June 2004. Clinicopathologic data, including patient’s age, sex, TNM staging18 and histological grade are shown in Table I. Oral normal mucosa was obtained from volunteers without OSCC during non neoplastic or preprosthetic surgical procedures (control samples). The age, sex and smoking habits of the healthy volunteers were matched with patients. The present study was approved by the local Ethics Committee, and a signed informed consent was obtained from all patients as well as normal volunteers. In each case, a portion of the tumor was resected, immediately snap frozen and stored at −80°C. Genomic DNA was extracted as previously described.19 For immunohistochemistry, a portion of the tissue was fixed in 10% buffered formalin and paraffin embedded.
TABLE I.
CLINICO-PATHOLOGICAL CHARACTERISTICS OF THE TUMORS
| Tumors | Sex | Age (y) | TNM1 | Histology2 |
|---|---|---|---|---|
| #CA2 | F | 41 | T3N0M0 | 2 |
| #CA3 | M | 59 | T4N0M0 | 1 |
| #CA4 | M | 42 | T1N0M0 | 1 |
| #CA5 | M | 55 | T4N1M0 | 3 |
| #CA7 | M | 37 | T4N0M0 | 3 |
| #CA8 | M | 49 | T2N0M0 | 2 |
| #CA9 | M | 59 | T2N0M0 | 2 |
| #CA10 | M | 74 | T4N0M0 | 1 |
| #CA11 | F | 50 | T2N0M0 | 1 |
| #CA12 | M | 79 | T3N0M0 | 1 |
| #CA13 | M | 73 | T2N0M0 | 1 |
| #CA14 | M | 72 | T4N3M0 | 2 |
| #CA16 | M | 53 | T4N2M0 | 2 |
| #CA17 | M | 61 | T2N0M0 | 1 |
| #CA18 | M | 68 | T3N0M0 | 1 |
| #CA19 | F | 57 | T2N0M0 | 1 |
| #CA20 | M | 57 | T2N0M0 | 2 |
| #CA21 | F | 72 | T2N2M0 | 2 |
| #CA22 | M | 73 | T4N0M0 | 2 |
| #CA24 | M | 63 | T2N0M0 | 2 |
TNM classification18.
Histological grading: 1 – well differentiated; 2 – moderate; 3 – undifferentiated.
Reverse transcription–PCR analysis
Total RNA was extracted from tumor cells with Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA), according to the manufacturer’s recommendations and treated with DNAse (Invitrogen Life Technologies, Carlsbad, CA). First-strand cDNA was prepared from 1 μg of total RNA treated with DNAse, using the Superscript first-strand synthesis system (Invitrogen Life Technologies). After reverse transcription, the cDNA was used as a template for PCR amplification of the human WWOX cDNA. The first and second amplifications were performed with nested primers, as previously described.17 Both reactions were carried out in a volume of 25 μl containing 10 pmol of each primer, 2.5 mM MgCl2, 1.5 mM dNTP mix, 1× PCR buffer, and 1.25 unit of Taq DNA Polymerase, Recombinant (Invitrogen Life Technologies). Amplifications were carried out in a Mastercycler gradient thermocycler (Eppendorf AG) as follows: an initial denaturation for 8 min at 95°C followed by 35 cycles of 94°C for 30 sec, 57°C for 30 sec, 72°C for 1 min, and a final extension for 5 min at 72°C. One microliter of the amplification product from the first reaction was used for the second reaction under the same conditions stated earlier. Glyceraldehyde-3-phosphate dehydrogenase cDNA was amplified as a control for cDNA quality.20 The amplified products were submitted to electrophoresis on a 6.5% polyacrylamide gel, followed by silver staining. DNA bands corresponding to the normal and abnormal size transcripts were purified using the GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Piscataway, NJ), and sequenced on the ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). The 2 primer sets of the second PCR amplification that amplify the whole open reading frame and 2 other primers designed to facilitate sequencing of the open reading frame (WW1FOR 5′-CGGCAAAGATAC GACGGCAG-3′, exon 4 and WW2FOR 5′-ACTTTTGCTCT ACCCTGG-3′, exon 7) were used.
Mutation screening of the WWOX gene
Genomic DNA isolated from the tumors, normal oral mucosas and peripheral blood was used to amplify all exons of the WWOX gene. PCR amplifications were performed using primers previously described.5 The reaction conditions were: 100–300 ng of genomic DNA template; 10 pmol of each primer; 1.5 mM MgCl2, 1.5 mM dNTP mix, 1× PCR buffer and 1.25 unit of Taq DNA Polymerase, Recombinant (Invitrogen Life Technologies) in a 25 μl final volume in a Mastercycler gradient thermocycler (Eppendorf AG, Hamburg). Amplifications were carried out as follows: an initial denaturation for 8 min at 95°C, followed by 35 cycles of 94°C for 30 sec, 57°C for 30 sec, 70°C for 40 sec and a final extension for 5 min at 72°C. All products were analyzed by electrophoresis on a 6.5% polyacrylamide gel, followed by silver staining, purified using the GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences) and sequenced on the ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
Western blot analyses
Total OSCC and normal oral mucosa protein lysates were made using RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% SDS) containing protease inhibitor cocktail (Chemicon International, Temecula, CA). OSCC and normal tissue were washed twice with 1× PBS, homogenized and then lysed by tissue grinder (Kontes, Vineland, NJ) in RIPA buffer. The lysates were centrifuged at 14,000g for 40 min to remove tissue debris. For Western blotting, 50 μg of total protein was separated by 4–15% SDS-PAGE gel and transferred to Hybond-PVDF membrane (Amersham Bioscience). Blots were blocked in PBS buffer with 5% skim milk for 45 min at room temperature. Wwox protein was detected using affinity purified antiWwox polyclonal primary antiserum.21 This antiserum has no detectable crossreactivity with other cellular proteins as demonstrated by immunoblotting of total protein extracts from cell lines containing homozygous deletions of the WWOX gene.21 In addition, preadsorption of the antiserum to a GST fusion protein containing the WWOX WW domains completely eliminated immunohistochemical reactivity. Primary antibody exposure was performed for 2 h at room temperature with rabbit polyclonal anti-Wwox (1:500) and conjugated antirabbit secondary antibody at dilution 1:20,000 (Molecular Probes, Eugene, Oregon). The detection was performed by chemiluminescence ECL plus Kit (Amersham Bioscience). Actin was detected using mouse monoclonal anti-β-actin antibody (Santa Cruz Biotechnology, San Diego, CA) diluted 1:1,000 and conjugated antimouse secondary antibody; 1:2,500 (Molecular Probes, Eugene, OR).
WWOX immunohistochemistry
Tissue sections from OSCC and oral normal mucosa were stained with Wwox antiserum.21 Briefly, 4 μm paraffin-embedded sections were dewaxed in xylene and hydrated with graded ethanol. Endogenous peroxidase activity was blocked with 3% H2O2 in water for 10 min. Heat-induced epitope retrieval was performed with 1 mM EDTA buffer pH 8.0 for 30 min in a steamer at 96°C. Primary polyclonal rabbit antiWwox antiserum (140 μg/ml) was used at a 1:100 dilution (in BSA 0.5%) for 18 h at 4°C. This was followed by incubation with the labeled streptavidin–biotin (LSAB) Kit (DakoCytomation California Inc, Carpinteria, CA). Peroxidase activity was developed with DAB (Sigma, St Louis, MI) with timed monitoring using a positive control sample. The sections were then counterstained with hematoxylin, dehydrated and mounted. Two experienced independent pathologists examined multiple fields and scored tissue sections for extent of staining, regardless of staining intensity (+1, 0–10% positive cells; +2, 11–50% positive cells; +3, greater than 50% positive cells). OSCC scored as +3 was considered as having normal expression to Wwox (oral normal mucosas were all +3).
Results
Aberrant WWOX gene transcripts
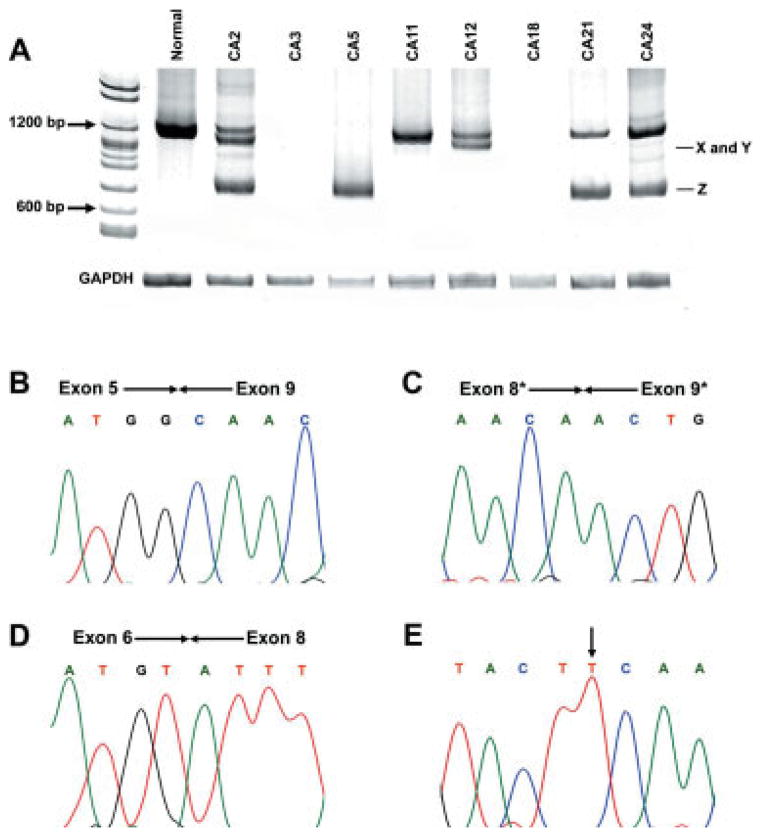
RT-PCR amplification was performed to analyze WWOX mRNA expression. Seven of 20 cases (35%) showed altered or absent expression of WWOX gene. Of these 7 tumors, 5 showed transcripts with total or partial loss of exons. One tumor sample (#CA5) did not exhibit a normal size transcript. Furthermore, in 2 tumor samples (#CA3 and #CA18) no transcript was detected. Both samples were positive for Cytokeratin 19 that was used as a marker of epithelial cells.22 All normal mucosa had normal size transcripts without aberrant transcripts (Fig. 1a). Sequence analysis of the RT-PCR products showed loss of exons 6–8 (#CA2, #CA5, #CA21 and #CA24) (Fig. 1b), and 2 new WWOX transcripts were detected, with loss of exon 7 (#CA2) (Fig. 1d) and partial loss of exons 8–9 (#CA12) (Fig. 1c). Interestingly, tumor #CA2 showed 2 transcript variants, with the loss of exons 6–8 and deletion of exon 7. The remaining tumors showed only normal-size transcript. Representative results of nested RT-PCR analysis are show in Figure 1.
Figure 1.
RT-PCR amplifications of the WWOX cDNA of OSCC. (a) Electrophoresis in polyacrylamide gel 6.5% was used to detect wild-type transcript (1284 bp) and alterations in the WWOX mRNA transcripts. Absence of transcript was seen in 2 tumors (#CA3 and #CA18) as well as 3 different types of aberrant transcripts: X (#CA2), Y (#CA12) and Z (#CA2, #CA5, #CA12, #CA21 and #CA24). (b–d), representative sequences from OSCC showing loss of exons (* represents a partial loss of the exon). (b), #CA5; (c), #CA12; (d) #CA2. (d) frameshift alteration in the open reading frame. (e) somatic mutation (C→T) of tumor #CA12, causing a S329F alteration.
Expression of Wwox protein
To determine whether WWOX aberrant transcripts were translated into protein, we performed Western blot analysis of lysates from all 20 cases of OSCC as well as oral normal mucosa. We detected Wwox using affinity purified polyclonal antiserum raised against Wwox amino acid residues 12–94 that recognizes conserved domains of the Wwox protein.21 Normal oral mucosa samples detected the 46 kDa protein band corresponding to Wwox. Products of the aberrant transcripts present in OSCC (#CA2, #CA5, #CA12, #CA21 and #CA24) were not seen, suggesting that these aberrant transcripts were not translated into protein or, less probably, that the proteins synthesized were being quickly degraded, making them undetectable (Fig. 2).
Figure 2.
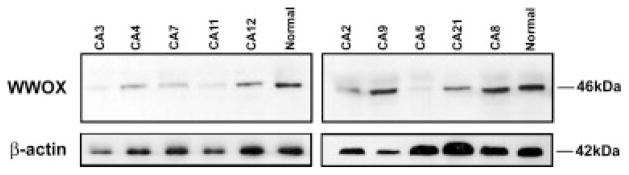
Western blot analysis of Wwox protein in OSCC. Wwox polyclonal antiserum was used to detect WWOX protein in total protein extracts. Only wild-type Wwox protein with 46 kDa was detected.
Wwox expression in normal oral mucosa and OSCC was also confirmed by immunohistochemistry. Epithelial cells in normal oral mucosa displayed a cytoplasmic staining for Wwox protein as shown in Figure 3. All normal tissue showed a score +3 for Wwox (Fig. 3a). Of the 20 OSCC tumors, 8 tumors (40%) showed a reduced expression to Wwox. Five of these (#CA2, #CA10, #CA11, #CA12 and #CA18) were score +2 (Fig. 3c) and 3 (#CA3, #CA5 and #CA16) showed a score +1 (Fig. 3b).
Figure 3.
Immunohistochemical detection of Wwox protein in normal oral mucosa and OSCC. Representative immunostaining results of Wwox protein expression. (a) Normal oral mucosa showing (strong) cytoplasmic staining in all layers of the epithelium (score +3). (b) OSCC representative tumor (score +1) with absence of expression of the Wwox protein (#CA5). (c) OSCC tumor demonstrating a few tumor cells expressing the Wwox protein, score +2 (#CA12) and (d) tumor showing score 3 expression of Wwox protein (#CA24); 400×.
Taken together, the results obtained by RT-PCR and immunohistochemistry showed that the majority of tumors (5/7) with altered transcripts had a marked reduction in the expression of Wwox protein (Table II). Furthermore, tumor #CA5 that had only an aberrant transcript did not stain for Wwox at all (Fig. 3b).
TABLE II.
RT-PCR AND IMMUNOHISTOCHEMISTRY (IH) SUMMARY
| Tumors | RT-PCR1 | Aberrant product | IH score2 |
|---|---|---|---|
| #CA2 | N and aberrant | Exon 6–8 deletion Exon 7 deletion |
+2 |
| #CA3 | A | – | +1 |
| #CA4 | N | – | +3 |
| #CA5 | Aberrant | Exon 6–8 deletion | +1 |
| #CA7 | N | – | +3 |
| #CA8 | N | – | +3 |
| #CA9 | N | – | +3 |
| #CA10 | N | – | +2 |
| #CA11 | N | – | +2 |
| #CA12 | N and aberrant | Exon 8–9 deletion3 | +2 |
| #CA13 | N | – | +3 |
| #CA14 | N | – | +3 |
| #CA16 | N | – | +1 |
| #CA17 | N | – | +3 |
| #CA18 | A | – | +2 |
| #CA19 | N | – | +3 |
| #CA20 | N | – | +3 |
| #CA21 | N and aberrant | Exon 6–8 deletion | +3 |
| #CA22 | N | – | +3 |
| #CA24 | N and aberrant | Exon 6–8 deletion | +3 |
RT-PCR: N, wild type transcript; A, absence of transcript.
Immu-nohistochemistry scores; +1, 0–10% positive cells; +2, 11–50% positive cells; +3, >51% positive cells. Normal mucosas were used as positive controls, all score +3.
Partial exon deletion.
WWOX gene sequence mutation and variants
To determine the presence of somatic or germline mutations of the WWOX gene in OSCC and normal tissue, direct sequencing of PCR products of all coding regions of the WWOX exons was performed in both tissue and peripheral blood from all samples. A unique somatic missense mutation (homozygous) was detected in tumor #CA12. There was a C to T transition at the second nucleotide of codon 329 (exon 8) in the SDR domain, that resulted in the substitution of Serine to Phenylalanine (Fig. 1e). To confirm the somatic mutation, DNA from peripheral blood from this OSCC patient was examined and a wild-type sequence was present (data not shown). Sequencing analyses of all other tumors demonstrated the occurrence of 5 previously described nucleotide variations; C746G, A660G, C969G, C1442T and T1497G.23 None of these alterations affect critical residues of the WW domains or the SDR domain and represent common polymorphisms. In addition, analysis of germline DNA from 30 unrelated subjects was performed to demonstrate that the C329T was not a polymorphism.
Discussion
In this study, molecular alterations of the WWOX gene in 20 OSCC tumors were assessed. Previous reports have described WWOX alterations based either on genomic DNA and RT-PCR or protein expression by immunohistochemistry and Western blot. The present study is the first to analyze a large series of OSCCs and normal oral mucosas, using all of the methods mentioned earlier. We demonstrated that WWOX gene expression is commonly altered in OSCC, suggesting that WWOX is involved in the process of carcinogenesis of the oral squamous epithelium. Analysis of DNA, RNA and protein data showed that WWOX is altered in 50% of OSCC tumors. Among the primary OSCC tumors studied, RT-PCR showed loss or aberrant transcripts in 35%. In addition, mutation analysis revealed one novel missense mutation, located in the SDR domain. To date, only one somatic mutation, also located in exon 8, has been described.17 By immunohistochemistry, the Wwox protein expression was reduced in 40% (8/20). We also provided, by Western blot, evidence that the aberrant transcripts are not translated into protein. The possibility of rapid degradation of the instable product protein must also be considered. This pattern of aberration affecting WWOX is similar to that of the FHIT gene in human tumors24,25 and could be caused by its location at a fragile site in the human genome.26 It has been known for some time that a fragile site replicates late during the cell cycle and common fragile sites are susceptible to and preferentially targeted by the same carcinogenic agents. It is conceivable that breakage at WWOX/FRA16D and FHIT/FRA3B loci can be inflicted concordantly.27,28
In one tumor (#CA18), immunohistochemistry showed moderate (score +2) expression, but no transcript was detected by RT-PCR. As the analysis at the molecular level and immunohistochemical analysis were done on separate pieces of tumor, strong (score +3) protein expression in 2 cases (#CA21, #CA24) with loss of exons 6–8 might be explained by the occurrence of heterogeneous expression. Three cases with reduced or missing protein expression (#CA10, #CA11 and #CA16) did not exhibit any abnormality at genetic level, so besides heterogeneous expression, the possibility of epigenetic abnormalities such as WWOX methylation can be contemplated.29,30 In addition, the presence of alterations in mRNA transcription correlated with the reduced expression of Wwox protein in 5 cases (#CA2, #CA3, #CA5, #CA12 and #CA18).
In summary, in this report, we demonstrate a reduction in the expression of WWOX gene in 50% of the OSCC tumors compared with the normal mucosa as well as a novel mutation of the WWOX gene. These results show that the WWOX gene is frequently altered in OSCC and contributes to the carcinogenesis processes in oral cancer.
Acknowledgments
Grant sponsor: CNPq, PRONEX and FAPEMIG, Brazil; Grant sponsor: NCI RO1 CA102444.
References
- 1.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of eighteen major cancers. Int J Cancer. 1993;54:594–606. doi: 10.1002/ijc.2910540413. [DOI] [PubMed] [Google Scholar]
- 2.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, Dermody JJ. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Kresty LA, Mallery SR, Knobloch TJ, Song H, Lloyd M, Casto BC, Weghorst CM. Alterations of p16INK4a and p14ARF in patients with severe oral epithelial dysplasia. Cancer Res. 2002;62:5295–300. [PubMed] [Google Scholar]
- 4.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, Baker E, Richards RI. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651–63. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 5.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24. 1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–5. [PubMed] [Google Scholar]
- 6.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–73. [PubMed] [Google Scholar]
- 7.Croce CM, Sozzi G, Huebner K. Role of FHIT in human cancer. J Clin Oncol. 1999;17:1618–24. doi: 10.1200/JCO.1999.17.5.1618. [DOI] [PubMed] [Google Scholar]
- 8.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 9.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM. The FHIT gene, spanning the chromosome 3p14. 2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–97. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 10.Mineta H, Miura K, Takebayashi S, Misawa K, Ueda Y, Suzuki I, Ito M, Wennerberg J. Low expression of fragile histidine triad gene correlates with high proliferation in head and neck squamous cell carcinoma. Oral Oncol. 2002;39:56–63. doi: 10.1016/s1368-8375(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 11.Tai SK, Lee JI, Ang KK, El-Naggar AK, Hassan KA, Liu D, Lee JJ, Ren H, Hong WK, Mao L. Loss of Fhit expression in head and neck squamous cell carcinoma and its potential clinical implication. Clin Cancer Res. 2004;10:5554–7. doi: 10.1158/1078-0432.CCR-04-0208. [DOI] [PubMed] [Google Scholar]
- 12.Richards RI. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–45. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 13.Sozzi G, Sard L, De Gregorio L, Marchetti A, Musso K, Buttitta F, Tornielli S, Pellegrini S, Veronese ML, Manenti G, Incarbone M, Chella A, et al. Association between cigarette smoking and FHIT gene alterations in lung cancer. Cancer Res. 1997;57:2121–3. [PubMed] [Google Scholar]
- 14.Mori M, Mimori K, Shiraishi T, Alder H, Inoue H, Tanaka Y, Sugimachi K, Huebner K, Croce CM. Altered expression of FHIT in carcinoma and precarcinomatous lesions of the esophagus. Cancer Res. 2000;60:1177–82. [PubMed] [Google Scholar]
- 15.Stein CK, Glover TW, Palmer JL, Glisson BS. Direct correlation between FRA3B expression and cigarette smoking. Genes Chromosomes Cancer. 2002;34:333–40. doi: 10.1002/gcc.10061. [DOI] [PubMed] [Google Scholar]
- 16.Canto MT, Devesa SS. Oral cavity and pharynx cancer incidence rates in the United States, 1975–1998. Oral Oncol. 2002;38:610–7. doi: 10.1016/s1368-8375(01)00109-9. [DOI] [PubMed] [Google Scholar]
- 17.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, Croce CM. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62:2258–60. [PubMed] [Google Scholar]
- 18.Greene FL, Page DL, Fritz A, Balch CM, Haller DG, Marrow M, editors. AJCC cancer staging manual. 6. New York: Springer-Verlag; 2002. [Google Scholar]
- 19.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheimvan Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier JA, Voulalas P, Roeder D, Maciag T. Extension of the life-span of human endothelial cells by an interleukin-1 α antisense oligomer. Science. 1990;249:1570–4. doi: 10.1126/science.2218499. [DOI] [PubMed] [Google Scholar]
- 21.Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford M, Aldaz CM. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet Genome Res. 2003;100:101–10. doi: 10.1159/000072844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aihara T, Noguchi S, Ishikawa O, Furukawa H, Hiratsuka M, Ohigashi H, Nakamori S, Monden M, Imaoka S. Detection of pancreatic and gastric cancer cells in peripheral and portal blood by amplification of keratin 19 mRNA with reverse transcriptase-polymerase chain reaction. Int J Cancer. 1997;72:408–11. doi: 10.1002/(sici)1097-0215(19970729)72:3<408::aid-ijc6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 23.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JE. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98:11417–22. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, Huebner K, Edmonds P, Croce CM. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res. 2004;10:3053–8. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 25.Ishii H, Vecchione A, Furukawa Y, Sutheesophon K, Han SY, Druck T, Kuroki T, Trapasso F, Nishimura M, Saito Y, Ozawa K, Croce CM, et al. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res. 2003;1:940–7. [PubMed] [Google Scholar]
- 26.Yendamuri S, Kuroki T, Trapasso F, Henry AC, Dumon KR, Huebner K, Williams NN, Kaiser LR, Croce CM. WW domain containing oxidoredutase gene expression is altered in non-small cell lung cancer. Cancer Res. 2003;63:878–81. [PubMed] [Google Scholar]
- 27.Park SW, Ludes-Meyers J, Zimonjic DB, Durkin ME, Popescu NC, Aldaz CM. Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br J Cancer. 2004;91:753–9. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guler G, Uner A, Guler N, Han S, Iliopoulos D, Hauck WW, McCue P, Huebner K. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100:1605–14. doi: 10.1002/cncr.20137. [DOI] [PubMed] [Google Scholar]
- 29.Kuroki T, Yendamuri S, Trapasso F, Matsuyama A, Aqeilan RI, Alder H, Rattan S, Cesari R, Nolli ML, Williams NN, Mori M, Kanematsu T, et al. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin Cancer Res. 2004;10:2459–65. doi: 10.1158/1078-0432.ccr-03-0096. [DOI] [PubMed] [Google Scholar]
- 30.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Fragile genes as bio-markers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–33. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]