Summary
WWOX is a cancer gene, spanning the common chromosomal fragile site 16D. Genomic and expression aberrations affecting this gene and locus are common in various neoplasias including breast cancer. The aim of the present study was to evaluate the relationship between WWOX expression at the protein level with respect to clinico-pathological characteristics. We performed immunohistochemical analyses on breast specific tissue microarrays representing, human normal breast epithelium (n=16), ductal carcinoma in situ (n=15) and invasive breast cancer cases (n=203). Staining intensity measurements were objectively determined utilizing an image analysis system. Western blot analyses were also performed on an independent set of 23 invasive breast carcinomas. All normal breast epithelial samples express WWOX protein abundantly while 34% (69/203 cases) of invasive breast carcinomas were ‘completely negative’ for WWOX expression and an additional 26% (52/203) of cases expressed WWOX very weakly. For DCIS samples five out of 15 (33%) were negative or weak for WWOX staining. Interestingly, we found a statistically significant correlation between WWOX expression and estrogen receptor (ER) status, 27% of ER+ breast carcinomas were completely negative for WWOX expression versus 46% for ER−cases ( p = 0.0054). Furthermore, when negative plus weakly WWOX stained cases were considered the difference became more significant with 51% of ER+ cases and 73% for the ER − group, with a p=0.003. These data indicate that loss of WWOX expression is a common event in breast cancer. It is unclear at this point whether loss of WWOX expression is a consequence of tumor progression or represents a subclass of breast carcinomas. The strong association of WWOX expression with ER status reinforces the suggested role of this protein as an enzyme involved in sex steroid metabolism.
Keywords: breast cancer, estrogen receptor, FRA16D, tumor suppressor, WWOX
Introduction
Genomic and gross chromosomal abnormalities in breast cancer affecting the long arm of chromosome 16 have been frequently reported over the years [1–4]. It was further observed that allelic losses affecting various regions of 16q already occur at pre-invasive stages of carcinoma development [4–6]. In studies from our laboratory we identified the ch16q23.1–24.1 region as having the highest incidence of allelic losses in DCIS lesions [6]. We followed up these studies by the cloning of WWOX, the gene spanning the second most common chromosomal fragile site, FRA16D [7]. Since then, abnormalities affecting WWOX at the genomic and expression level were reported in numerous neoplasias including, breast, ovarian, esophageal, liver and other carcinomas and leukemias [7–10]. We also observed that ectopic WWOX expression was able to inhibit anchorage independent growth and in vivo tumorigenicity of highly aggressive breast carcinoma lines, suggesting a putative tumor supressor role for this novel protein [11].
WWOX encodes a 46 kD protein that contains two WW domains and a short chain oxidoreductase (SDR) domain. Little is known of the biochemical function(s) of WWOX. The SDR domain is predicted to be involved in sex-steroid metabolism and the WW domains are likely involved in protein-protein interactions [7, 12, 13].
In this report we analyzed WWOX protein expression levels by means of Western blot and tissue microarray (TMA) immnuhistochemistry analyses on breast specimens correlating with clinico-pathological features.
Material and methods
Breast tissue microarrays
Breast TMAs were obtained from the Fox Chase Cancer Center and a Breast Progression TMA from the Cooperative Breast Cancer Tissue Resource. By pooling both TMAs we were able to analyze a total of 234 cases including 16 normal breast tissues (10 normal tissues from individuals without breast cancer and six normal tissues adjacent to invasive breast cancers), 203 primary invasive ductal carcinoma (IDCA) specimens and 15 DCIS cases. The histologic grade considered applied only to the invasive component of the tumor. Grade was determined by the Elston and Ellis approach [14] to the Scarff Bloom Richardson method [15]. Stage at time of diagnosis was based on the TNM classification (American Joint Committee on Cancer, 1992). The breakdown of cases analyzed in terms of TNM staging and nuclear grade is shown in Table 1. Estrogen receptor (ER) status was available for all 203 IDCA cases. Progesterone receptor (PR) status was not available for 16 of those 203 cases.
Table 1.
Clinico-pathological characteristics of cases studied
| Characteristics | Cases |
|---|---|
| TNM status | |
| T1-3 N0 M0 | 62 |
| T1-3 N1-3 M0 | 81 |
| T1-3 N1-3 M1 | 60 |
| Nuclear grade | |
| Grade I | 34 |
| Grade II | 97 |
| Grade III | 72 |
| Total number of IDCA cases | 203 |
Immunostaining method
Prior to anti-WWOX immunostaining, endogenous per-oxidase activity was blocked with 3% H2O2 in water for 10 min. Heat-induced epitope retrieval was performed with 1.0 mM EDTA Buffer pH 8.0 for 10 min in a microwave oven followed by a 20 min cool down. In order to block non-specific antibody binding, tissue sections were incubated with 10% goat serum in PBS for 30 min. Primary polyclonal rabbit WWOX antibody (140 μg/ml) was used at a 1:50 dilution overnight at 4 °C. This was followed by incubation with Envision plus labeled polymer, anti-rabbit-HRP antibody (Dako) for 30 min at room temperature. Staining development was performed with DAB with timed monitoring using a positive control sample. The slides were then counterstained with hematoxylin, dehydrated, cleared and mounted.
Evaluation of anti-WWOX immunohistochemical staining
Anti-WWOX staining intensity was measured using a Chromavision Automated Cellular Imaging System (ACIS®). For performing such measurements we utilized the generic DAB software application provided by ACIS®. The area of each individual TMA core was considered for the measurements in its totality. The software determines brown intensity (i.e. positive stained cells) regardless of the area covered by the positive cells. The cutoff to establish positive and negative staining was determined to be at a mean intensity of 63 (arbitrary units) over a total of 255 (color saturation scale). Therefore, cores with values ≤63 were ‘completely negative’ for WWOX immunostaining, no brown detected. Between 63 and ≤70 values were determined to fall in the ‘weak’ staining category. Cores with staining intensity values >70 were considered to fall in the ‘strong’ staining category.
Western blot analysis
Total protein extracts were prepared from a set of 23 human breast IDCA samples obtained from the cooperative human tissue network (CHTN). As normal controls we used protein extracts from human breast epithelial organoids, obtained from mammoplasty specimens (CHTN). As negative control we used protein extracts from the ovarian cell line PEO1, known not to express WWOX due to a homozygous deletion affecting this gene (8). Total cell protein lysates were made from frozen tissues using RIPA buffer (50 mM Tris pH7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% SDS) containing protease inhibitor cocktail (Roche, Mannheim, Germany). For western blotting 50 μg of total protein was separated by 12.5% SDS–PAGE and transferred to PVDF membranes (Millipore, Billerica, MA). Immunodetection was performed using Protein Detector™ (KPL, Gaithersburg, MD) western blotting reagents as described by the manufacturer. WWOX protein was detected using affinity purified anti-WWOX polyclonal primary antibodies (280 ng/ml) (12) and HRP conjugated anti-rabbit secondary antibody (KPL, 1:2000) followed by chemiluminescence autoradiography. Actin was detected using monoclonal anti-actin antibody (ICN biomedicals, Burlingame, CA, 1:1000) and HRP conjugated anti-mouse secondary antibody (KPL, 1:5000). Quantitation of western blot autoradiographs was done using a Kodak digital science Image Station 440CF.
Statistical methods
Univariate analysis of clinical–pathological parameters based on WWOX staining was determined by χ2 test with Yates’ correction. Ordinal-by-ordinal association was assessed by Kendall’s tau-b test. Pearson’s test was used to analyze the correlation between WWOX staining intensity and number of lymph node status and patient age at diagnosis for DCIS and IDCA lesions. The basic significance level was fixed at p < 0.05 and all data was analyzed using SPSS® statistical software (Version 11.0; SPSS Inc., Chicago, IL).
Results
Loss of WWOX expression in pre-invasive and invasive breast cancer
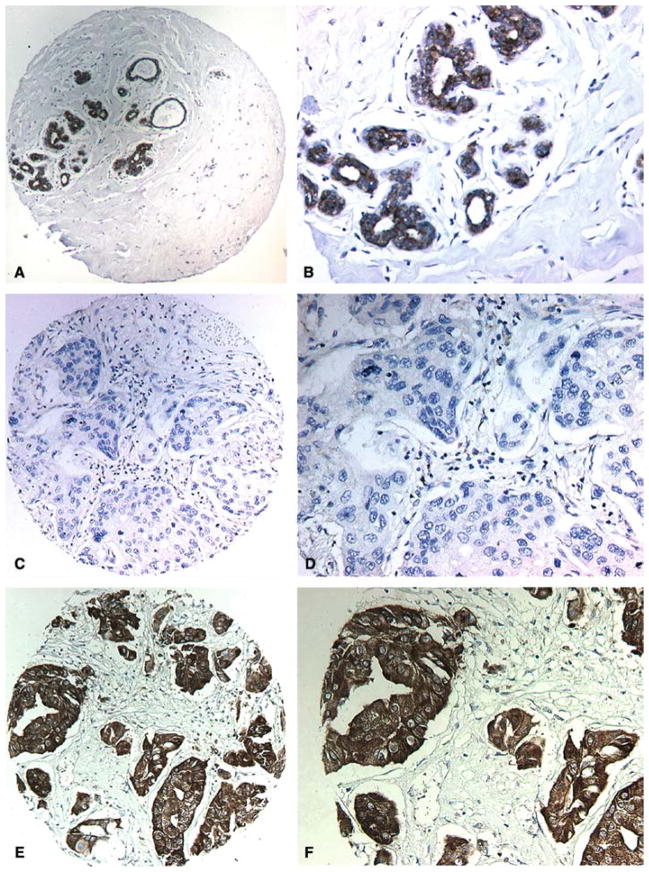
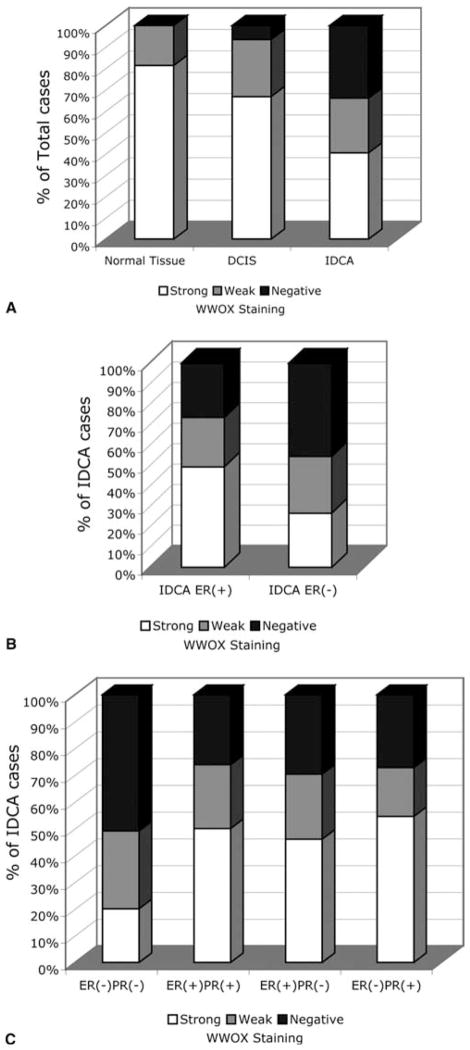
WWOX staining was detected in all normal mammary epithelial samples (16 out of 16). The pattern of WWOX cytoplasmic staining was uniformly strong both for luminal as well as myoepithelial cells. Only three samples stained weaker for WWOX than the rest, i.e. 81% (13 cases) of normal samples stained very strongly indicating high levels of WWOX expression, a representative example is shown in Figure 1A and B. Among DCIS samples strong WWOX staining was detected in 10 out of 15 cases (67%) while five samples lack or had very weak WWOX staining (33%). A total of 69 out of 203 (34%) breast invasive ductal carcinoma cases were ‘completely negative’ for WWOX staining and 52 (26%) additional cases displayed only very weak WWOX staining, the remaining IDCA samples stained quite strongly for WWOX (representative microphotographs are shown in Figure 1C–F). The observed decrease or increase of WWOX staining was uniform to all tumor epithelial cells, showing a homogeneous and diffuse cytoplasmic pattern of protein expression. In summary, a total of 121 IDCA cases (i.e. 60% of 203) demonstrated loss or reduced levels of WWOX staining when compared with normal samples (p=0.003). These data are illustrated in Figure 2 and the statistical significant differences are indicated in Table 2. As can be observed, both in Figure 2A and Table 2, there is a trend for the reduction of WWOX expression from in situ carcinoma lesions to the more advanced fully invasive lesions. However, no statistically significant associations were detected between levels of WWOX expression and other predictors of tumor aggressiveness such as lymph node status, metastasis status, clinical staging or nuclear grade. In addition, no association was found with age of the patient at time of diagnosis.
Figure 1.
WWOX immunohistochemical staining in normal and breast cancer tumor samples. (A) Representative low power photomicrograph from a TMA core of normal resting breast displaying strong staining in epithelial cells. (B) High power photomicrograph showing WWOX inmunostaining localizing to the cytoplasm of luminal epithelial cells. (C) TMA core representing IDCA completely negative for WWOX staining. (D) High power image of C. (E) TMA core representing IDCA displaying strong WWOX inmunostaining. (F) High power photomicrograph showing in detail strong cytoplasmic inmunostaining in nests and cords of epithelial malignant cells.
Figure 2.
(A) Distribution of total cases (normal and tumor) according to WWOX expression levels. (B) Distribution of IDCA cases according to WWOX expression level and ER status. (C) WWOX staining in IDCA according to ER–PR status.
Table 2.
WWOX staining according to histopathological characteristics and ER–PR status
| Histopathological characteristics | WWOX staining
|
Total | p-Value | ||
|---|---|---|---|---|---|
| Negative | Weak | Strong | |||
| Histology | |||||
| Normal tissue | 0 (0%) | 3 (18.7%) | 13 (81.3%) | 16 | |
| DCIS | 1 (6.6%) | 4 (26.7%) | 10 (66.7%) | 15 | |
| IDCA | 69 (34%) | 52 (25.6%) | 82 (40.4%) | 203 | p = 0.003a |
| IDCA ER status | |||||
| ER (−) | 36 (45.6%) | 22 (27.8%) | 21 (26.6%) | 79 | |
| ER (+) | 33 (26.6%) | 30 (24.2%) | 61 (49.2%) | 124 | p = 0.003 |
| IDCA ER/PR status* | |||||
| ER(−) PR(−) | 28 (50.9%) | 16 (29.1%) | 11 (20%) | 55 | |
| ER(+) PR(+) | 22 (26.2%) | 20 (23.8%) | 42 (50%) | 84 | |
| ER(+) PR(−) | 11 (29.7%) | 9 (24.3%) | 17 (46%) | 37 | p = 0.013b |
| ER(−) PR(+) | 3 (27.3%) | 2 (18.2%) | 6 (54.5%) | 11 | |
Not included in this four way analysis 16 cases in which PR status was unavailable.
p-Value refers to difference between Normal and IDCA.
p-Value refers to difference between ER(−) PR(−) group versus the three other groups.
It is worth reporting that in every normal and tumor sample analyzed by immunostaining in which WWOX expression was detected, the staining was always cytoplasmic, not a single case out of the 235 samples analyzed showed nuclear WWOX staining. These observations are in sharp contrast with a previous report in which WWOX cytoplasmic and nuclear staining was observed in some cells from normal mammary epithelium [16]. The reason for this discrepancy is unclear but in that report the number of normal cases analyzed was not indicated and only one microphotograph of one mammary lobular structure was shown with the majority of epithelial cells displaying only cytoplasmic staining.
WWOX expression correlates with ER status
When WWOX staining and ER status were considered we observed an interesting statistically significant correlation between levels of WWOX expression and ER status, 27% of ER+ breast carcinomas were completely negative for WWOX expression versus 46% for ER− cases ( p=0.0054) (Table 2). This correlation was even more significant when the WWOX negative category plus the weakly stained category were considered as a single group compared to the strongly staining category. In this case the sum of both categories increased to 73% of cases (i.e. negative + weak) for the ER− group and 51% for the ER+ group respectively (p=0.003) (Figure 2B, Table 2).
We also performed the same type of analysis but now incorporating the PR status, thus determining four groups due to all possible combination on the status of both receptors. The ER− PR− group showed the highest number of WWOX negative cases amounting to 51% (28 out of 55), increasing to a dramatic 80% of cases (44 out of 55) if we include the weak-staining category. This group was significantly different from the three other groups with a p=0.013 (Figure 2C, Table 2). Interestingly, the ER− PR+ group did not behave statistically differently than the ER+ groups. However the small number of samples of this specific group (11 cases) did not allow us to draw strong conclusions on this observation (Figure 2C). Representative microphotographs illustrating the association of WWOX expression with ER expression are shown in Figure 3.
Figure 3.
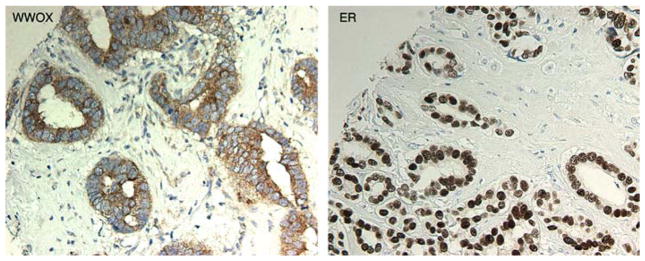
WWOX expression in association with ER+ expression. (Left panel). Photomicrograph showing positive WWOX staining in a breast carcinoma. (Right panel). Photomicrograph of the same tumor shown on the (left panel) displaying ER+ nuclear staining in 100% of cells.
Western blot analysis of WWOX expression in invasive breast carcinomas
Western blot analyses were performed using protein extracts obtained from 23 invasive ductal breast carcinoma samples, as control normal epithelial organoids were isolated from normal mammoplasty specimens. The normal epithelial organoids express very high levels of WWOX protein as can be observed in Figure 4, samples B30, B32 and B42. This is consistent with the immunohistochemical observations described above (Figure 1A and B). Breast carcinomas demonstrated variable levels of WWOX expression with some tumors expressing significant amounts of WWOX protein, e.g. T82 in Figure 4, while other tumors displayed barely detectable, if any, WWOX protein. The relative intensity of each reactive WWOX band was normalized to Actin, used as loading control. The values obtained were in turn expressed relative to the levels detected in the breast epithelial organoids. We concluded that 20 out of the 23 tumors (87%) expressed WWOX protein at levels lower than 50% that of WWOX protein in the normal mammary epithelial organoid preparations. This analysis is in agreement with the immunohistochemical studies, indicating that a high proportion of breast carcinomas expressed significantly reduced levels of WWOX protein. Importantly, the western blot analysis also demonstrates that the antibody used in our studies detects only full-length WWOX protein in both normal and tumors.
Figure 4.
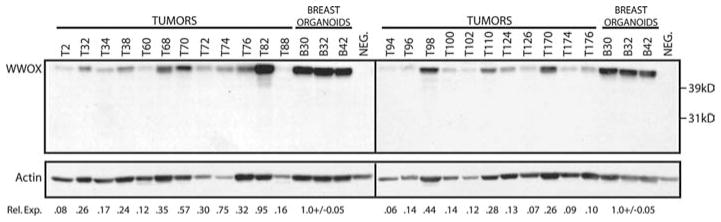
WWOX protein levels in normal and tumor breast tissue. Immunodetection of WWOX in human tissue extracts. Human breast protein extracts were separated by 12.5% SDS–PAGE and transferred to PVDF membranes. WWOX protein was detected using affinity purified rabbit polyclonal anti-WWOX antibody. The membrane was then probed with mouse monoclonal anti-actin antibody for normalization of differences in protein loading. Three normal human protein extracts prepared from breast organoids were included on each gel. WWOX protein levels in 23 breast tumors showed significant differences compared to normal breast WWOX levels. The full-length WWOX protein is indicated. NEG. – negative control extract (PEO1 cell line) that does not express WWOX protein. Rel. Exp.=Relative WWOX Expression. WWOX expression in each sample was first normalized to actin to correct for loading differences. WWOX expression in tumors relative to WWOX levels in normal organoids was then determined using the actin normalized values.
Discussion
In the original report describing the cloning of WWOX, we observed that expression of this gene at the RNA level was highly variable among breast carcinoma lines. Some breast cancer lines showed no expression of the WWOX transcript while other lines over expressed WWOX as detected by Northern and RT-PCR analyses [7, 11]. Interestingly the breast cancer line that expresses by far the highest levels of WWOX mRNA and protein was the ER+ breast cancer line MCF7. The T47D ER+ breast cancer line also expresses WWOX protein, while the ER− and aggressive lines such as MDA435 and MDA231 showed practically undectable levels of this protein [12]. This early data already appeared to indicate a possible association between WWOX expression and ER status.
We also previously reported that WWOX is highly expressed in organs with high production of sex steroid hormones such as testis, prostate and ovaries [7]. In our original analysis of the short chain dehydrogenase domain of WWOX, we observed that this oxidoreductase enzyme displays a putative signature suggestive of function, which is a serine residue 12 amino acids upstream of the YNRSK substrate-binding motif. This serine is at a nearly identical location to that observed in steroid dehydrogenases (usually position −13 from Tyr), which is suggested to play an important role in the catalysis of steroid substrates [17]. Based on these observations we speculated that WWOX might play a role in sex-steroid metabolism.
In this study we report a striking statistically significant correlation between WWOX expression and ER status in breast carcinomas. A higher proportion of ER+ breast carcinomas express more WWOX than the ER− counterparts. In our studies, due to the limited number of ER− PR+ breast carcinoma samples, we could not rule out that PR+ status alone correlates to high WWOX expression.
These findings further strengthen the hypothesis that WWOX plays a role in sex-steroid metabolism and very likely functions in a pathway or pathways associated with the estrogen–progesterone axis. Interestingly, it has been demonstrated that estradiol concentrations are much higher in breast carcinomas than in normal breast tissue and approximately 50 to 100-fold higher than circulating levels of this hormone in post-menopausal women. It is also known that for the most part the higher tumor estradiol levels are the result of in situ synthesis [Reviewed in 18]. Perhaps the higher levels of WWOX protein found mostly in ER+ tumors and estradiol dependent breast cancer cell lines is somehow associated to the aforementioned tumor in situ estradiol synthesis phenomenon.
WWOX was originally cloned due to its mapping to a locus affected by a high frequency of gross genomic rearrangements including, losses of heterozygosity as a consequence of large deletions, recombination or chromosomal translocations [7]. However other investigators have shown in leukemias that epigenetic mechanisms could also play a role in loss of expression of this protein [10].
This locus was identified to be that of the second most common chromosomal fragile site, FRA16D [7, 19, 20]. Hemi and homozygous genomic losses have been traditionally considered a hallmark of tumor suppressor activity at the loci of origin. In spite of the fact that the majority of currently existent breast cancer lines are hemizygous for WWOX we failed to identify mutations in the remaining active alleles, we only observed homozygous losses affecting intronic regions and lack of expression in aggressive breast cancer lines [7, 12]. However, homozygous losses affecting WWOX were later demonstrated in ovarian, lung carcinoma cell lines and other tumor cell types [8]. We demonstrated that when ectopically expressed in aggressive breast cancer lines, WWOX behaves as an inhibitor of anchorage independent growth and of in vivo tumorigenesis [11]. Thus, WWOX could also be classified as a ‘putative’ tumor suppressor gene.
In this report we demonstrate that WWOX is abundantly expressed in normal human breast epithelium both by immunohistochemistry and Western analysis. In agreement with the behavior of classical tumor suppressor genes, we observed a complete loss of expression in over a third (34%) of invasive breast carcinomas. This percentage increases up to 60% of cases when we include carcinomas with very weak WWOX expression. We also determined that reduced WWOX expression is also observed in a third of preinvasive lesions (DCIS).
A main question arising from these studies is whether the loss of WWOX expression is causatively associated with tumor development or a consequence of tumor progression. The fact that we find a tendency for lower WWOX expression in invasive lesions when compared to DCIS makes the second alternative more likely (i.e. expression loss occurring during tumor progression). However, the fact that 80% of ER negative cases demonstrate loss or reduced expression of WWOX also raises the possibility that at least some of those tumors may represent a subclass of lesions in which WWOX loss could be a causative event. While this manuscript was in preparation Guler et al. [21] reported similar observations of WWOX protein expression in a study of 97 breast carcinomas. It is still an issue of controversy whether ER− status simply represents a stage of progression or that subsets of breast carcinomas originate as ER− negative lesions.
In summary, two main conclusions can be drawn from these studies, first that loss of WWOX expression is a frequent event in breast cancer and second that a strong correlation with WWOX expression and ER status is observed. Further studies will be necessary to elucidate whether loss of WWOX expression is an early causative event at least in a subset of tumors, or whether this event predominantly occurs during breast carcinoma progression.
Acknowledgments
Studies supported by NIH-NCI RO1 CA102444.
References
- 1.Dutrillaux B, Gerbault-Seureau M, Zafrani B. Characterization of chromosomal anomalies in human breast cancer. A comparison of 30 paradiploid cases with few chromosome changes. Cancer Genet Cytogenet. 1990;49:203–217. doi: 10.1016/0165-4608(90)90143-x. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, Tanigami A, Yamakawa K, Akiyama F, Kasumi F, Sakamoto G, Nakamura Y. Allelotype of breast cancer: cumulative allele losses promote tumor progression in primary breast cancer. Cancer Res. 1990;50:7184–7189. [PubMed] [Google Scholar]
- 3.Tsuda H, Callen D, Fukutomi T, Nakamura Y, Hirohashi S. Allele loss on chromosome 16q24..2-qter occurs frequently in breast cancer irrespectively of differences in phenotype and extent of spread. Cancer Res. 1994;54:513–517. [PubMed] [Google Scholar]
- 4.Aldaz CM, Chen T, Sahin A, Cunningham J, Bondy M. Comparative allelotype of in situ and invasive human breast cancer: high frequency of microsatellite instability in lobular breast carcinomas. Cancer Res. 1995;55:3976–3981. [PubMed] [Google Scholar]
- 5.Radford D, Fair K, Phillips N, Ritter J, Steinbrueck T, Holt M, Donis-Keller H. Allelotyping of ductal carcinoma in situ of the breast: deletion of loci on 8p, 13q, 16q, 17p, and 17q. Cancer Res. 1995;55:3399–3405. [PubMed] [Google Scholar]
- 6.Chen T, Sahin A, Aldaz C. Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res. 1996;56:5605–5609. [PubMed] [Google Scholar]
- 7.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 8.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JE. WWOX, a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, Croce CM. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62:2258–2260. [PubMed] [Google Scholar]
- 10.Ishii H, Vecchione A, Furukawa Y, Sutheesophon K, Han SY, Druck T, Kuroki T, Trapasso F, Nishimura M, Saito Y, Ozawa K, Croce CM, Huebner K. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res. 2003;1:940–947. [PubMed] [Google Scholar]
- 11.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 12.Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford M, Aldaz CM. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet Genome Res. 2003;100:101–110. doi: 10.1159/000072844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene. 2004;23:5049–5055. doi: 10.1038/sj.onc.1207680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloom HJG, Richardson WW. Histological grading and prognosis in breast cancer. A study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer. 1957;11:359–377. doi: 10.1038/bjc.1957.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. The value of histological grade in breast cancer. Experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe A, Hippo Y, Taniguchi H, Iwanari H, Yashiro M, Hirakawa K, Kodama T, Aburatani H. An opposing view on WWOX protein function as a tumor suppressor. Cancer Res. 2003;63:8629–8633. [PubMed] [Google Scholar]
- 17.Duax WL, Ghosh D. Structure and function of steroid dehydrogenases involved in hypertension, fertility, and cancer. Steroids. 1997;62:95–100. doi: 10.1016/s0039-128x(96)00166-3. [DOI] [PubMed] [Google Scholar]
- 18.Purohit A, Newman SP, Reed MJ. The role of cytokines in regulating estrogen synthesis: implications for the etiology of breast cancer. Breast Cancer Res. 2002;4:65–69. doi: 10.1186/bcr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paige AJ, Taylor KJ, Stewart A, Sgouros JG, Gabra H, Sellar GC, Smyth JF, Porteous DJ, Watson JE. A 700-kb physical map of a region of 16q23.2 homozygously deleted in multiple cancers and spanning the common fragile site FRA16D. Cancer Res. 2000;60:1690– 1697. [PubMed] [Google Scholar]
- 20.Krummel KA, Roberts LR, Kawakami M, Glover TW, Smith DI. The characterization of the common fragile site FRA16D and its involvement in multiple myeloma translocations. Genomics. 2000;69:37–46. doi: 10.1006/geno.2000.6321. [DOI] [PubMed] [Google Scholar]
- 21.Guler G, Uner A, Guler N, Han SY, Iliopoulos D, Hauck WW, McCue P, Huebner K. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100(8):1605–1614. doi: 10.1002/cncr.20137. [DOI] [PubMed] [Google Scholar]