Abstract
Although the water-soluble metabolite profile of human mesenchymal stem cells is known, the lipid profile still needs further investigation. In this study, methanol-chloroform was used to extract pid-soluble metabolites and perchloric acid was used to extract water-soluble metabolites. Furthermore, a dual phase extraction method using methanol-chloroform and water was used to obtain both water and lipid fractions simultaneously. All metabolite extractions were analyzed on a 9.4T high-resolution nuclear magnetic resonance spectrometer. Metabolite resonance peaks were assigned in the acquired spectra according to the chemical shift, and the extraction efficiency of ferent methods was compared. Results showed that in the spectra of water-soluble extracts, major metabolites comprised low molecular weight metabolites, including lactate, acetic acid, fatty acids, threonine, glutamic acid, creatine, choline and its derivatives, while in the spectra of lipid-soluble extracts, most metabolites were assigned to fatty acids. Among the different extraction procedures, perchloric acid was more efficient in extracting water-soluble metabolites and methanol-chloroform was efficient in extracting organic components compared with the dual phase extraction method. Nuclear magnetic resonance spectroscopy showed that as low as 0.7 mg organic yield was enough to obtain clear resonance peaks, while about 6.0 mg water-soluble yield was needed to obtain relatively favorable spectral lines. These results show that the efficiency of extracting water and lipid fractions is higher using perchloric acid and methanol-chloroform compared with dual phase extraction and that nuclear magnetic resonance spectroscopy is highly sensitive for analyzing lipid-soluble extracts.
Keywords: neural regeneration, nuclear magnetic resonance spectroscopy, mesenchymal stem cells, metabolite profiles, extraction method, optimization, water-soluble, lipid-soluble, perchloric acid, methanol-chloroform, grants-supported paper, neuroregeneration
Research Highlights
(1) The water-soluble metabolite profile of human mesenchymal stem cells is known, but the lipid profile remains unclear. This study utilized 9.4T high-resolution nuclear magnetic resonance spectroscopy to analyze water- and lipid-soluble metabolite profiles of mesenchymal stem cells.
(2) Nuclear magnetic resonance spectroscopy showed that as low as 0.7 mg organic yield was ficient to obtain clear resonance peaks, while approximately 6.0 mg water-soluble yield was required to obtain relatively favorable spectral lines.
(3) The results demonstrate that the efficiency of extracting water- and lipid-soluble fractions is higher using perchloric acid and methanol-chloroform compared with dual phase extraction.
INTRODUCTION
Mesenchymal stem cells are pluripotent progenitors that can differentiate into a lineage of specialized cell types under appropriate culture conditions[1]. The observation that stem cells transplanted into injured or diseased organs can restore function has provoked great interest in stem cell study. Human umbilical cord-derived mesenchymal stem cells help to avoid ethical issues and offer great prospects for regenerative medicine and cell therapy[2,3]. To monitor molecular and cellular changes in mesenchymal stem cells, a full understanding of its metabolite profile is necessary. The newly-developed technique of nuclear magnetic resonance spectroscopy provides an accurate and non-invasive way to monitor metabolic changes over time, both in vivo and in vitro [4]. There are two kinds of cell metabolites based on their solubility and extraction solvents used; water-soluble and lipid-soluble metabolites. The water-soluble metabolite profile of mesenchymal stem cells has been elucidated; however, little is known about its lipid-soluble metabolite profile[5,6]. In studying cell metabolites, cell extraction is an ideal way for biochemical analysis and quantification. An optimal extraction method should be of high efficiency and low variability. The classic perchloric acid method fulfills this requirement in obtaining water-soluble metabolites, and the methanol-chloroform method can successfully extract lipid-soluble metabolites[7,8]. More recently, Bligh and Dye developed a dual phase extraction procedure which can obtain both fractions[9,10]. The main advantage of the dual phase extraction procedure is that it enables simultaneous extraction of lipids and aqueous metabolites from a single cell sample, with a high efficiency in extracting water-soluble metabolites[11,12]. However, it remains unknown whether the dual phase extraction method is superior in extracting lipid-soluble metabolites, or whether it can reduce the quantity of cells required for extraction to minimize cell culture.
In the present study, we first sought to provide a more detailed understanding of metabolic profiles by analyzing both water- and lipid-soluble extracts of human mesenchymal stem cells. Second, we compared the extraction efficiencies of perchloric acid, methanol-chloroform and the dual phase extraction methods by calculating the net wet outputs of different procedures, and we evaluated the detection sensitivity for aqueous and organic extraction on a 400 MHz nuclear magnetic resonance device. Our aim is to optimize the extraction protocol to permit the efficient use of cells in nuclear magnetic resonance study.
RESULTS
Metabolic profiles of human mesenchymal stem cells
The metabolite extraction of human mesenchymal stem cells was divided into aqueous and organic components, based on the solubility and extraction solvents used. The aqueous component was extracted using perchloric acid and the organic component was extracted using methanol-chloroform. Each was analyzed on a 9.4 T nuclear magnetic resonance device, and 1H-magnetic resonance spectra were acquired. In the spectra of metabolites for human mesenchymal stem cells, we assigned the main resonance peaks based on chemical shifts reported in previous studies[13,14], and we quantitatively analyzed the concentration of the main metabolites. The resonance assignments of the main metabolites and their chemical shifts are shown in Table 1, and the typical spectra of water-and lipid-soluble metabolites are shown in Figures 1 and 2. The absolute concentrations of the main components are shown in Tables 2 and 3. The aqueous and organic metabolites of human mesenchymal stem cells exhibited different spectral properties. In the water-soluble extract profiles, major metabolites were assigned to lactic acid, choline, glutamic acid and some amino acids. The peak at 1.28 ppm indicates that unsaturated fatty acid is also present. In the lipid-soluble extract profiles, the main metabolites were assigned to fatty acids. There is a sharp and significant peak at 1.28 ppm in the lipid spectra, whereas the lactic acid peak is not visible in the lipid profiles.
Table 1.
Major metabolites of human mesenchymal stem cells in the 1H-nuclear magnetic resonance spectrum
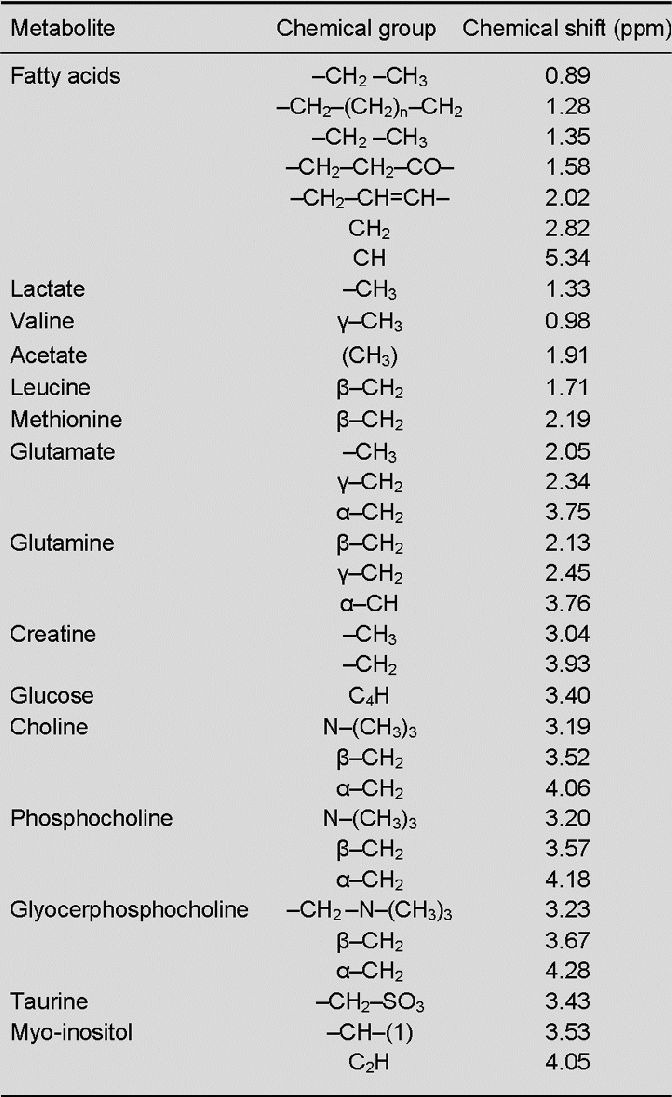
Figure 1.
Typical 1H-nuclear magnetic resonance spectrum of the water-soluble metabolites of human umbilical cord mesenchymal stem cells.
Cell extracts were prepared with perchloric acid (upper) and dual phase extraction (lower) methods. The concentrations of fatty acids (FA) and acetate (Ace) extracted with the perchloric acid method were higher than those obtained with the dual phase extraction method, while the concentrations of glutamate (Glu), choline (Cho) and methionine (Meth) were lower than those obtained with the dual phase extraction method (with statistical significance).
Myo: Myo-inositol; Cr: Creatine; Suc: succinate; TMSP: (trimethylsilyl) propionate acid sodium salt; PCA: perchloric.
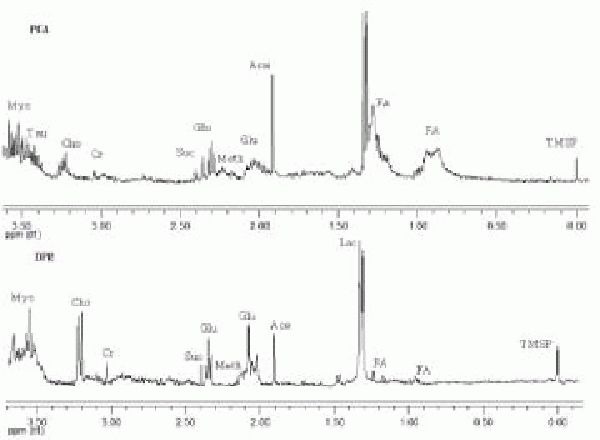
Figure 2.
Typical 1H-nuclear magnetic resonance spectrum of the lipid-soluble metabolites of human umbilical cord mesenchymal stem cells. Cell extracts were prepared using the methanol-chloroform (upper) and dual phase extraction (lower) methods. The concentration of glutamate (Glu) extracted with the dual phase extraction method was higher than that obtained with the methanol-chloroform method (with statistical significance).
FA: Fatty acids; TMS: tetramethylsilane; DPE: dual phase extraction.
Table 2.
Water-soluble metabolite concentrations (μmol/g of yield) obtained using the perchloric acid and dual phase extraction methods
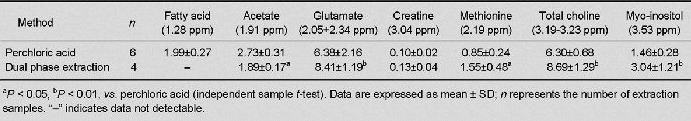
Table 3.
Lipid-soluble metabolite concentrations (μmol/g of yield) obtained using the methanol-chloroform and dual phase extraction methods
Extraction yields of the different methods
Extraction yields of each method were assessed and recorded. For each procedure, 12 samples were used. For the water-soluble extracts, 1.0 × 107 cells from each sample were used for both the dual phase and perchloric acid extraction methods. The lipid yield of the dual phase extraction method was compared with that of the methanol-chloroform procedure, for which 1.5 × 106 cells per sample were used. The net weight of extraction yields was calculated in milligrams. Significant differences in aqueous yields were found between the perchloric acid method (9.64 ± 0.65 mg) and the dual phase method (2.95 ± 0.35 mg) (P < 0.01). For lipid fractions, no significant difference was found between the methanol-chloroform method (2.52 ± 0.30 mg) and the dual phase method (2.83 ± 0.43 mg; P > 0.05). However, when taking cell quantity into consideration, the methanol-chloroform method was a more efficient procedure for extracting lipid metabolites, in that about one-tenth of the cell number was sufficient to obtain results comparable with the dual phase extraction procedure. For water-soluble extracts, the classic perchloric acid method produced a greater total yield.
Optimal quantities for nuclear magnetic resonance spectroscopy detection
Different net weights of the various extract yields were analyzed to explore the optimal net weights for nuclear magnetic resonance spectrum acquisition. Desirable spectral lines had clear resonance peaks and a smooth baseline.
Results showed that as low as 0.7 mg organic yield was enough to obtain clear resonance peaks, while more than 5.0 mg water-soluble yield was needed to obtain relatively favorable spectral lines (Figures 3, 4). No organic fractions less than 0.7 mg were further analyzed, since this extraction yield results from 0.5 × 106 cells, which can be cultured practically in a 25 cm2 flask.
Figure 3.
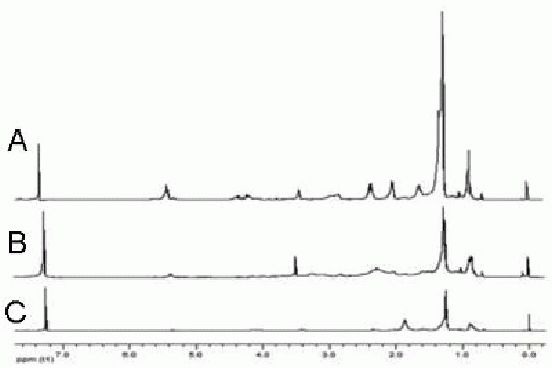
Lipid-soluble metabolic spectra of different net weights for detection.
A: 3.2 mg; B: 1.5 mg; C: 0.7 mg. The spectral peaks increase with increasing net weight.
Figure 4.
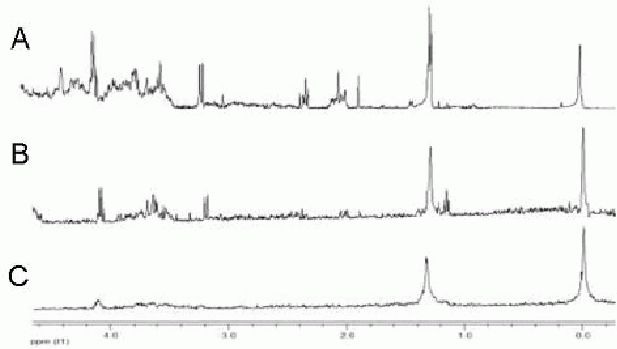
Water-soluble metabolic spectra of different net weights for detection.
A: 5.6 mg; B: 3.2 mg; C: 1.4 mg. The spectral peaks increase with increasing net weight.
DISCUSSION
The increased interest in stem cell research has highlighted the importance of having a thorough understanding of metabolite profiles. Metabolites can be divided into aqueous and organic, based on their solubility. Jacobus identified water-soluble metabolites in murine embryonic stem cells and neural stem cells in an in vitro nuclear magnetic resonance spectroscopy study[5]. However, few reports have discussed the lipid-soluble metabolite profiles of stem cells. The lipid profiles, which reflect the metabolism of the phospholipid bilayer of the cell membrane and of intracellular lipids, can give more information on cell physiopathological status[15]. The present study is the first to investigate the lipid-soluble metabolite profile of stem cells. In previous studies, the fatty acid peak at 1.28 ppm was considered to be a specific biomarker of neural progenitor cells; however, this has been disproved by the finding that the 1.28 ppm peak is also visible in mesenchymal stem cells as well as non-stem cell lines[16,17]. In this study, we showed that in human mesenchymal stem cells, the fatty acid peak can also be seen in the profiles and is most apparent in the lipid spectra. The combined analysis of both water- and lipid-soluble metabolites may provide more comprehensive data for stem cell researchers.
The results from our study showed completely different characteristics of aqueous and organic metabolites with 1H nuclear magnetic resonance spectroscopy. In the lipid-soluble profile, main metabolites are saturated and unsaturated fatty acids, while in the water-soluble metabolite profile, most resonance peaks were assigned to low molecular weight metabolites, including lactic acid, acetic acid, succinic acid, glutamic acid, creatine, myo-inositol, choline and its derivatives. Lactate is the end product of anaerobic glycolysis and an increased concentration often indicates hypoxia or ischemia[18,19]. Other metabolites representing key cellular functions include choline, creatine, glutamine and myo-inositol. Choline and its derivatives, phosphocholine and glycerophosphocholine, are generally considered to be related to cell membrane metabolism, and changes in their concentration have often been taken to indicate cell proliferation or growth arrest[20,21]. The peak at ˜3.04 ppm is composed of creatine and phosphocreatine, which play important roles in cellular energy metabolism[22,23,24]. Glutamate is involved in neurotransmission and synthesis in the central nervous system[25]. In stem cells, glutamate is the major respiratory fuel, and decreased concentrations can induce metabolic stress and make cells susceptible to infection[26]. Myo-inositol has been considered to be a glial marker and functions as an osmolyte, whereas in stem cells, myo-inositol is a key precursor of membrane phosphatidylinositol and myelin sheet structures. Increased concentrations may indicate cell membrane turnover or damage to myelin sheets[27]. An in vivo nuclear magnetic resonance spectroscopy investigation, examining mesenchymal stem cells as they differentiate into adipocytes, Shi discovered not only an increase in the concentration of methylene at 1.28 ppm and methyl at 0.89 ppm, but also a significant decrease in intracellular metabolites, including choline, creatine, glutamate and myo-inositol[12]. This may indicate that these metabolites may also be biomarkers of stem cell differentiation. However, because few reports have examined lipid-soluble profiles, the functions of fatty acids in the lipid-soluble metabolite fraction remain unknown.
By comparing the extraction yields of different methods, we found that the perchloric acid procedure was more efficient in generating total aqueous extracts compared with the dual phase extraction method, and the classic methanol-chloroform method was efficient in extracting organic metabolites. Although the dual phase extraction method is able to obtain both fractions simultaneously and has the advantage of being a simple procedure, our results show that its extraction efficiency for either fraction was inferior to perchloric acid or methanol-chloro-form. Our finding is somewhat inconsistent with the results of Le Belle and colleagues, who dissolved their extract in D2O and mainly focused on water-soluble metabolite profiles[11]. However, we are strongly in agreement that the dual phase extraction method is superior to the perchloric acid method in extracting glutamate and choline-containing compounds. Since metabolites may have different solubilities in different solvents, the discrepancies between the different methods may result from differences in the solvents used. However, we calculated the extraction yield using dry net weight rather than wet weight of protein because macromolecules can interfere with the detection of small metabolites. In addition, we quantitatively calculated the metabolite concentration using the ratio of the spectral area to the internal standard and normalized by dividing by the net weight of the yield to decrease errors caused by variations in extraction yield.
It was unexpected that nuclear magnetic resonance spectroscopy could be so sensitive. It was capable of detecting minor lipid-soluble metabolites, and less than 1 mg extraction yields were enough to obtain good spectral quality. Most resonance peaks in lipid metabolites were fatty acids. This is due to the high solubility of these lipids and their high content of hydrogen protons[28]. As for water-soluble extracts, the detection threshold was much higher. Although there was no significant difference between the lipid yields of the methanol-chloroform and the dual phase extraction methods, it is important to note that the water-soluble fraction separated from the dual phase extraction procedure was not enough to obtain desirable spectral lines. If a greater water-soluble yield is required from the dual phase extraction method, more cells must be utilized. In contrast, the perchloric acid and methanol-chloroform methods can produce good spectra using a smaller number of cells compared with the dual phase extraction method. In this regard, we propose a strategy for cell extraction that is economical in cell usage and labor-saving. When lipid metabolites are the focus of study, the methanol-chloroform method may be the preferred choice because only a small quantity of cells is required and because of the ease of extraction. For water-soluble metabolites, the perchloric acid method may be appropriate for its high total extraction yield. A combination of the perchloric acid and methanol-chloroform methods enables the simultaneous extraction of both aqueous and organic metabolites from a much smaller quantity of cells. Although this approach adds additional steps for cell extraction, it minimizes cell culture and avoids unnecessary waste. However, it should be noted that the dual phase extraction method has a higher efficiency in extracting glutamate and choline-containing components, and if tissue samples are to be extracted, the dual phase extraction method may also be preferred since tissue samples can be frequently obtained more readily.
In conclusion, we provide a more detailed understanding of human mesenchymal stem cell metabolic profiles by combined analysis of both aqueous and organic extracts using 1H nuclear magnetic resonance spectroscopy. Furthermore, we propose strategies for selecting the optimal extraction methods. Because of technical limitations, only one-dimensional spectra were acquired. Our findings should be confirmed using two-dimensional spectrum analysis.
MATERIALS AND METHODS
Design
An in vitro comparative study.
Time and setting
The experiments were performed in the Research Center for Reproductive Medicine of Shantou University Medical College, China from March to October 2011.
Materials
Mesenchymal stem cells were isolated from Wharton's jelly tissue of newborn umbilical cord, and then cultured. According to the Administrative Regulations on Medical Institutions published by the State Council of China[29], written informed consent was obtained from all parents.
Inclusion criteria: full-term healthy male fetus was born from caesarean operation with an Apgar score no less than 9 at 1 and 5 minutes. The exclusion criteria for all subjects included: mother during pregnancy was older than 35 years or younger than 20; mother has any psychiatric illness (including bipolar disorder, schizophrenia, dementia, alcohol or drug abuse or dependence) or presence of any physical illness (including hypertension, ischemic heart disease, diabetes, Cushing's disease or obesity).
Methods
Culture of human mesenchymal stem cells
Mesenchymal stem cells were isolated from Wharton's jelly tissue and cultured according to previous reports with some modifications. After dissection from the placenta[30,31], fresh umbilical cord was obtained in the operating room and stored in serum-free low-glucose Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) before processing and carried to the laboratory as soon as possible. After removal of blood vessels, the mesenchymal tissue was carefully dissected from Wharton's jelly with a scalpel and washed in PBS for 3 minutes. The harvested tissues were then mechanically cut into 1 mm3 pieces and placed in 75-cm2 culture flasks (Corning, Steuben County, NY, USA). Finally, the tissues were cultured in F12-Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and stored in a 37°C incubator under 5% CO2 (Thermo Fisher Scientific, Boston, USA). After 1 week, fibroblast-like cells were visible around the tissue. When they reached 70–80% confluence, the cells were trypsinized and transferred to new flasks for expansion. Culture medium was changed every 3 days. Identification by immunophenotyping and determination of differentiation capacities are described in our previous work[32,33]. All cells used were at passage 4.
Different methods for extracting cell metabolites
Perchloric acid method for extracting water-soluble metabolites: The water-soluble metabolites were extracted according to previous reports with some modifications[8]. After removing the culture medium, culture flasks were washed twice with cold PBS and cells were scraped with a rubber policeman. Approximately 1 × 107 cells from each sample were collected and centrifuged at 2 000 r/min (Eppendorf, Hamburg, Germany) for 10 minutes at room temperature, and the clear supernatant was discarded. To each tube, 3 mL ice-cold 4% perchloric acid was added. The mixture was sonicated on an ice bath for 5 minutes (BILON99-11DL Instruments, BiLang Company, Shanghai, China) and then centrifuged at 13 000 r/min for 20 minutes at 0°C (Eppendorf). The supernatant was carefully separated and neutralized with 1 mol/L KOH. The precipitated salt was removed by centrifugation. The clear supernatant was collected in a 5-mL centrifuge tube and placed in a low-pressure lyophilizer (LYO-8, Tofflon, Shanghai, China) for 48 hours until all fluid had evaporated and only a dry powder was left. The dried sample was then weighed carefully and the data were recorded. Every sample was weighed three times and an average was obtained. The net weights of the extracts were expressed in milligrams. All samples were then stored at –20°C before nuclear magnetic resonance analysis.
Methanol-chloroform method for extracting lipid-soluble metabolites: Lipid-soluble metabolites were extracted as by Folch, with slight modification[7]. Briefly, 1.5 × 106 cells from each sample were collected and centrifuged. After discarding the supernatant, 1 mL ice-cold methanol was added to each tube and the mixture was vigorously vibrated for 1 minute. After a 15-minute incubation in the first solvent, 2 mL ice-cold chloroform and 0.5 mL 0.9% saline solution were added to each sample; the final ratio of chloroform/methanol was 2:1 (v/v). The mixed solvents were left overnight at 4°C for phase separation. The lower chloroform phase, which contained the lipid-soluble metabolites, was carefully separated and dried under a nitrogen stream. The dried sample was weighed and then stored at –20°C until nuclear magnetic resonance spectroscopy (400 MHz; Bruker Avance, Switzerland).
Dual-phase extraction method for extracting both metabolites: The dual-phase extraction was performed according to Bligh and Dye with some modifications[9,10,11]. Approximately 1 × 107 cells from each sample were collected and centrifuged. Then, 1.5 mL ice-cold methanol and chloroform in a ratio of 2:1 (v/v) were added to the cell pellets, and the mixture was sonicated for 5 minutes. Then another 1.5 mL chloroform and distilled water in a ratio of 1:2 (v/v) was added to the first solvent to form an emulsion. The solvents were centrifuged at 13 000 r/min (0°C, 20 minutes) for phase separation. Aqueous metabolites-containing upper phase (methanol and water) was carefully separated from the lower chloroform phase, which contained the organic metabolites. The water phase was lyophilized, while the lipid fraction was dried under a nitrogen stream. The dried sample was weighed and then stored at –20°C until nuclear magnetic resonance spectroscopy (400 MHz, Bruker Avance).
Analysis of cell extracts using 1H nuclear magnetic resonance spectroscopy
1H nuclear magnetic resonance spectra were acquired on a high-resolution nuclear magnetic resonance spectroscopy system (400 MHz; Bruker Avance). The lyophilized aqueous powder was dissolved in 0.5 mL D2O, in which a known amount of 3-trimethylsilyl propionic acid-d4 sodium salt was added as an internal standard. The organic samples were dissolved in 0.5 mL CDCl3 in which tetramethylsilane was added as internal standard. All samples were transferred into 5-mm nuclear magnetic resonance test tubes. One-dimensional 1H nuclear magnetic resonance spectra were acquired using the ZGPR sequence. Parameters used were: sweep width = 5 kHz, TR = 20 seconds, number of scans = 64 and data points = 32 K. Original data were pre-processed with Fourier transformation, phase correction and baseline correction, and then outputted. The peak areas of main metabolites were calculated by integration using Mestrec4.7 software (incorporated in the Bruker nuclear magnetic resonance system). Individual metabolites were assigned according to chemical shifts and the absolute concentration of main metabolites were calculated by the ratio of spectral area to the internal standard and corrected according to proton numbers contributing to the resonances according to the following formula[34]: Cm = 9 × (Im/Nm) × CTMSP or TMS × V/(ITMSP or TMS × W). Cm = metabolite concentration, Im = the integral value of the resonance peak of interest, Nm = the number of protons of the metabolites, CTMSP or TMS = the concentration and V = the volume of the internal standard added, ITMSP or TMS = the integral value of the internal standard and W = the net weight of the dried extract.
Statistical analysis
The net weights of the extracts were expressed in milligrams, and the final metabolite concentrations were expressed as mol/g. Data were expressed as mean ± SD and the statistical significance level was P < 0.05 using independent sample t-test analyzed by SPSS 17.0 software (SPSS, Chicago, IL, USA).
Footnotes
Funding: This study was supported by the Key Program of the National Natural Science Foundation of China, No. 30930027; the National Natural Science Foundation of China, No. 60971075; and the Foundation for Basic and Clinical Medicine (2010) of Shantou University Medical College, China.
Conflicts of interest: None declared.
Ethical approval: The experiment was approved by the Ethics Committee of Shantou University Medical College, China.
(Reviewed by Patel B, Raye Z, Zhong SJ, Wang XL, Sun HR)
(Edited by Wang J, Su LL, Li CH, Song LP, Liu WJ, Zhao M)
REFERENCES
- [1].Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- [2].Wang HS, Hung SC, Peng ST, et al. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells. 2004;22(7):1330–1337. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- [3].Weiss ML, Medicetty S, Bledsoe AR, et al. Human umbilical cord matrix stem cells: preliminary characterization and effect of transplantation in a rodent model of Parkinson's disease. Stem Cells. 2006;24(3):781–792. doi: 10.1634/stemcells.2005-0330. [DOI] [PubMed] [Google Scholar]
- [4].Lim TS, Hong YH, Lee HY, et al. Metabolite investigation in both anterior and posterior cingulate gyri in alzheimer's disease spectrum using 3-tesla MR spectroscopy. Dement Geriatr Cogn Disord. 2012;33(2-3):149–155. doi: 10.1159/000338177. [DOI] [PubMed] [Google Scholar]
- [5].Jansen JF, Shamblott MJ, van Zijl PC, et al. Stem cell profiling by nuclear magnetic resonance spectroscopy. Magn Reson Med. 2006;56(3):666–670. doi: 10.1002/mrm.20968. [DOI] [PubMed] [Google Scholar]
- [6].Xu ZF, Pan AZ, Yong F, et al. Human umbilical mesenchymal stem cell and its adipogenic differentiation: Profiling by nuclear magnetic resonance spectroscopy. World J Stem Cells. 2012;4(4):21–27. doi: 10.4252/wjsc.v4.i4.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- [8].Glonek T, Kopp SJ, Kot E, et al. P-31 nuclear magnetic resonance analysis of brain: the perchloric acid extract spectrum. J Neurochem. 1982;39(5):1210–1219. doi: 10.1111/j.1471-4159.1982.tb12557.x. [DOI] [PubMed] [Google Scholar]
- [9].Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- [10].Tyagi RK, Azrad A, Degani H, et al. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magn Reson Med. 1996;35(2):194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- [11].Le Belle JE, Harris NG, Williams SR, et al. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed. 2002;15(1):37–44. doi: 10.1002/nbm.740. [DOI] [PubMed] [Google Scholar]
- [12].Shi C, Wang X, Wu S, et al. HRMAS 1H-NMR measured changes of the metabolite profile as mesenchymal stem cells differentiate to targeted fat cells in vitro: implications for non-invasive monitoring of stem cell differentiation in vivo. J Tissue Eng Regen Med. 2008;2(8):482–490. doi: 10.1002/term.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sitter B, Sonnewald U, Spraul M, et al. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002;15(5):327–337. doi: 10.1002/nbm.775. [DOI] [PubMed] [Google Scholar]
- [14].Martinez-Bisbal MC, Marti-Bonmati L, Piquer J, et al. 1H and 13C HR-MAS spectroscopy of intact biopsy samples ex vivo and in vivo 1H MRS study of human high grade gliomas. NMR Biomed. 2004;17(4):191–205. doi: 10.1002/nbm.888. [DOI] [PubMed] [Google Scholar]
- [15].Skar-Gislinge N, Simonsen JB, Mortensen K, et al. Elliptical structure of phospholipid bilayer nanodiscs encapsulated by scaffold proteins: casting the roles of the lipids and the protein. J Am Chem Soc. 2010;132(39):13713–13722. doi: 10.1021/ja1030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Manganas LN, Zhang X, Li Y, et al. Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain. Science. 2007;318(5852):980–985. doi: 10.1126/science.1147851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Loewenbruck KF, Fuchs B, Hermann A, et al. Proton MR spectroscopy of neural stem cells: does the proton-NMR peak at 1.28 ppm function as a biomarker for cell type or state? Rejuvenation Res. 2011;14(4):371–381. doi: 10.1089/rej.2010.1102. [DOI] [PubMed] [Google Scholar]
- [18].Yang ZX, Huo SS, Cheng XF, et al. Quantitative multivoxel proton MR spectroscopy study of brain metabolites in patients with amnestic mild cognitive impairment: a pilot study. Neuroradiology. 2012;54(5):451–458. doi: 10.1007/s00234-011-0900-0. [DOI] [PubMed] [Google Scholar]
- [19].Aydin H, Ozgul E, Agildere AM. Acute necrotizing encephalopathy secondary to diphtheria, tetanus toxoid and whole-cell pertussis vaccination: diffusion-weighted imaging and proton MR spectroscopy findings. Pediatr Radiol. 2010;40(7):1281–1284. doi: 10.1007/s00247-009-1498-9. [DOI] [PubMed] [Google Scholar]
- [20].Yue Q, Shibata Y, Isobe T, et al. Absolute choline concentration measured by quantitative proton MR spectroscopy correlates with cell density in meningioma. Neuroradiology. 2009;51(1):61–67. doi: 10.1007/s00234-008-0461-z. [DOI] [PubMed] [Google Scholar]
- [21].Valonen PK, Griffin JL, Lehtimaki KK, et al. High-resolution magic-angle-spinning 1H NMR spectroscopy reveals different responses in choline-containing metabolites upon gene therapy-induced programmed cell death in rat brain glioma. NMR Biomed. 2005;18(4):252–259. doi: 10.1002/nbm.955. [DOI] [PubMed] [Google Scholar]
- [22].Dezortova M, Jiru F, Petrasek J, et al. 1H MR spectroscopy as a diagnostic tool for cerebral creatine deficiency. MAGMA. 2008;21(5):327–332. doi: 10.1007/s10334-008-0137-z. [DOI] [PubMed] [Google Scholar]
- [23].Walecki J, Barcikowska M, Cwikla JB, et al. N-acetyl-aspartate, choline, myoinositol, glutamine and glutamate (glx) concentration changes in proton MR spectroscopy (1H MRS) in patients with mild cognitive impairment (MCI) Med Sci Monit. 2011;17(12):105–111. doi: 10.12659/MSM.882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barger AV, Campeau NG, Port JD, et al. MRS is the test of choice for detecting and monitoring disorders of creatine metabolism. Pediatr Neurol. 2009;40(5):408–410. doi: 10.1016/j.pediatrneurol.2008.12.012. [DOI] [PubMed] [Google Scholar]
- [25].Choi C, Dimitrov IE, Douglas D, et al. Improvement of resolution for brain coupled metabolites by optimized (1)H MRS at 7T. NMR Biomed. 2010;23(9):1044–1052. doi: 10.1002/nbm.1529. [DOI] [PubMed] [Google Scholar]
- [26].Mates JM, Perez-Gomez C, Nunez de Castro I, et al. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int J Biochem Cell Biol. 2002;34(5):439–458. doi: 10.1016/s1357-2725(01)00143-1. [DOI] [PubMed] [Google Scholar]
- [27].Haris M, Cai K, Singh A, et al. In vivo mapping of brain myo-inositol. Neuroimage. 2011;54(3):2079–2085. doi: 10.1016/j.neuroimage.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mahrous EA, Lee RB, Lee RE. Lipid profiling using two-dimensional heteronuclear single quantum coherence NMR. Methods Mol Biol. 2009;579:89–102. doi: 10.1007/978-1-60761-322-0_5. [DOI] [PubMed] [Google Scholar]
- [29].State Council of the People's Republic of China. Administrative Regulations on Medical Institution. 1994 Sep 01; [Google Scholar]
- [30].Wang HS, Hung SC, Peng ST, et al. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells. 2004;22(7):1330–1337. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- [31].Salehinejad P, Alitheen NB, Ali AM, et al. Comparison of different methods for the isolation of mesenchymal stem cells from human umbilical cord Wharton's jelly. In Vitro Cell Dev Biol Anim. 2012;48(2):75–83. doi: 10.1007/s11626-011-9480-x. [DOI] [PubMed] [Google Scholar]
- [32].Ma L, Feng XY, Cui BL, et al. Human umbilical cord Wharton's Jelly-derived mesenchymal stem cells differentiation into nerve-like cells. Chin Med J (Engl) 2005;118(23):1987–1993. [PubMed] [Google Scholar]
- [33].Huang P, Lin LM, Wu XY, et al. Differentiation of human umbilical cord Wharton's jelly-derived mesenchymal stem cells into germ-like cells in vitro. J Cell Biochem. 2010;109(4):747–754. doi: 10.1002/jcb.22453. [DOI] [PubMed] [Google Scholar]
- [34].Cao Z, Wu LP, Li YX, et al. Change of choline compounds in sodium selenite-induced apoptosis of rats used as quantitative analysis by in vitro 9.4T MR spectroscopy. World J Gastroenterol. 2008;14(24):3891–3896. doi: 10.3748/wjg.14.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]


