Abstract
Wallerian degeneration is a subject of major interest in neuroscience. A large number of genes are differentially regulated during the distinct stages of Wallerian degeneration: transcription factor activation, immune response, myelin cell differentiation and dedifferentiation. Although gene expression responses in the distal segment of the sciatic nerve after peripheral nerve injury are known, differences in gene expression between the proximal and distal segments remain unclear. In the present study in rats, we used microarrays to analyze changes in gene expression, biological processes and signaling pathways in the proximal and distal segments of sciatic nerves undergoing Wallerian degeneration. More than 6,000 genes were differentially expressed and 20 types of expression tendencies were identified, mainly between proximal and distal segments at 7–14 days after injury. The differentially expressed genes were those involved in cell differentiation, cytokinesis, neuron differentiation, nerve development and axon regeneration. Furthermore, 11 biological processes were represented, related to responses to stimuli, cell apoptosis, inflammatory response, immune response, signal transduction, protein kinase activity, and cell proliferation. Using real-time quantitative PCR, western blot analysis and immunohistochemistry, microarray data were verified for four genes: aquaporin-4, interleukin 1 receptor-like 1, matrix metalloproteinase-12 and periaxin. Our study identifies differential gene expression in the proximal and distal segments of a nerve during Wallerian degeneration, analyzes dynamic biological changes of these genes, and provides a useful platform for the detailed study of nerve injury and repair during Wallerian degeneration.
Keywords: nerve regeneration, peripheral nerve injury, Wallerian degeneration, sciatic nerve injury, microarray, expression profiling, biological process, rat, NSFC grant, neural regeneration
Introduction
Peripheral nerve injuries are common in modern society. Unlike the central nervous system, which cannot regenerate, injured peripheral nerves have the ability to regenerate on their own. Many preclinical and clinical studies of peripheral nerve injury have made significant progress in understanding peripheral nerve injury. Moreover, clinicians possess a clear understanding of the response of the peripheral nervous system to trauma, and therapeutic methods. A series of pathophysiologic events occur after injury in neuronal cell bodies and axons, both of them interdependent in the recovery process (Lee et al., 2000; Costigan et al., 2002; Bosse et al., 2006; Webber et al., 2010; Sheth et al., 2011). During peripheral nerve regeneration, neuronal survival and target reinnervation depend on the intrinsic regenerative capacity of peripheral axons and an appropriate growth microenvironment. After peripheral nerve injury, Wallerian, or anterograde, degeneration occurs, arising from the fracture of distal axons and proximal nerve fibers. Other neurons interacting with the cell body facilitate the regeneration of axons and the myelin sheath, and reconnection with the distal side (Huang et al., 2007; Viader et al., 2011; Gao et al., 2013; Li et al., 2013; Yao et al., 2013).
Previous studies have identified many genes that are up- or downregulated after peripheral nerve injury, and some new treatment strategies target the regulation of key genes (Bosse et al., 2006; Sheth et al., 2011; Li et al., 2013; Yao et al., 2013). However, systemic knowledge of gene expression changes and related biological processes during peripheral nerve injury and regeneration is still lacking. To find preventative approaches and effective treatments for peripheral nerve injury, we need to have a deeply integrated insight into the molecular mechanisms regulating peripheral nerve regeneration from a much broader perspective. Microarray techniques have given us the capacity to analyze biological changes in thousands of genes, many of which are already targeted in other conditions, and have been used successfully to monitor gene expression profile changes in many systemic studies (Yao et al., 2012; Gao et al., 2013; Hu et al., 2013; Li et al., 2013; Akane et al., 2014). By detecting the spectrum of genes expressed during peripheral nerve injury and regeneration, DNA microarray allows us to see changing trends in the expression of thousands of genes at once, known as expression profiling. Previous studies have confirmed certain changes in gene expression and biological processes in the proximal segment of injured sciatic nerves at early time points using DNA microarray analysis (Huang et al., 2007; Viader et al., 2011; Gao et al., 2013; Hu et al., 2013; Li et al., 2013; Yao et al., 2013; Akane et al., 2014).
The present study was designed to identify systemic gene expression changes following peripheral nerve injury and to uncover the potential involvement of biological processes by comparing proximal and distal segments of the nerve. We used DNA microarray technology to detect mRNA expression changes in the sciatic nerve at 0, 4, 7, 14, 21 and 28 days after transection in adult rats. We then used gene ontology enrichment analysis to identify the biological processes associated with the differentially expressed genes. Expression trends identified in the microarray were confirmed using real-time quantitative PCR (RT-qPCR), western blot analysis and fluorescent immunohistochemistry.
Materials and Methods
Animal models and tissue collection
432 male Sprague-Dawley rats weighing 180–220 g were provided by the Experimental Animal Center of Nantong University, China, and divided equally between the microarray, RT-qPCR, Western blot and immunohistochemistry assay (36 animals per assay, with each assay performed three times). All tests were conducted in accordance with NIH Guidelines for the Care and Use of Laboratory Animals (Yao et al., 2013) and were ethically approved by the Administration Committee of Experimental Animals, Jiangsu Province, China.
Rats were further divided into 6 groups: 0, 4, 7, 14, 21, 28 days (6 per group) to undergo sciatic nerve resection. An incision was made on the lateral aspect of the mid-thigh in the right hind limb. The sciatic nerve was identified and lifted. Two segments (each 0.5 cm in length) of the sciatic nerve, proximal and distal to the division of the tibial and common peroneal nerves, were excised, and the incision sites were then closed. The rats were housed in high temperature- and humidity-controlled cages with sawdust bedding after surgery, and allowed free access to food and water. The day 0 group of rats was sacrificed immediately, and the other groups at 4, 7, 14, 21 and 28 days following surgery. Proximal and distal sciatic nerve tissues (0.5 cm) were collected separately.
RNA isolation and microarray experiments
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) from the proximal and distal nerve stumps according to the manufacturer's protocols. RNA quality, amplification, labeling and hybridization of each sample were carried out using the Affymetrix GeneChip Hybridization Oven 640 and Gene Array Scanner 3000 (Affymetrix, Santa Clara, CA, USA). The arrays were scanned and the subsequent data compiled using GeneChip Scan Control software (Affymetrix). All steps, from RNA amplification to final scanner output, were conducted by the National Engineering Center for Biochip, Shanghai, China. Three biological replicates were performed at each time point. Differentially-expressed genes were identified by GeneChip scanner at each time point, with absorbance ratios set for different genes.
Data analysis
Gene screening and bioinformatic analyses were performed by the Bioinformatics Center at the Key Laboratory of Systems Biology, Shanghai Institute for Biological Sciences, China. Series Test Cluster of Gene Ontology analysis was used to determine the main functions of the differentially expressed genes according to gene ontology, the key functional classification of NCBI. Pathway analysis was used to determine the significant pathways of the differential genes according to the Kyoto Encyclopedia of Genes and Genomes. Key factors, networks and signal flow were calculated according to gene expression fold-change and gene interaction in pathway (Li et al., 2013; Yao et al., 2013).
RT-qPCR assays
The nerve stumps of rats killed at 0, 4, 7, 14, 21, and 28 days after injury were dissected and then suspended in RNA Stabilization Reagent (Qiagen, Valencia, CA, USA). The total RNA fraction was prepared from stored tissue specimens using an RNeasy Mini Kit (Qiagen), according to the manufacturer's protocol. The relative expression values of each mRNA were calculated using comparative CT and were normalized using glyceraldehyde phosphate dehydrogenase (GAPDH) mature mRNA for each data point. The primers used in the RT-qPCR assays are as follows:
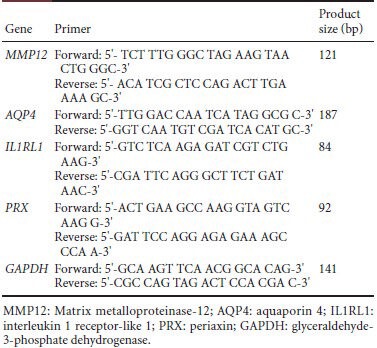
Table 1.
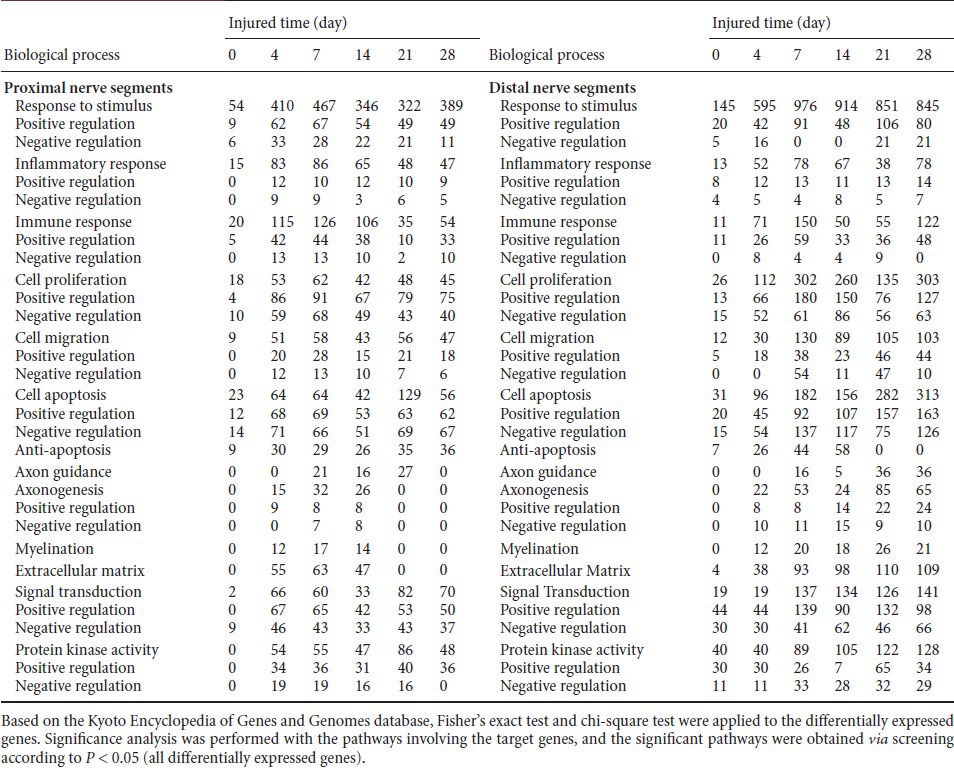
Biological processes of differentially expressed genes in proximal and distal nerve segments at 0, 4, 7, 14, 21 and 28 days after sciatic nerve injury (number of differentially expressed genes)
Western blot analysis
Sciatic nerve tissue obtained at various time points following injury was lysed immediately, and protein content was tested using the bicinchoninic acid kit (Yao et al., 2013) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The membranes were incubated with mouse anti-AQP4 monoclonal antibody (1:200; Santa Cruz Biotechnology), mouse anti-IL1RL1 monoclonal antibody (1:500; Santa Cruz Biotechnology), rabbit anti-MMP12 polyclonal antibody (1:500; Santa Cruz Biotechnology), or rabbit anti-PRX polyclonal antibody (1:500; Santa Cruz Biotechnology) at 4°C, overnight, and visualized with IRDye 800-conjugated affinity purified goat anti-mouse/rabbit IgG (1:5,000; Santa Cruz Biotechnology) at room temperature for 2 hours. Protein content was normalized using mouse anti-beta-actin monoclonal antibody (Santa Cruz Biotechnology) as an internal control for each data point. The image was scanned with a GS800 Densitometer Scanner (Bio-Rad, Hercules, CA, USA), and absorbance values were analyzed using PDQuest 7.2.0 software (Bio-Rad).
Immunohistochemistry
The distal and proximal nerve stumps were fixed in freshly prepared 4% paraformaldehyde. Samples were incubated with mouse monoclonal antibodies against AQP4 (1:200 dilution; Santa Cruz Biotechnology), IL1RL1, MMP12 or PRX (all 1:500 dilution; Santa Cruz Biotechnology) and visualized with Dylight 488-labeled goat anti-mouse IgG (1:250 dilution; Sigma-Aldrich, St Louis, MO, USA) at room temperature for 2 hours. The nucleus was stained with Hoechst 33342 before samples were mounted and observed with a confocal laser scanning microscope (TCS SP2, Leica Microsystems, Q550IW, Germany).
Statistical analysis
Differentially expressed genes were selected in a logical sequence according to the random variance model (REM) corrective analysis of variance. The expression model profiles were correlated with the actual or expected number of genes assigned to each model profile. Significant profiles demonstrated a higher probability than expected by Fisher's exact test and multiple comparison tests. Fisher's exact test and chi-square test were employed to analyze significant pathways. All data were expressed as mean ± SD and analyzed by one-way analysis of variance and Scheffe's post-hoc test with SPSS 19.0 for Windows software (SPSS, Chicago, IL, USA). P < 0.05 was considered statistically significant.
Results
Gene expression profiling of proximal and distal nervous tissue after sciatic nerve injury in rats
DNA microarray analysis revealed that all biological replicates represented significant homology in gene expression. To understand global tendencies of gene expression changes, they were classified by fold-change: 2–5, 5–10, 10–50, and >50. In the proximal segments, upregulation mostly occurred within 7 days, before downregulation (P < 0.05). The data suggested that nerve transection might stimulate the upregulation of a few genes, which initiated the expression of more genes in a circular pattern. In the distal segments, expression tendencies differed from those in the proximal segments, with expression of most genes being low in the early stages (P < 0.05). An advantage was achieved in the expression of downregulated genes, which may be characteristic of Wallerian degeneration. According to gene expression profiling, we drew simple diagrams of the expression changes in four selected genes: AQP4, IL1RL1, MMP12 and PRX (Figure 1). To our knowledge, there have been no reports on the involvement of these four genes in the repair of peripheral nerve injury.
Figure 1.

Bioinformatics analysis of expression trends for AQP4, IL1RL1, MMP12 and PRX genes in the distal and proximal sciatic nerve stumps of rats at various time points after nerve resection.
Based on the Kyoto Encyclopedia of Genes and Genomes database, significant pathways were identified using Fisher's exact test and the chisquare test (P < 0.05). X axis represents time after injury (days); Y axis represents relative gene expression. AQP4: Aquaporin-4; MMP12: matrix metalloproteinase-12; IL1RL1: interleukin 1 receptor-like 1; PRX: periaxin.
Biological process analysis of differential gene expressions in proximal and distal nervous tissue of rats after sciatic nerve injury
Bioinformatics analysis identified differential expression of more than 6,000 genes in proximal and distal nervous tissue after sciatic nerve injury, including those involved in neuronal differentiation, development, and regeneration, as well as cytokine synthesis. These differentially expressed genes are mainly involved in responses to stimuli, cell apoptosis, inflammatory response, immune response, signal transduction, cell migration, cell proliferation, axon guidance and axonogenesis, myelination, extracellular matrix, and protein kinase activity. Some genes might be involved in more than one biological process.
Differential expression of AQP4, IL1RL1, MMP12 and PRX genes in proximal and distal nervous tissue after sciatic nerve injury
To verify the microarray data, RT-qPCR analysis was performed for four of the differentially expressed genes: AQP4, IL1RL1, MMP12 and PRX (Figure 2). Changes observed were consistent with the results from the microarray analysis.
Figure 2.
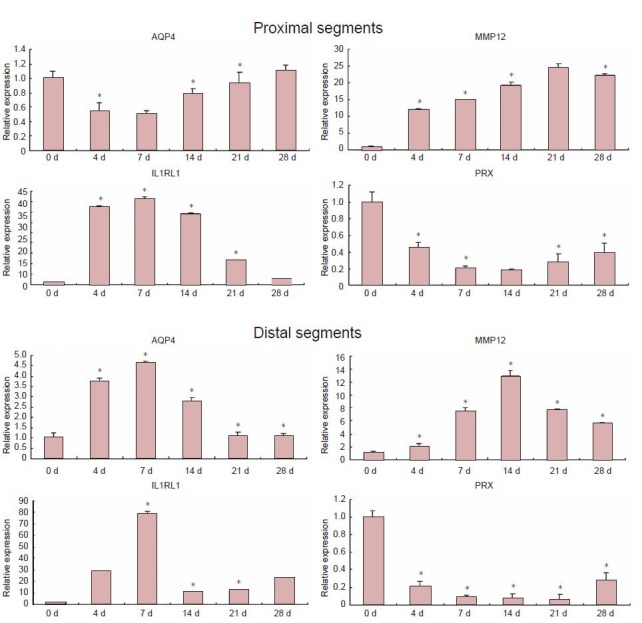
AQP4, IL1RL1, MMP12 and PRX gene expression of distal and proximal sciatic nerve stumps of rats at 0, 4, 7, 14, 21, 28 days (d) after sciatic nerve injury (real-time quantitative PCR analysis).
Relative expression values of each mRNA were calculated using the Comparative CT method and normalized against GAPDH mature mRNA at each data point. *P < 0.05 vs. day 0. Data are expressed as mean ± SD and analyzed by one-way analysis of variance and Scheffe's post-hoc test. AQP4: Aquaporin-4; MMP12: matrix metalloproteinase-12; IL1RL1: interleukin 1 receptor-like 1; PRX: periaxin.
Differential expression of AQP4, IL1RL1, MMP12 and PRX proteins in proximal and distal nervous tissue after sciatic nerve injury
To further confirm the data of microarray and RT-qPCR analyses, western blots were performed to analyze the expression of AQP4, IL1RL1, MMP12 and PRX proteins (Figure 3). Expression of all four proteins changed with time post-injury. Protein expression trends were consistent with the results obtained with RT-qPCR and microarray bioinformatics analyses.
Figure 3.
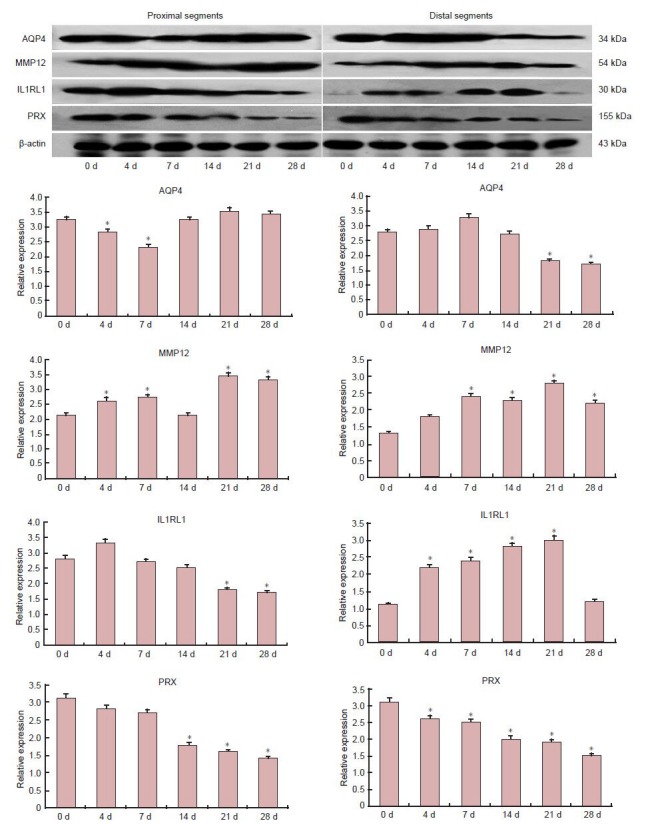
Western blot analysis of total protein lysates from distal and proximal sciatic nerve stumps of rats at 0, 4, 7, 14, 21 and 28 days (d) after sciatic nerve injury.
Anti-AQP4, MMP12, IL1RL1 and PRX content (upper) and relative expressions (down). *P < 0.05, vs. day 0 (one-way analysis of variance and Scheffe's post-hoc test). Data are expressed as mean ± SD. AQP4: Aquaporin-4; MMP12: matrix metalloproteinase-12; IL1RL1: interleukin 1 receptor-like 1; PRX: periaxin.
Localization of AQP4, IL1RL1, MMP12 and PRX using immunohistochemistry
Finally, immunohistochemistry was used to localize the expression of AQP4, IL1RL1, MMP12 and PRX (Figure 4). All four proteins showed changes in expression over time post-injury that were consistent with the results obtained from the microarray, RT-qPCR and western blot analyses.
Figure 4.
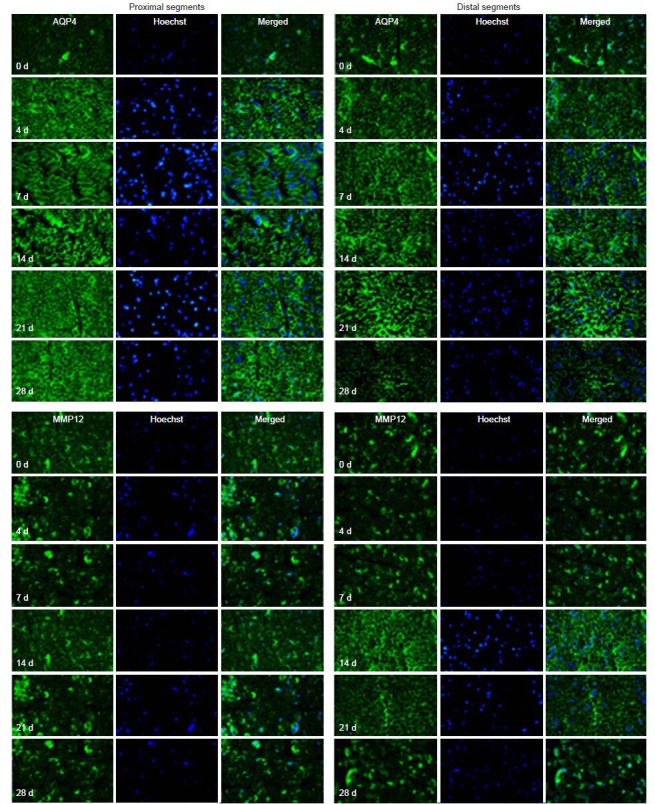
Immunohistochemistry (AQP4 and MMP12) of the distal and proximal sciatic nerve stumps of rats at 4, 7, 14, 21, and 28 days (d) after sciatic nerve injury (× 63).
Double-immunostaining for Hoechst 33342 (blue) and AQP4 or MMP12 (green) and their overlay showed that the proteins were localized in the nucleus. The results were consistent with real-time PCR and western blot analysis. AQP4: Aquaporin-4; MMP12: matrix metalloproteinase-12.
Figure 5.
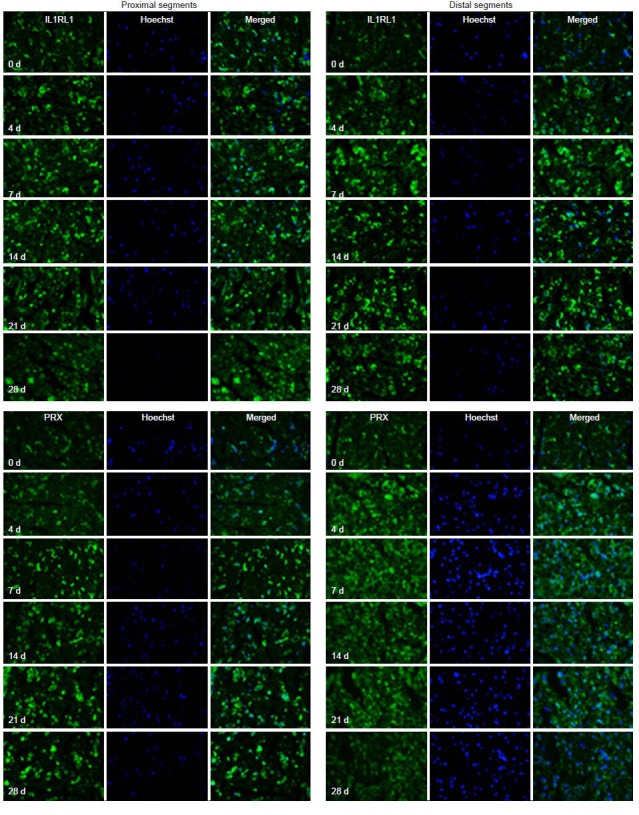
Immunohistochemistry (IL1RL1 and PRX) of the distal and proximal sciatic nerve stumps of rats at 4, 7, 14, 21, and 28 days (d) after sciatic nerve injury (× 63).
Double-immunostaining for Hoechst 33342 (blue) and IL1RL1 or PRX (green) and their overlay showed that the proteins were localized in the nucleus. The results were consistent with real-time PCR and western blot analysis. IL1RL1: Interleukin 1 receptor-like 1; PRX: periaxin.
Discussion
Wallerian degeneration, a process that occurs before nerve regeneration, remains a subject of major interest in modern neurobiology. A large number of genes are differentially regulated during the distinct stages of Wallerian degeneration. Nerve regeneration is largely a recapitulation of nerve injury and repair accompanied by a series of biological reactions and cellular interactions (Kawai et al., 2006; Hu et al., 2007; Vargas et al., 2010; Benowitz et al., 2011; Tang et al., 2013; Yao et al., 2014). Peripheral nerve repair is a result of reactivated regeneration mechanisms in combination with newly activated injury-dependent reactions. In the initial stages of nerve injury and repair, genetic responses feature a number of degeneration and regeneration-specific genes. Therefore, understanding the factors that regulate the rapid macrophage responses in the peripheral nervous system during Wallerian degeneration, and comparing the differences in the expression of these molecules in the injured peripheral nervous system, may provide insights into the reasons for slow Wallerian degeneration (Koenig et al., 2000; Stepanek et al., 2005; Hu et al., 2007; Belanger et al., 2011; Wang et al., 2012).
To verify the microarray data obtained in the present study, we analyzed four differentially expressed genes (AQP4, IL1RL1, MMP12 and PRX) which have not previously been associated with nerve repair after nerve injury. AQP4 belongs to the aquaporin family of integral membrane proteins that conduct water through cell membranes. AQP4 is expressed in astrocytes and is upregulated by direct stimulus to the central and peripheral nervous systems. IL1RL1 is a member of the Toll-like receptor superfamily. However, unlike other members of the family, IL1RL1 does not induce an inflammatory response through activation of nuclear factor-κB, although it activates and adjusts mitogen-activated protein kinases. MMP12 is secreted as an inactive protein. MMP family proteins are involved in the breakdown of the extracellular matrix in normal physiological processes, such as reproduction and tissue remodeling, as well as in disease processes. PRX is a protein found in humans. The PRX gene encodes L- and S-periaxin that codes for proteins involved in Schwann cell myelination.
In the present study, we used microarray technology to investigate the expression patterns of various genes after peripheral nerve injury, and identified trends in the changes of differentially expressed genes. We also performed gene ontology analysis to determine how the distinctively expressed genes regulate different biological processes after peripheral nerve injury during their regeneration. In multicellular organisms, complex and dynamic networks are composed of many genes, and their functions are dependent on their expression mechanism and microenvironment (Hunter 1991; Ronnett et al., 2002; Michel et al., 2010).
In summary, knowledge of the dynamic changes in differentially expressed genes and related biological processes will provide a deeper insight into the role of genetic specificity in regulation and control. Here, we analyzed the trends in the changes of 11 classes of biological processes and presented the expression profiles of related but differentially expressed genes, comparing the differentiation of proximal segments with distal segments in the gene expression profiles. Our study sheds light on the undiscovered roles of these differentially expressed genes, especially some key regulatory genes, and provides a useful platform for the detailed study of peripheral nerve injury and regeneration at the molecular level. Detailed examination of gene expression profiles will provide new insight into the molecular mechanisms and signal cascades underlying nerve degeneration and regeneration.
Footnotes
Funding: This study was supported by the National Natural Science Foundation of China, No. 81370982, 31170946; Key Program, Grant No. 81130080; the Priority Academic Program Development of Jiangsu Higher Education Institutions in China.
Conflicts of interest: None declared.
Copyedited by Slone-Murphy J, Haase R, Wang J, Qiu Y, Li CH, Song LP, Zhao M
References
- 1.Akane H, Saito F, Shiraki A, Imatanaka N, Akahori Y, Itahashi M, Wang L, Shibutani M. Gene expression profile of brain regions reflecting aberrations in nervous system development targeting the process of neurite extension of rat offspring exposed developmentally to glycidol. J Appl Toxicol. 2014;1:7. doi: 10.1002/jat.2971. [DOI] [PubMed] [Google Scholar]
- 2.Bélanger E, Henry FP, Vallée R, Randolph MA, Kochevar IE, Winograd JM, Lin CP, Côté D. In vivo evaluation of demyelination and remyelination in a nerve crush injury model. Biomed Opt Expess. 2011;2:2698–2708. doi: 10.1364/BOE.2.002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benowitz LI, Popovich PG. Inflammation and axon regeneration. Curr Opin Neurol. 2011;24:577–583. doi: 10.1097/WCO.0b013e32834c208d. [DOI] [PubMed] [Google Scholar]
- 4.Bosse F, Hasenpusch-Theil K, Kury P, Müller HW. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J Neurochem. 2006;96:1441–1457. doi: 10.1111/j.1471-4159.2005.03635.x. [DOI] [PubMed] [Google Scholar]
- 5.Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao B, Sun W, Wang X, Jia X, Ma B, Chang Y, Zhang W, Xue D. Whole genome expression profiling and screening for differentially expressed cytokine genes in human bone marrow endothelial cells treated with humoral inhibitors in liver cirrhosis. Int J Mol Med. 2013;32:1204–1214. doi: 10.3892/ijmm.2013.1495. [DOI] [PubMed] [Google Scholar]
- 7.Hu P, Bembrick AL, Keay KA, McLachlan EM. Immune cell involvement in dorsal root ganglia and spinal cord after chronic constriction or transection of the rat sciatic nerve. Brain Behav Immun. 2007;21:599–616. doi: 10.1016/j.bbi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Hu X, Zhang S, Chen G, Lin C, Huang Z, Chen Y, Zhang Y. Expression of SHH signaling molecules in the developing human primary dentition. BMC Dev Biol. 2013;13:11. doi: 10.1186/1471-213X-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter T. Protein kinase classification. Methods Enzymol. 1991;200:3–37. doi: 10.1016/0076-6879(91)00125-g. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 12.Koenig HL, Gong WH, Pelissier P. Role of progesterone in peripheral nerve repair. Rev Reprod. 2000;5:189–199. doi: 10.1530/ror.0.0050189. [DOI] [PubMed] [Google Scholar]
- 13.Lee SK, Wolfe SW. Peripheral nerve injury and repair. J Am Acad Orthop Surg. 2000;8:243–252. doi: 10.5435/00124635-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Guo W, Zhang P, Li H, Gu X, Yao D. Signal flow and pathways in response to early Wallerian degeneration after rat sciatic nerve injury. Neurosci Lett. 2013;536:56–63. doi: 10.1016/j.neulet.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Liu Q, Wang Y, Gu Y, Liu D, Wang C, Ding G, Chen J, Liu J, Gu X. Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One. 2013;8:e57000. doi: 10.1371/journal.pone.0057000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel G, Thierry T, Scornet D, Cock JM, Kloareg B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytologist. 2010;188:82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- 17.Ronnett GV, Moon C. G proteins and olfactory signal transduction. Annu Rev Physiol. 2002;64:189–222. doi: 10.1146/annurev.physiol.64.082701.102219. [DOI] [PubMed] [Google Scholar]
- 18.Sheth S, Li X, Binder S, Dry SM. Differential gene expression profiles of neurothekeomas and nerve sheath myxomas by microarray analysis. Mod Pathol. 2011;24:343–354. doi: 10.1038/modpathol.2010.203. [DOI] [PubMed] [Google Scholar]
- 19.Stepanek L, Stoker AW, Stoeckli E, Bixby JL. Receptor tyrosine phosphatases guide vertebrate motor axons during development. J Neurosci. 2005;25:3813–3823. doi: 10.1523/JNEUROSCI.4531-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang X, Wang Y, Zhou S, Qian T, Gu X. Signaling pathways regulating dose-dependent dual effects of TNF-α on primary cultured Schwann cells. Mol Cell Biochem. 2013;378:237–246. doi: 10.1007/s11010-013-1614-x. [DOI] [PubMed] [Google Scholar]
- 21.Vargas ME, Watanabe J, Singh SJ, Robinson WH, Barres BA. Endogenous antibodies promote rapid myelin clearance and effective axon regeneration after nerve injury. Proc Natl Acad Sci USA. 2010;107:11993–11998. doi: 10.1073/pnas.1001948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viader A, Chang LW, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J Neurosci. 2011;31:17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Tang X, Yu B, Gu Y, Yuan Y, Yao D, Ding F, Gu X. Gene network revealed involvements of birc2, birc3, and tnfrsf1a in anti-apoptosis of injured peripheral nerves. PLoS One. 2012;7:e43436. doi: 10.1371/journal.pone.0043436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webber C, Zochodne D. The nerve regenerative microenvironment: early behavior and partnership of axons and Schwann cells. Exp Neurol. 2010;223:51–59. doi: 10.1016/j.expneurol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- 25.Yao D, Li M, Shen D, Ding F, Lu S, Zhao Q, Gu X. Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci Bull. 2013;29:321–332. doi: 10.1007/s12264-013-1340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao L, Cao J, Sun H, Guo A, Li A, Ben Z, Zhang H, Wang X, Ding Z, Yang X, Huang X, Ji Y, Zhou Z. FBP1 and p27kip1 expression after sciatic nerve injury: implications for Schwann cellsproliferation and differentiation. J Cell Biochem. 2014;115:130–140. doi: 10.1002/jcb.24640. [DOI] [PubMed] [Google Scholar]


