Abstract
This study investigated the possible involvement of microRNAs in the regulation of genes that participate in peripheral neural regeneration. A microRNA microarray analysis was conducted and 23 microRNAs were identified whose expression was significantly changed in rat dorsal root ganglia after sciatic nerve transection. The expression of one of the downregulated microRNAs, microRNA-214, was validated using quantitative reverse transcriptase-PCR. MicroRNA-214 was predicted to target the 3′-untranslated region of Slit-Robo GTPase-activating protein 3. In situ hybridization verified that microRNA-214 was located in the cytoplasm of dorsal root ganglia primary neurons and was downregulated following sciatic nerve transection. Moreover, a combination of in situ hybridization and immunohistochemistry revealed that microRNA-214 and Slit-Robo GTPase-activating protein 3 were co-localized in dorsal root ganglion primary neurons. Western blot analysis suggested that Slit-Robo GTPase-activating protein 3 was upregulated in dorsal root ganglion neurons after sciatic nerve transection. These data demonstrate that microRNA-214 is located and differentially expressed in dorsal root ganglion primary neurons and may participate in regulating the gene expression of Slit-Robo GTPase-activating protein 3 after sciatic nerve transection.
Keywords: nerve regeneration, peripheral nerve injury, sciatic nerve injury, Slit-Robo GTPaseactivating protein 3, microRNA-214, dorsal root ganglia, gene expression, microarray, bioinformatics, NSFC grant, neural regeneration
Introduction
Peripheral sensory neurons can regenerate after nerve injury (Woolf et al., 1990; Bergman et al., 1999); however, the molecular mechanism that regulates neural regeneration is not well understood. The Slit-Roundabout (Robo) signaling pathway is one modulator of peripheral neural regeneration (Yi et al., 2006; Zhang et al., 2010). The Slit family of proteins plays a crucial role in neurite outgrowth and axon pathfinding. Slits bind to Robo, which is a transmembrane receptor that determines the cellular response to Slit (Bashaw and Goodman, 1999). In our previous study, we observed that Robo1 and Robo2 are both expressed in neurons of adult rat dorsal root ganglia, and nerve injury can induce upregulation of these proteins (Yi et al., 2006). Treatment of cultured dorsal root ganglion neurons with recombinant, soluble Slit1 protein promoted neurite outgrowth and elongation. In contrast, treatment with a recombinant human Robo2/Fc chimera inhibited neurite outgrowth and elongation (Zhang et al., 2010). Moreover, we observed that the expression pattern of Slit-Robo GTPase-activating protein 3 was similar to that of Robo2, and that both are co-expressed in dorsal root ganglion neurons (Chen et al., 2012). Slit-Robo GTPase-activating protein 3 contains a Rho GAP domain that regulates the activities of Rho family GTPases and affects actin polymerization, which influences dendrite elaboration, neurite outgrowth and axon guidance (Yamashita et al., 1999). To investigate the influence of Slit-Robo GTPase-activating protein 3 on peripheral neural regeneration, it is necessary to elucidate the molecular mechanism underlying regulating Slit-Robo GTPase-activating protein 3 in dorsal root ganglion neurons.
The regulation of neuronal signaling pathways, as part of a coordinated response to nerve injury, can occur at the transcriptional and post-transcriptional levels. The majority of mammalian genes are now thought to be regulated, in part, by post-transcriptional gene silencing (Tam Tam et al., 2011). MicroRNAs are approximately 22 nucleotide, endogenous, noncoding RNAs that post-transcriptionally repress the expression of protein-coding genes by base-pairing with the 3′-untranslated regions of target mRNAs. Approximately two-thirds of human genes are estimated to be regulated by microRNAs (Friedman et al., 2009). MicroRNAs have emerged as ubiquitous regulators of developmental timing and cellular differentiation (Flynt and Lai, 2008). To date, however, few reports are available on changes in microRNAs expression in dorsal root ganglia after sciatic nerve injury. This study was designed to investigate the changes in Slit-Robo GTPase-activating protein 3-associated microRNA expression in dorsal root ganglia during peripheral neural regeneration following sciatic nerve transection.
Materials and Methods
Establishment of a model of sciatic nerve transection
Wistar rats (80 ± 5 g) were purchased from the Dongchuang Laboratory Animal Center (Changsha, Hunan Province, China). Rats were intraperitoneally anesthetized with 10% chloral hydrate (0.3 mL/100 g). The right sciatic nerve was exposed under sterile conditions and was transected approximately 3 mm proximal to its division into the tibial and common peroneal nerves. The wounds were closed and rats were allowed to recover for 1, 3, 5 or 7 days. The experiments were performed in accordance with the guidelines for experimental animal use established by the ethics committee of Central South University, China.
Tissue collection
The control dorsal root ganglia and sciatic nerves were taken from the contralateral (left) side. The injured sciatic nerve sample (approximately 5 mm) was taken from the proximal stump of the injured sciatic nerve. For the following experiments, the injured ipsilateral nerves were called injured nerves and the ipsilateral dorsal root ganglia were called injured dorsal root ganglia. The contralateral nerves were called control nerves and dorsal root ganglia from the contralateral side were called control dorsal root ganglia. Nerves and dorsal root ganglia from intact animals were called intact nerves and intact dorsal root ganglia, respectively. After specified time periods, animals were euthanized and dorsal root ganglia and proximal sciatic nerve stumps were quickly dissected out. For the microarray analysis, quantitative reverse transcription-PCR and western blot assays, control and injured L4–5 dorsal root ganglia were collected and immediately frozen. For anatomical studies, animals (n = 6 per time point) were anesthetized with an overdose of sodium pentobarbital (100 mg/kg, i.p.) and perfused transcardially. Dorsal root ganglia and proximal sciatic nerve stumps were dissected out, post-fixed, dehydrated and embedded in paraffin. Paraffin-embedded tissue was kept at 4°C in a dry environment before sectioning.
Total RNA isolation from rat dorsal root ganglia
After sciatic nerve transection, the injured and control L4–5 dorsal root ganglia from Wistar rats (n = 10 for microarray analysis and n = 6 for quantitative reverse transcription-PCR) were collected 3 days after nerve transection. Total RNA was harvested using TRIzol (Invitrogen, Carlsbad, CA, USA) and the RNeasy mini kit (QIAGEN, Shanghai, China) according to the manufacturer's instructions. The quality of the purified RNA was assessed using a BioAnalyzer 2,100 (Agilent Technology, Santa Clara, CA, USA). RNA samples were stored at −80°C. The small RNA fraction from each of the total RNA samples was enriched using the Pure Link™ miRNA Isolation Kit (Invitrogen).
MicroRNA labeling and microarray hybridization
MicroRNA samples were quantified using a Nanodrop instrument, and then labeled using the miRCURY™ Hy3™/Hy5™ Power labeling kit and hybridized on the miRCURY™ LNA Array (v.11.0) (Exiqon Inc., Woburn, MA, USA). The miRCURY LNA microRNA Array used contains more than 1,700 capture probes, covering all microRNAs annotated in miRBase 11.0. The samples were hybridized on a hybridization station. Scanning was performed with the Axon GenePix 4000B microarray scanner (Molecular Devices Inc., Sunnyvale, CA, USA). GenePix pro V6.0 (Molecular Devices, Inc.) was used to read the raw intensity of the image. The ratio of red signal to green signal was calculated after background subtraction and normalization using the global Lowess (Locally Weighted Scatter plot Smoothing) regression algorithm (MIDAS, TIGR Microarray Data Analysis System (Dana-Farber Cancer Institute, Boston, MA, USA)). The threshold value we used to classify differentially expressed microRNAs was fold change ≥ 1.5 or ≤ 0.67.
Bioinformatic prediction and analysis
The potential targets of significantly changed microRNAs were predicted using the bioinformatic software TargetScan Human 6.0 (http://www.targetscan.org), and the targets of each microRNA were imported to the Database for Annotation, Visualization and Integrated Discovery Version 6.7 (http://david.abcc.ncifcrf.gov/) (Huang da et al., 2009) for Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses.
Quantitative reverse transcription-PCR
cDNAs were synthesized with 1 μg total RNA from each sample using an M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA) and microRNA-specific primers (Sangon Biotech (Shanghai), Shanghai, China). PCR was conducted in 20 μL using an identical amount of cDNA per reaction and 0.5 μmol/L forward and reverse primers. The PCR reaction included 40 cycles of 95°C for 15 seconds, 65°C for 30 seconds, and 72°C for 32 seconds. The quantitative reverse transcription-PCR reactions were carried out in triplicate for each cDNA sample using Express SybrGreenER qPCR SuperMix Universal (Invitrogen). Relative quantitation of gene and microRNA expression were normalized against the reference gene S12 using a 2−ΔΔCT method (Paz et al., 2007). All experiments were carried out three times independently.
Cell culture, transfections and dual-luciferase assays
293T cells (ATCC# CRL-1573) were incubated in Dulbecco's modified Eagle's medium (DMEM) with 1.5 g/L sodium bicarbonate, 10% fetal bovine serum (Sigma, St. Louis, MO, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma) and incubated at 37°C, in a 5% CO2 atmosphere. All transfections were conducted with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The luciferase reporter assay was carried out using the 3′-untranslated region of Slit-Robo GTPase-activating protein 3, which was PCR amplified from Rattus norvegicus genomic DNA. The product was inserted into the MluI and HindIII sites of psiCHECKTM-2 (Promega) immediately downstream from the luciferase stop codon. 293 T cells were transfected with 0.4 μg of firefly luciferase reporter vector in the presence or absence of 0.5 μg of microRNA-214, and the cells were seeded at 70% confluence in 96-well plates. PRLTK vector (Promega) served as an internal control for transfection efficiency. Luciferase activity was measured 24 hours after transfection, using GloMaxTm20/20 dual-luciferase assays (Promega).
In situ hybridization and immunohistochemistry
Following sciatic nerve transection, L4–5 dorsal root ganglia of six Wistar rats were collected after 1, 3, 5 and 7 days for in situ hybridization using the Digoxigenin-labeled miRCURY LNA microRNA detection probe kit (Exiqon, Vedbaek, Denmark). A scrambled microRNA probe was used as a negative control. U6, a small nuclear RNA was used as a positive control. Paraffin-embedded sections (4 μm thick) of dorsal root ganglia and sciatic nerve stumps were mounted on microslides. The sections were deparaffinized and digested with proteinase K. The sections were prehybridized for 1 hour and then hybridized with microRNA LNA probe in hybridization buffer (20 nmol/L, Roche, Nutley, NJ, USA) overnight (approximately 8 hours). After 3 consecutive washes, the sections were treated with blocking buffer for 1 hour and then incubated with alkaline phosphatase-conjugated polyclonal sheep anti-digoxigenin fragment antigen-binding fragments (1:500; Roche) for 2 hours. The sections were then incubated with alkaline phosphatase substrate for 2 hours. After incubation with AP stop solution (KTBT buffer), the slides were rinsed, dehydrated and mounted. Light microscopy (Nikon, Tokyo, Japan) images were taken on the subsequent day. The sections used to localize microRNA-214 were counterstained using nucleic red after in situ hybridization.
We combined in situ hybridization with immunohistochemistry to compare microRNA and Slit-Robo GTPase-activating protein 3 localization patterns in single dorsal root ganglion neurons. In brief, in situ hybridization was conducted as described above. After blocking, the sections were incubated with AP-conjugated polyclonal sheep anti-digoxigenin-fragment antigen-binding fragments and Slit-Robo GTPase-activating protein 3-specific polyclonal antibody (1:300; Proteintech, Rocky Hill, NJ, USA) for 2 hours. The sections were then incubated with an FITC-conjugated AffiniPure goat anti-rabbit IgG (1:300; Jackson ImmunoResearch Laboratories., West Grove, PA, USA) for 2 hours. After the immunohistochemical labeling was imaged with a fluorescence microscope, the remaining steps of in situ hybridization were carried out. In situ hybridization images were captured in the same immunofluorescence fields.
Immunohistochemical labeling and in situ hybridization sections were imaged on a fluorescence microscope equipped with a digital camera (Nikon 90I; Nikon Instruments, Melville, NY, USA). For densitometry, five randomly selected images at 20 × magnification were taken from each animal. For each image, the mean absorbance value was calculated using Image Pro Plus 6.0 (Media Cybernetics, Inc., Bethesda, MD, USA).
Western blot assay
After sciatic nerve transection, the injured and control L4–5 dorsal root ganglia from Wistar rats (n = 6 per time point) were collected at 1, 3 and 7 days. Dorsal root ganglia were homogenized with ice-cold radioimmunoprecipitation assay buffer (1:10, w/v) containing 25 mmol/L Tris (pH 7.5), 1 mmol/L ethylenediaminetetraacetic acid, 2 mmol/L MgCl2, 50 mmol/L KCl, and 5 mmol/L dithiothreitol (Boster, Wuhan, Hubei Province, China) and then centrifuged at 9,000 × g for 15 minutes at 4°C. Proteins for western blot analysis were quantified using BioRad reagent. 20 μg of solubilized proteins were loaded per lane on sodium dodecyl sulfate gels and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were then transferred to nitrocellulose membranes. After incubating the membranes with 5% non-fat milk, the membranes were incubated with rabbit anti-Slit-Robo GTPase-activating protein 3 polyclonal antibody (1:800; Proteintech) at 4°C overnight. The appropriate secondary antibodies were then incubated with the membranes for 2 hours at room temperature and bound antibody was visualized. The images were analyzed using Image Pro Plus 6.0 software. α-Tubulin was detected as a loading control.
Statistical analysis
The numerical data for immunohistochemistry and in situ hybridization are expressed as mean ± SEM. One-way analysis of variance and Dunnett's t-test or two-tailed t-tests were used for the statistical analyses of means between groups, using SPSS 17.0 software (SPSS, Chicago, IL, USA). P values less than 0.05 were considered statistically significant.
Results
Different expression of microRNAs in injured dorsal root ganglia
To investigate expression profiles of microRNAs in dorsal root ganglia, microarray analyses were performed. The analysis used total RNA collected from L4–5 dorsal root ganglia of rats. A total of 23 microRNAs showed significant changes in expression between the injured dorsal root ganglia and the control dorsal root ganglia (P < 0.01; Figure 1A). Of these, 18 were significantly downregulated, while five were upregulated.
Figure 1.
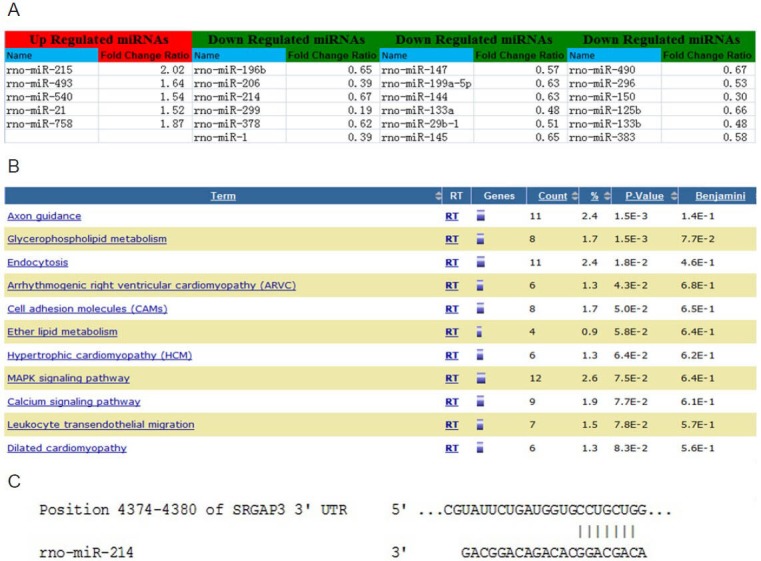
Microarray and bioinformatic analysis.
(A) Following sciatic nerve transection, the expression of 23 miRs was significantly changed in injured dorsal root ganglia versus control dorsal root ganglia. These miRs had expression levels greater than or equal to a 1.5-fold change (increase or decrease) in expression. (B) Potential targets of miR-214 were significantly enriched in the axon guidance pathway. (C) MiR-214 targets the 3’-UTR of srGAP3. miRs: MicroRNAs; srGAP3: Slit-Robo GTPase-activating protein 3.
Prediction of potential microRNA targets
The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis showed that potential microRNA-214 targets were significantly enriched in the axon guidance pathway (P = 0.0015; Figure 1B). MicroRNA-214 was predicted to have a binding site in the 3′-UTR of Slit-Robo GTPase-activating protein 3, a molecule in the axon guidance pathway (Figure 1C).
Validation of microRNA-214 expression level in dorsal root ganglia
In addition to microarray profiling, quantitative reverse transcription-PCR was used to quantify microRNA-214 in dorsal root ganglia. The downregulation of microRNA-214 was validated in injured dorsal root ganglia (Figure 2A).
Figure 2.
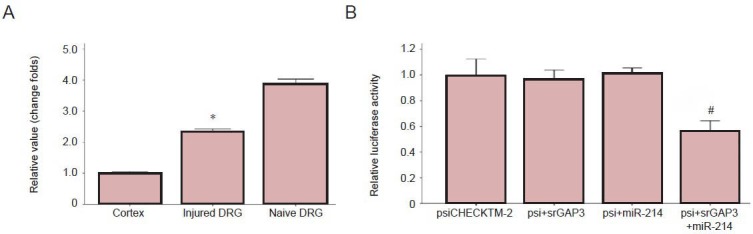
Quantitative reverse transcription-PCR and luciferase reporter assays validated the expression level of miR-214 in injured DRG.
(A) The downregulation of miR-214 in DRGs after sciatic nerve transection was validated using quantitative reverse transcription-PCR. The relative quantity of miR-214 was calculated using the equation RQ = 2−ΔΔCT. The S12 gene served as an internal reference. (B) The relative luciferase activity of psiCHECKTM-2, the psiCHECKTM-2 vector containing the 3’-untranslated region of srGAP3 (psi + srGAP3), the psiCHECKTM-2 vector containing miR-214 oligonucleotide (psi + miR-214), the psiCHECKTM-2 vector containing the 3’-untranslated region of srGAP3 and the miR-214 oligonucleotide (psi + srGAP3 + miR-214). (A, B) The results are expressed as mean ± SEM from independent experiments replicated in triplicate; *P < 0.01, vs. control DRG; #P < 0.01, vs. other groups using two-tailed t-test. miR: MicroRNA; srGAP3: Slit-Robo GTPase-activating protein 3; DRG: dorsal root ganglia.
MicroRNA-214 targeted Slit-Robo GTPase-activating protein 3
To determine whether microRNA-214 can target the 3′-UTR of Slit-Robo GTPase-activating protein 3, luciferase reporter assays were carried out using the psiCHECKTM-2 microRNA luciferase reporter vector. The results consistently showed that the luciferase activity of psiCHECKTM-2 vectors co-transfected with the 3′-UTR of Slit-Robo GTPase-activating protein 3 and microRNA-214 was significantly decreased (Figure 2B).
Localization of microRNA-214 in dorsal root ganglion neurons and sciatic nerves
To localize microRNA-214 in dorsal root ganglia and sciatic nerves, in situ hybridization was performed. Cytosolic labeling for microRNA-214 was observed in dorsal root ganglia neurons. MicroRNA-214 was also located in the axonal plasma of the sciatic nerve. The distribution of microRNA-214 was interesting: the signal was distributed in a parallel manner under the axolemma in longitudinal sections, while in transverse sections the signal had a circular shape (Figure 3).
Figure 3.
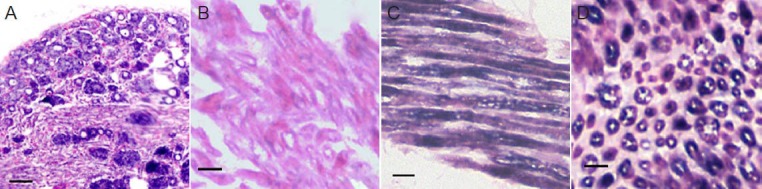
Localization of miR-214 in dorsal root ganglia neurons and sciatic nerve axon (in situ hybridization).
(A) MiR-214 was distributed mainly in the cytoplasm of primary dorsal root ganglion neurons. (C, D) In the sciatic nerve, miR-214 was located mainly in the axoplasm. There was no miR-214 labeling in the scramble probe control (B). Bars in A: 20 μm; in B–D: 5 μm. miR: MicroRNA.
MicroRNA-214 was differentially expressed in rat dorsal root ganglion neurons responding to injury
The expression levels of microRNA-214 in injured and control dorsal root ganglia were compared using in situ hybridization. In injured dorsal root ganglion neurons at 1, 3, 5 and 7 days after injury, microRNA-214 was significantly downregulated at day 1 and kept on decreasing thereafter (P < 0.05; Figure 4).
Figure 4.
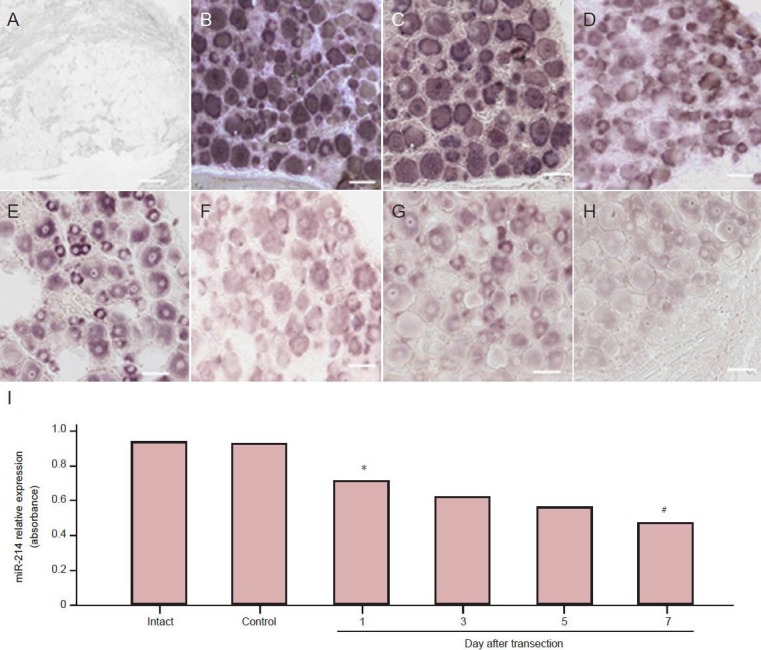
Changes in miR-214 relative expression in dorsal root ganglion neurons of sciatic nerve transected rats (in situ hybridization).
There was no specific labeling for the scrambled control probe (A). A high level of labeling was observed in dorsal root ganglion neurons for the positive control probe (B). MiR-214 was moderately expressed in intact dorsal root ganglia (C) and control dorsal root ganglia at 3 days after sciatic nerve transection (D). Significant downregulation of miR-214 was observed in injured dorsal root ganglion neurons at 1 day (E), 3 days (F), 5 days (G) and 7 days (H) after sciatic nerve transection. Bars: 20 μm. (I) Quantitative expression of miR-214. The results are expressed as the mean and there were six rats in each group at each time point. *P < 0.05, vs. control dorsal root ganglia; #P < 0.05, vs. 1 day after sciatic nerve transection us-ing one-way analysis of variance and Dunnett's t-tests. miR: MicroRNA.
Comparison of early microRNA-214 and Slit-Robo GTPase-activating protein 3 changes in response to sciatic nerve transection
A previous study showed that Slit-Robo GTPase-activating protein 3 is upregulated in dorsal root ganglion neurons following peripheral nerve injury (Chen et al., 2012). Immunohistochemistry showed that the expression of Slit-Robo GTPase-activating protein 3 in dorsal root ganglion neurons at 1, 3 and 7 days after injury (Figure 5B–F) was significantly higher than in control dorsal root ganglion neurons (Figure 5A). This result was consistent with western blot analysis (Figure 5G). Compared to the downregulation of microRNA-214, the level of Slit-Robo GTPase-activating protein was significantly upregulated after injury and kept increasing with time (P < 0.05; Figure 5H).
Figure 5.
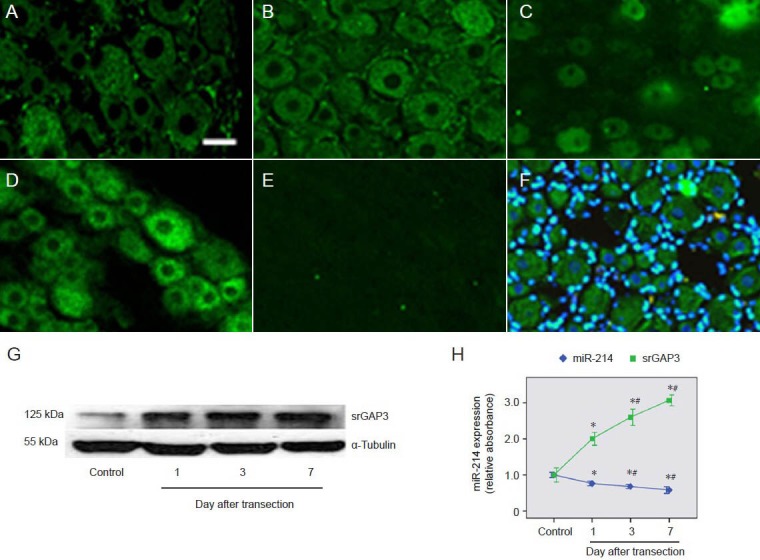
Comparison of changes in srGAP3 and miR-214 levels in dorsal root ganglion neurons.
(A–F) srGAP3 detected by immunohistochemistry (scale bar: 20 μm). The level of srGAP3 was significantly upregulated in primary dorsal root ganglion neurons at 1 day (B), 3 days (C) and 7 days (D) after sciatic nerve transection, compared with levels in control dorsal root ganglion neu-rons (A). There was no specific labeling of srGAP3 in negative control sections (E). Overlays of srGAP3 (green) and DAPI (blue) staining showed cytoplasmic labeling of srGAP3 (F). (G) Western blot analysis of srGAP3 protein levels in control dorsal root ganglia and ipsilateral dorsal root ganglia at 1, 3 and 7 days after sciatic nerve transection. (H) Variation trends of miR-214 and srGAP3 levels in dorsal root ganglion neurons. Mean absorbance values were normalized using values for α-tubulin. The results are expressed as mean ± SEM. The quotient of the mean absorbance value/mean absorbance of each group (n = 6) and that of the control dorsal root ganglion neurons was calculated as a relative value. *P < 0.05, vs. control dorsal root ganglia; #P < 0.05, vs. 1 day after sciatic nerve transection using one-way analysis of variance and Dunnett's t-tests. srGAP3: Slit-Robo GTPase-activating protein 3; miR: microRNA.
Co-expression of microRNA-214 and Slit-Robo GTPase-activating protein 3 in single dorsal root ganglion neurons
To compare microRNA-214 and Slit-Robo GTPase-activating protein 3 localization patterns in single dorsal root ganglion neurons, we combined in situ hybridization with immunohistochemistry. Co-expression of microRNA-214 and Slit-Robo GTPase-activating protein 3 was observed in control dorsal root ganglion neurons and in injured dorsal root ganglion neurons (Figure 6). Accompanying the downregulation of microRNA-214, Slit-Robo GTPase-activating protein 3 expression was significantly upregulated in nearly all dorsal root ganglion neurons at 3 days after injury.
Figure 6.
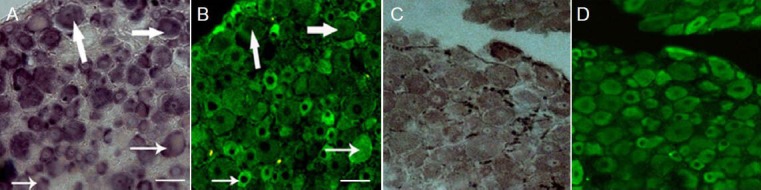
Co-expression of miR-214 and srGAP3 in primary dorsal root ganglion neurons.
The arrow-marked cells show that in control dorsal root ganglion neurons with high miR-214 labeling, the level of srGAP3 was minimal and that the level of srGAP3 was high in dorsal root ganglion neurons with low miR-214 labeling. In injured dorsal root ganglion neurons, downregulated miR-214 labeling and upregulated srGAP3 labeling were observed in nearly all dorsal root ganglion neurons at 3 days after sciatic nerve transec-tion. (A) In situ hybridization in the control dorsal root ganglions; (B) immunofluorescence in the control dorsal root ganglions; (C) in situ hy-bridization in the injured dorsal root ganglions; (D) immunofluorescence in the injured dorsal root ganglions. Scale bars: 20 μm. miR: MicroRNA; srGAP3: Slit-Robo GTPase-activating protein 3.
Discussion
MicroRNAs are important for neurogenesis and neuronal differentiation (Aranha et al., 2010; Loven et al., 2010). They are highly expressed in the nervous system and modulate neuronal gene expression (Lau and Hudson, 2010); however, few studies have attached importance to the role of microRNAs in dorsal root ganglia after nerve transaction (Wu et al., 2011). Although sciatic nerve regeneration has been studied for many decades, many studies have focused on the roles of Schwann cells or the way peripheral nerves repair after crush injuries (Tomita et al., 2009; Wu et al., 2011). Much less is known about nerve regeneration after complete transection (Zhou et al., 2011). During regeneration, studies have shown that a number of changes occur in gene expression within 3 days of nerve injury (Adilakshmi et al., 2012). Furthermore, the protein level of Slit-Robo GTPase-activating protein 3 has been shown to increase 3 days after nerve injury (Chen et al., 2012). To identify microRNAs that regulate Slit-Robo GTPase-activating protein 3 expression in dorsal root ganglia, a 3-day sciatic nerve transection rat model was chosen for microarray analysis.
Microarray-based microRNA profiling is an effective method for screening a large number of microRNAs (Lim et al., 2005; Chen et al., 2010). The development of a new microarray platform called miChip that accurately and sensitively monitors microRNAs expression was based on the use of modified nucleotides called locked nucleic acids (LNAs) (Castoldi et al., 2006). An LNA is a synthetic RNA analog and is capable of increasing the melting temperature of an LNA/RNA hybrid. As a consequence of this property, a uniform temperature can be applied to a genome-wide set of probes (Castoldi et al., 2007), allowing the establishment of normalized hybridization conditions. The microarray analysis was conducted using injured and control dorsal root ganglia from the 3 day model of sciatic nerve transection. As a result, significant changes in expression of 23 microRNAs, involving both downregulation and upregulation, were detected. Studies have shown that microRNA-21, microRNA-223, microRNA-455-5p, microRNA-431 and microRNA-18 were significantly increased, while microRNA-138, microRNA-483 and microRNA-383 were significantly decreased at 7 days following nerve transaction (Strickland et al., 2011). In another study, complete Freund's adjuvant-induced inflammation significantly reduced microRNA-1 and microRNA-16 expression in dorsal root ganglia, whereas microRNA-206 was downregulated in a time dependent manner. After sciatic nerve partial ligation, microRNA-1 and microRNA-206 were downregulated in dorsal root ganglia. On the other hand, axotomy increases the relative expression of microRNA-1, microRNA-16, and microRNA-206 in a time-dependent fashion (Kusuda et al., 2011). These results indicate that the microRNA profile in dorsal root ganglia was influenced by the type of injury or stimulus. Even for same injury model, variation in experimental conditions and temporal differences might also affect microRNA expression. These data provided an overview of microRNA expression in the early stages of neuronal injury.
To identify microRNAs that might target Slit-Robo GTPase-activating protein 3 and be associated with the axon guidance pathway, target prediction was carried out for the differentially expressed microRNAs from the microarray analysis. Kyoto Encyclopedia of Genes and Genomes pathway analysis was then conducted using the predicted target genes of each microRNA (Hua et al., 2009; Fei et al., 2012). Among the 23 microRNAs, we found that the axon guidance signaling pathway was most significantly enriched by the predicted targets of microRNA-214. Meanwhile, microRNA-214 was predicted to have one binding site in the 3′-UTR of Slit-Robo GTPase-activating protein 3. The microarray results showed that microRNA-214 was downregulated in dorsal root ganglia after sciatic nerve transection and quantitative reverse transcription-PCR confirmed this result. Subsequently, a luciferase reporter assay was conducted. Luciferase activity of psiCHECKTM-2 vectors co-transfected with the 3′-UTR of Slit-Robo GTPase-activating protein 3, and microRNA-214 was significantly decreased. This result indicated a specific interaction between microRNA-214 and the 3′-UTR of Slit-Robo GTPase-activating protein 3. Thus, Slit-Robo GTPase-activating protein 3 was confirmed to be a target of microRNA-214.
MicroRNAs in animals are transcribed as primary microRNAs (pri-miRs) and subsequently cleaved by a microprocessor complex consisting of the RNase III enzyme Drosha and the double-stranded RNA-binding protein DGCR8/Pasha (Han et al., 2006). This process yields a pre-miR that is then exported to the cytoplasm by Exportin-5 (Lund et al., 2004). In the cytoplasm, pre-miR is processed into a 21-23-nt mature microRNA duplex by the RNAse III enzyme Dicer (Bernstein et al., 2001; Hutvagner et al., 2001). Previous microarray and quantitative reverse transcription-PCR results showed a decrease in microRNA-214 expression in dorsal root ganglia. However, the cellular location of microRNA-214 was not known. Therefore, we determined the distribution of microRNA-214 in control dorsal root ganglia. Using in situ hybridization, we observed the existence of microRNA-214 in the predicted location: the cytoplasm of primary dorsal root ganglion neurons.
The microRNA pathway is an important layer of post- transcriptional gene regulation (Jackson et al., 2010). In the cytoplasm, mature microRNAs trigger formation of the RNA-induced silencing complex, which is implicated in a sequence-specific translational repression called RNA interference (Bagga et al., 2005). Interestingly, components of the RNA-induced silencing complex have been detected in severed sciatic nerve fibers and regenerating dorsal root ganglion axons in vitro (Murashov et al., 2007). A list of microRNAs residing within the distal axonal domain of superior cervical ganglia has recently been reported (Natera-Naranjo et al., 2010). Thus, current observations suggest that microRNAs may play an important regulatory role in peripheral nerve health even after development. In this study, though we did not acquire quantitative data, in situ hybridization identified microRNA-214 in axoplasm. More precise localization requires further confirmation, such as by electron microscopic in situ hybridization. It is well established that microRNAs are common and effective modulators of gene expression in many eukaryotic cells (Lykken and Li, 2011). However, only a small amount of work has been published regarding the expression of microRNAs in dorsal root ganglia after nerve injury. MicroRNA-96, microRNA-182 and microRNA-183 showed a small but significant downregulation in ipsilateral dorsal root ganglia after sciatic nerve ligation (Aldrich et al., 2009). MicroRNA-21 was upregulated by 7-fold in dorsal root ganglia at 7 days post-axotomy, and it directly downregulated the expression of Sprouty2 (Strickland et al., 2011). To investigate the expression change of microRNA-214 and its target Slit-Robo GTPase-activating protein 3 in dorsal root ganglion neurons following sciatic nerve transection, in situ hybridization, immunohistochemistry and western bolt assays were conducted. MicroRNA-214 was significantly downregulated at 1–7 days after injury. Interestingly, consistent with our previous hypothesis, Slit-Robo GTPase-activating protein 3 was significantly upregulated at 1 day after injury and kept increasing thereafter. The inverse change in levels of microRNA-214 and its target Slit-Robo GTPase-activating protein 3 indicate de-repression of Slit-Robo GTPase-activating protein 3 in dorsal root ganglia following sciatic nerve transection.
Moreover, intracellular co-expression of microRNA-214 and Slit-Robo GTPase-activating protein 3 was identified using a combination of in situ hybridization and immunohistochemistry. In control dorsal root ganglion neurons with high levels of microRNA-214, Slit-Robo GTPase-activating protein 3 levels were low, and vice versa. The possible inverse correlation between microRNA-214 and Slit-Robo GTPase-activating protein 3 was also shown in dorsal root ganglion neurons 3 days after injury with downregulation of microRNA-214 and upregulation of Slit-Robo GTPase-activating protein 3 in most injured dorsal root ganglion neurons. An explanation for the inverse expression of microRNA-214 and Slit-Robo GTPase-activating protein 3 is that there is intracellular repression of Slit-Robo GTPase-activating protein 3 under physiological conditions, while the trauma-induced de-repression is both intracellular and intercellular.
The neuronal repair mechanism often involves molecules that govern early neural development (Costigan et al., 2002). As revealed by in situ hybridization, expression of the three members of the Slit-Robo GTPase-activating protein family in the rat brain generally overlaps with that of Robo1 and/or Robo2 receptors (Whitford et al., 2002). Robo2 is upregulated in dorsal root ganglia after peripheral nerve injury (Yi et al., 2006), and co-localization and parallel increases of Robo2 and Slit-Robo GTPase-activating protein 3 are found in dorsal root ganglia after sciatic nerve transection. These findings suggest that members of the Slit-Robo pathway may synergistically participate in a regenerating response to nerve injury. Given the inverse relationship between Slit-Robo GTPase-activating protein 3 expression and microRNA-214 expression, we suggest that microRNA-214 may participate in neural regeneration by regulating Slit-Robo GTPase-activating protein 3, a member of the Slit-Robo signaling pathway. The present study demonstrates that sciatic nerve transection results in a downregulation of several microRNAs, particularly microRNA-214. Bioinformatic analysis predicted that the 3′-UTRs of Slit-Robo GTPase-activating protein 3 may be a target of microRNA-214.
Together with the observation of upregulation of Slit-Robo GTPase-activating protein 3 expression in dorsal root ganglia after sciatic nerve transection, we suggest that the injury-induced downregulation of microRNA-214 may be the post injury response of dorsal root ganglia sensory neurons leading to a negative modulation of Slit-Robo GTPase-activating protein 3 translation, although the underlying mechanism has yet to be determined.
Footnotes
Funding: This work was supported by the National Natural Science Foundation of China, No. 81160158 and 30860290.
Conflicts of interest: None declared.
Copyedited by Allen J, Robens J, Yu J, Qiu Y, Li CH, Song LP, Zhao M
References
- 1.Adilakshmi T, Sudol I, Tapinos N. Combinatorial action of miR-NAs regulates transcriptional and post-transcriptional gene silencing following in vivo PNS Injury. PLoS One. 2012;7:e39674. doi: 10.1371/journal.pone.0039674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience. 2009;164:711–723. doi: 10.1016/j.neuroscience.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranha MM, Santos DM, Xavier JM, Low WC, Steer CJ, Sola S, Rodrigues CM. Apoptosis-associated micro RNAs are modulated in mouse, rat and human neural differentiation. BMC Genomics. 2010;11:514. doi: 10.1186/1471-2164-11-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arevalo MA, Rodriguez-Tebar A. Activation of casein kinase II and inhibition of phosphatase and tensin homologue deleted on chromosome 10 phosphatase by nerve growth factor/p75NTR inhibit glycogen synthase kinase-3beta and stimulate axonal growth. Mol Biol Cell. 2006;17:3369–3377. doi: 10.1091/mbc.E05-12-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atwal JK, Pinkston-Gosse J, Syken J, Stawicki S, Wu Y, Shatz C, Tessier-Lavigne M. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- 6.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Bashaw GJ, Goodman CS. Chimeric axon guidance receptors: the cytoplasmic domains of slit and netrin receptors specify attraction versus repulsion. Cell. 1999;97:917–926. doi: 10.1016/s0092-8674(00)80803-x. [DOI] [PubMed] [Google Scholar]
- 8.Bergman E, Carlsson K, Liljeborg A, Manders E, Hokfelt T, Ulfhake B. Neuropeptides, nitric oxide synthase and GAP-43 in B4-binding and RT97 immunoreactive primary sensory neurons: normal distribution pattern and changes after peripheral nerve transection and aging. Brain Research. 1999;832:63–83. doi: 10.1016/s0006-8993(99)01469-9. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Bertram J, Peacock JW, Tan C, Mui AL, Chung SW, Gleave ME, Dedhar S, Cox ME, Ong CJ. Inhibition of the phosphatidylinositol 3’-kinase pathway promotes autocrine Fas-induced death of phosphatase and tensin homologue-deficient prostate cancer cells. Cancer Res. 2006;66:4781–4788. doi: 10.1158/0008-5472.CAN-05-3173. [DOI] [PubMed] [Google Scholar]
- 11.Bhatt DH, Otto SJ, Depoister B, Fetcho JR. Cyclic AMP-induced repair of zebrafish spinal circuits. Science. 2004;305:254–258. doi: 10.1126/science.1098439. [DOI] [PubMed] [Google Scholar]
- 12.Boyd JG, Gordon T. Glial cell line-derived neurotrophic factor and brain-derived neurotrophic factor sustain the axonal regeneration of chronically axotomized motoneurons in vivo. Exp Neurol. 2003;183:610–619. doi: 10.1016/s0014-4886(03)00183-3. [DOI] [PubMed] [Google Scholar]
- 13.Castoldi M, Benes V, Hentze MW, Muckenthaler MU. miChip: a microarray platform for expression profiling of microRNAs based on locked nucleic acid (LNA) oligonucleotide capture probes. Methods. 2007;43:146–152. doi: 10.1016/j.ymeth.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) Rna. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Qian K, Tang ZP, Xing B, Liu N, Huang X, Zhang S. Bioinformatics and microarray analysis of microRNA expression profiles of murine embryonic stem cells, neural stem cells induced from ESCs and isolated from E8.5 mouse neural tube. Neurol Res. 2010;32:603–613. doi: 10.1179/174313209X455691. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZB, Zhang HY, Zhao JH, Zhao W, Zhao D, Zheng LF, Zhang XF, Liao XP, Yi XN. Slit-Robo GTPase-activating proteins are differentially expressed in murine dorsal root ganglia: modulation by peripheral nerve injury. Anat Rec (Hoboken) 2012;295:652–660. doi: 10.1002/ar.22419. [DOI] [PubMed] [Google Scholar]
- 17.Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30:9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fei J, Li Y, Zhu X, Luo X. miR-181a post-transcriptionally downregulates oncogenic RalA and contributes to growth inhibition and apoptosis in chronic myelogenous leukemia (CML) PLoS One. 2012;7:e32834. doi: 10.1371/journal.pone.0032834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 23.Hua YJ, Tang ZY, Tu K, Zhu L, Li YX, Xie L, Xiao HS. Identification and target prediction of miRNAs specifically expressed in rat neural tissue. BMC Genomics. 2009;10:214. doi: 10.1186/1471-2164-10-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 26.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi NR, Fan DP, Giehl KM, Bedard AM, Wiegand SJ, Tetzlaff W. BDNF and NT-4/5 prevent atrophy of rat rubrospinal neurons after cervical axotomy, stimulate GAP-43 and Talpha1-tubulin mRNA expression, and promote axonal regeneration. J Neurosci. 1997;17:9583–9595. doi: 10.1523/JNEUROSCI.17-24-09583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G. Differential expression of microRNAs in mouse pain models. Molecular Pain. 2011;7:17. doi: 10.1186/1744-8069-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau P, Hudson LD. MicroRNAs in neural cell differentiation. Brain Research. 2010;1338:14–19. doi: 10.1016/j.brainres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 31.Lin L, Rao Y, Isacson O. Netrin-1 and slit-2 regulate and direct neurite growth of ventral midbrain dopaminergic neurons. Mol Cell Neurosci. 2005;28:547–555. doi: 10.1016/j.mcn.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Pahlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci U S A. 2010;107:1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X, Richardson PM. Inflammation near the nerve cell body enhances axonal regeneration. J Neurosci. 1991;11:972–978. doi: 10.1523/JNEUROSCI.11-04-00972.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 35.Lykken EA, Li QJ. microRNAs at the regulatory frontier: an investigation into how microRNAs impact the development and effector functions of CD4 T cells. Immunol Res. 2011;49:87–96. doi: 10.1007/s12026-010-8196-4. [DOI] [PubMed] [Google Scholar]
- 36.MacLaren RE. Expression of myelin proteins in the opossum optic nerve: late appearance of inhibitors implicates an earlier non-myelin factor in preventing ganglion cell regeneration. J Comp Neurol. 1996;372:27–36. doi: 10.1002/(SICI)1096-9861(19960812)372:1<27::AID-CNE3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 37.Ming GL, Song HJ, Berninger B, Holt CE, Tessier-Lavigne M, Poo MM. cAMP-dependent growth cone guidance by netrin-1. Neuron. 1997;19:1225–1235. doi: 10.1016/s0896-6273(00)80414-6. [DOI] [PubMed] [Google Scholar]
- 38.Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murashov AK, Chintalgattu V, Islamov RR, Lever TE, Pak ES, Sierpinski PL, Katwa LC, Van Scott MR. RNAi pathway is functional in peripheral nerve axons. Faseb J. 2007;21:656–670. doi: 10.1096/fj.06-6155com. [DOI] [PubMed] [Google Scholar]
- 40.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. Rna. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, Krupsky M, Ben-Dov I, Cazacu S, Mikkelsen T, Brodie C, Eisenberg E, Rechavi G. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prinjha R, Moore SE, Vinson M, Blake S, Morrow R, Christie G, Michalovich D, Simmons DL, Walsh FS. Inhibitor of neurite outgrowth in humans. Nature. 2000;403:383–384. doi: 10.1038/35000287. [DOI] [PubMed] [Google Scholar]
- 43.Schwaiger FW, Hager G, Schmitt AB, Horvat A, Streif R, Spitzer C, Gamal S, Breuer S, Brook GA, Nacimiento W, Kreutzberg GW. Peripheral but not central axotomy induces changes in Janus kinases (JAK) and signal transducers and activators of transcription (STAT) Eur J Neurosci. 2000;12:1165–1176. doi: 10.1046/j.1460-9568.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- 44.Strickland IT, Richards L, Holmes FE, Wynick D, Uney JB, Wong LF. Axotomy-induced miR-21 promotes axon growth in adult dorsal root ganglion neurons. PLoS One. 2011;6:e23423. doi: 10.1371/journal.pone.0023423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tam Tam S, Bastian I, Zhou XF, Vander Hoek M, Michael MZ, Gibbins IL, Haberberger RV. MicroRNA-143 expression in dorsal root ganglion neurons. Cell Tissue Res. 2011;346:163–173. doi: 10.1007/s00441-011-1263-x. [DOI] [PubMed] [Google Scholar]
- 46.Tomita K, Hata Y, Kubo T, Fujiwara T, Yano K, Hosokawa K. Effects of the in vivo predegenerated nerve graft on early Schwann cell migration: quantitative analysis using S100-GFP mice. Neurosci Lett. 2009;461:36–40. doi: 10.1016/j.neulet.2009.05.075. [DOI] [PubMed] [Google Scholar]
- 47.Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chedotal A, Ghosh A. Regulation of cortical dendrite development by Slit-Robo interactions. Neuron. 2002;33:47–61. doi: 10.1016/s0896-6273(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 48.Woolf CJ, Reynolds ML, Molander C, O’Brien C, Lindsay RM, Benowitz LI. The growth-associated protein GAP-43 appears in dorsal root ganglion cells and in the dorsal horn of the rat spinal cord following peripheral nerve injury. Neuroscience. 1990;34:465–478. doi: 10.1016/0306-4522(90)90155-w. [DOI] [PubMed] [Google Scholar]
- 49.Wu D, Raafat M, Pak E, Hammond S, Murashov AK. MicroRNA machinery responds to peripheral nerve lesion in an injury-regulated pattern. Neuroscience. 2011;190:386–397. doi: 10.1016/j.neuroscience.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 51.Yi XN, Zheng LF, Zhang JW, Zhang LZ, Xu YZ, Luo G, Luo XG. Dynamic changes in Robo2 and Slit1 expression in adult rat dorsal root ganglion and sciatic nerve after peripheral and central axonal injury. Neurosci Res. 2006;56:314–321. doi: 10.1016/j.neures.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 52.Zhang HY, Zheng LF, Yi XN, Chen ZB, He ZP, Zhao D, Zhang XF, Ma ZJ. Slit1 promotes regenerative neurite outgrowth of adult dorsal root ganglion neurons in vitro via binding to the Robo receptor. J Chem Neuroanat. 2010;39:256–261. doi: 10.1016/j.jchemneu.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Zhou S, Yu B, Qian T, Yao D, Wang Y, Ding F, Gu X. Early changes of microRNAs expression in the dorsal root ganglia following rat sciatic nerve transection. Neurosci Lett. 2011;494:89–93. doi: 10.1016/j.neulet.2011.02.064. [DOI] [PubMed] [Google Scholar]


