Abstract
The apolipoprotein E gene ε4 allele is considered a negative factor for neural regeneration in late-onset Alzheimer's disease cases. The aim of this study was to establish a non-invasive, rapid method to genotype apolipoprotein E gene polymorphisms. Genomic DNA from mouth swab specimens was extracted using magnetic nanoparticles, and genotyping was performed by real-time PCR using TaqMan-BHQ probes. Genotyping accuracy was validated by DNA sequencing. Our results demonstrate 100% correlation to DNA sequencing, indicating reliability of our protocol. Thus, the method we have developed for apolipoprotein E genotyping is accurate and reliable, and also suitable for genotyping large samples, which may help determine the role of the apolipoprotein E ε4 allele in neural regeneration in late-onset Alzheimer's disease cases.
Keywords: nerve regeneration, neurodegeneration, late-onset Alzheimer's disease, apolipoprotein E gene, real-time PCR, DNA sequencing, risk factor, allele, neural regeneration
Introduction
Alzheimer's disease is the most common form of dementia related to aging, and is a progressive neurodegenerative disease affecting millions of people worldwide[1,2]. Clinically, Alzheimer's disease presents with anterograde episodic memory impairments, intellectual disturbances, language problems, and decline in other cognitive domains[3,4]. Neuropathologically, Alzheimer's disease is characterized by extracellular senile plaques (formed by aggregates of cleaved β-amyloid (Aβ) protein), intracellular neurofibrillary tangles (mainly composed of hyperphosphorylated tau protein), and neuronal loss[5,6,7,8]. Alzheimer's disease is typically divided into early-onset and late-onset forms. Early-onset Alzheimer's disease is rare, whereas late-onset Alzheimer's disease (with an onset age of over 65) accounts for approximately 95% of all Alzheimer's disease cases[9,10,11]. Early-onset Alzheimer's disease can be identified by genetic mutations in amyloid precursor protein, presenilin-1, and presenilin-2. Late-onset Alzheimer's disease lacks obvious genetic mutations[12,13,14], however many studies have shown that apolipoprotein E (APOE) ε4 gene is the most important known genetic risk factor[15,16]. Thus, is APOE ε4 a genetic maker of late-onset Alzheimer's disease?
APOE is polymorphic, and encodes a 34.2 kDa glycosylated protein, APOE[17]. APOE is located on chromosome 19q13.2, and comprised of four exons and three introns, covering 3,597 bp[18,19]. The two APOE single nucleotide polymorphisms associated with Alzheimer's disease are rs429358 and rs7412. The three common variants resulting from these polymorphisms are ε2 (rs429358, T; rs7412, T), ε3 (rs429358, T; rs7412, C) and ε4 (rs429358, C; rs7412, C). In humans, these variants result in three homozygous (ε2/ε2,ε3/ε3, and ε4/ε4), and three heterozygous (ε2/ε3, ε2/ε4, and ε3/ε4) phenotypes. APOE ε2, ε3, and ε4 determine the three major protein isoforms of APOE, specifically, APOE2, E3, and E4[20,21,22]. On the mature APOE polypeptide chain, APOE2 has cysteine residues at positions 112 and 158, APOE3 has a cysteine and arginine residue at positions 112 and 158, respectively, and APOE4 has arginine residues at both sites. Despite differing by only one or two amino acids at residues 112 and/or 158, the structure and function of APOE protein isoforms are profoundly altered[23,24,25]. Carriers of one or two APOE ε4 alleles have a three- or eleven-fold, respectively, increased risk for late-onset Alzheimer's disease[26]. As neuronal loss is one of the pathological characteristics of late-onset Alzheimer's disease, in recent years emphasis has been placed on neural regeneration research. The APOE ε4 allele is considered a negative factor for neural regeneration[27,28]. APOE4 exhibits greater neurotoxicity than APOE2 and APOE3, and affects neural regeneration mainly by generating neurotoxic fragments that lead to pathological mitochondrial dysfunction and cytoskeletal collapse[29]. However, the exact mechanism whereby the APOE ε4 allele inhibits neural regeneration in late-onset Alzheimer's disease is not completely understood[30,31].
APOE genotyping is crucial to APOE polymorphism analysis. Peripheral venous blood is the conventional tissue source for APOE polymorphism analysis[32,33]. Blood yields high-quality genomic DNA and can meet various research purposes. However, because of invasiveness, taking blood samples decreases compliance among the elderly, especially neuropsychiatric patients[34,35,36]. Moreover, blood specimens often need cold storage, thereby increasing the cost. In contrast, buccal mucosa sampling through mouth swabs is non-invasive and generates high-quality genomic DNA for single nucleotide polymorphism genotyping. Specimens can be stored at room temperature[37,38,39,40]. Ilveskoski et al.[41] reported successful APOE genotyping from buccal swabs using the restriction enzyme HhaI, although the process was labor- and time-consuming (taking a total of ten hours). Magnetic nanoparticles are novel materials for genomic DNA isolation in molecular biology. Wang and Su[42] have shown magnetic nanoparticles effectively and rapidly enrich trace amounts of DNA. Real-time PCR is a high-throughput method for single nucleotide polymorphism analysis[33]. Thus, we aimed to establish a method for genomic DNA extraction from mouth swab specimens using magnetic nanoparticles coupled with APOE genotyping by real-time PCR using TaqMan-BHQ probes.
Results
Rapid APOE genotyping by real-time PCR
Real-time PCR amplification took 50 minutes. The whole procedure was operated in closed tubes. After amplification, genotypes were automatically read by the Endpoint Genotyping module. Representative rs429358 and rs7412 genotyping results are shown (Figure 1).
Figure 1.
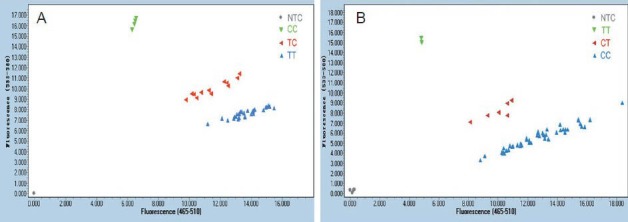
Results of rs429358 (A) and rs7412 (B) genotypings using the Roche LightCycler 480II system.
NTC: Negative template control.
APOE genotyping by DNA sequencing
DNA sequencing was performed by the Beijing Genomics Institute in China, using the ABI3730 DNA Sequencer (Applied Biosystems). Typical results are presented (Figures 2, 3).
Figure 2.
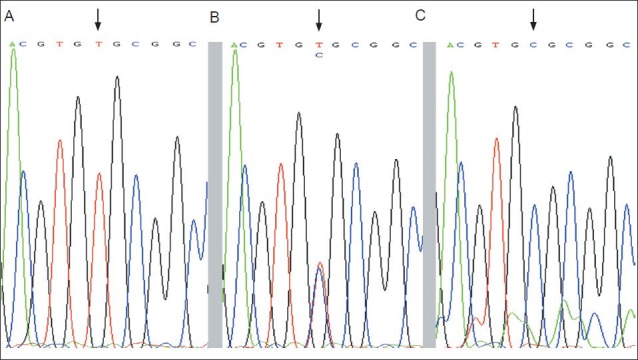
Screenshots of rs429358 genotyping obtained by DNA sequencing.
(A) rs429358 T/T; (B) rs429358 C/T; (C) rs429358 C/C. Arrows show polymorphic sites.
Figure 3.
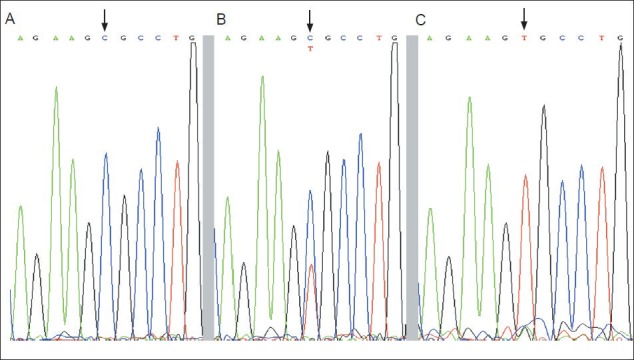
Screenshots of rs7412 genotyping obtained by DNA sequencing.
(A) rs7412 C/C; (B) rs7412 C/T; (C) rs7412 T/T. Arrows show polymorphic sites.
Genotype analysis of the APOE ε4 allele, a risk factor for late-onset Alzheimer's disease
APOE genotyping by real-time PCR using TaqMan-BHQ probes, showed 100% correlation with DNA sequencing results, demonstrating reliability of our protocol. Genotype and allele frequencies, and Hardy-Weinberg equilibrium tests are summarized (Table 1). Both cases and controls were in Hardy-Weinberg equilibrium (P > 0.05), indicating our subjects were genetically randomly selected. APOE genotype distributions between cases and controls were significantly different (P = 0.004). The APOE ε4 allele was associated with high risk (P = 0.001; odds ratio (OR) 3.958, 95% confidence interval (CI) 1.681–9.319) for developing late-onset Alzheimer's disease, supporting the APOE ε4 allele as a risk factor for late-onset Alzheimer's disease[16] (Tables 2, 3).
Table 1.
Genotype and allele frequencies of rs429358 and rs7412 and Hardy-Weinberg equilibrium (HWE) tests in cases and controls
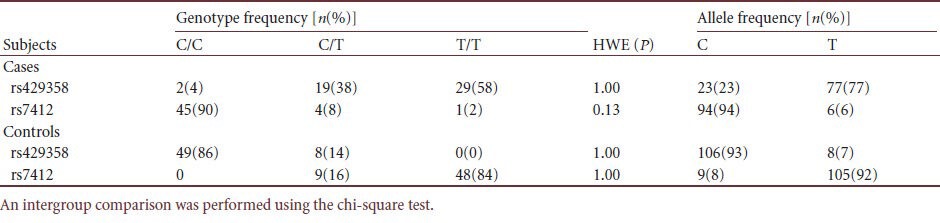
Table 2.
APOE phenotype distribution in cases and controls
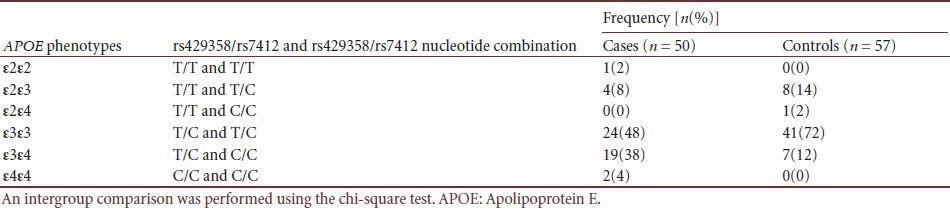
Table 3.
APOE genotype distribution in cases and controls
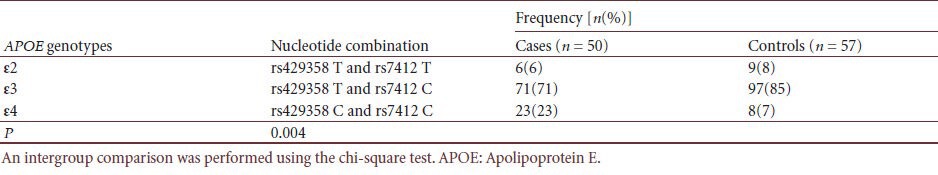
Discussion
The APOE ε4 allele is well known to play an important role in the pathological processes of late-onset Alzheimer's disease, in particular, Aβ generation or deposition, neurofibrillary tangle formation, lipid homeostasis, neuronal survival, and intracellular signal transduction[43,44,45]. In the central nervous system, APOE is considered important for repairing and maintaining myelin and neuronal membranes during growth and injury[46,47]. Of the three isoforms, APOE4 is the least stable, and less effective at accomplishing these functions. The APOE ε4 isoform shows higher affinity for binding Aβ and stabilizing amyloid fibrils, thereby enhancing Aβ accumulation in plaques. Late-onset Alzheimer's disease patients with one or two APOE ε4 alleles have more senile plaques and Aβ deposition[48,49,50,51]. APOE plays a fundamental role in regulating transport of cholesterol and phospholipids among cells[52]. APOE ε4 genotypes are associated with less APOE protein in plasma. Plasma low density lipoprotein levels are higher in APOE ε4 allele carriers[53,54,55,56]. Cholinergic signal transduction is impaired in the majority of late-onset Alzheimer's disease patients. Among these patients, APOE ε4 allele carriers have greater deficits in cholinergic activity in the hippocampus and cortex, and fewer cholinergic neurons[43,44].
APOE ε4 allele effects on neural regeneration in late-onset Alzheimer's disease cases are complex and can be summarized as follows: (1) APOE4 inhibits neurite outgrowth; (2) APOE4 leads to mitochondrial dysfunction including membrane potential alterations, mitochondrial motility reductions, and mitochondrial respiratory enzymes dysfunction; (3) APOE4 impairs the cytoskeleton and causes tau hyperphosphorylation; (4) APOE4 inhibits synaptogenesis, increases Aβ and lysosomal leakage, and causes neuronal apoptosis; and (5) animal experiments show APOE4 memory and learning impairments in mice[29,57].
Much remains to be determined to explain APOE ε4 allele effects on neural regeneration in late-onset Alzheimer's disease cases. Thus, APOE genotyping of large samples is indispensable. Besides compliance, efficiency and cost of sample collection need to be considered when screening large samples. Thomson and colleagues[58] recommended buccal cells as the first choice for PCR DNA analysis.
In our study, 107 mouth swab specimens were collected. The process of specimen collection was non-invasive and no discomfort was reported. The procedure of collecting mouth swab specimens was easy to manage, with no specialized facilities or even skilled collection staff required. It took only several seconds to collect one mouth swab specimen. Compared with blood specimens, the cost of mouth swab specimen collection was reduced. According to the manufacturer, one sterile mouth swab costs < 0.1 dollar. No anticoagulant was needed and specimens could be stored at room temperature for more than two weeks.
We used magnetic nanoparticles to extract genomic DNA from the mouth swab samples. Compared with traditional human genomic DNA extraction kits, magnetic nanoparticles have several advantages. They are highly efficient at enriching genomic DNA because of their large specific surface area, which can notably enlarge the reaction interface. Due to their superparamagnetic effects, magnetic nanoparticles are easy to use, enabling them to be moved and isolated with external magnetic fields supplied by magnetic stands or common magnetic irons[59]. Furthermore, magnetic nanoparticles degenerate easily, supporting environmental protection. In our assay, the time spent in extracting genomic DNA from one mouth swab specimen was just one hour.
PCR-restriction fragment length polymorphism analysis is the most common method applied to genotype APOE polymorphisms[60,61]. However, PCR-restriction fragment length polymorphism analysis is not an effective method, as it requires a number of reaction steps (i.e., PCR, overnight enzyme digestion, electrophoresis, and ultraviolet irradiation). Moreover, it is prone to insufficient restriction enzyme digestion resulting in inaccurate results. DNA sequencing is the most accurate method for APOE genotyping, but it is labor-consuming and requires expensive detection equipment that small laboratories can often not afford. Real-time PCR is a time- and labor-saving, high-throughput protocol for single nucleotide polymorphism analysis. To date, SYBR Green, hybridization probes, and TaqMan-MGB probes have been used in real-time PCR for APOE genotyping[32,33]. SYBR Green, a double stranded DNA-fluorescent dye, is inexpensive, however, it is not specific enough and tends to be affected by primer-dimer formation[61]. Hybridization and TaqMan-MGB probes are highly specific, although high price restricts their application. TaqMan-BHQ is a specific fluorescence probe that is cheaper than hybridization or TaqMan-MGB probes. TaqMan-BHQ probe application shows an increasing trend in recent years, especially in small laboratories. Our assay demonstrates that TaqMan-BHQ probes are accurate and reliable for APOE genotyping, with confirmation of our results by PCR with DNA sequencing. Genotyping with TaqMan-BHQ probes requires no post-PCR sample handling, reducing the chance of sample contamination and mix-up. We used negative template controls for real-time PCR amplification, and observed no contamination. Samples were rechecked on different days, with comparable results, showing good assay reproducibility. A single real-time PCR reaction took approximately 50 minutes, and the Roche LightCycler 480 II system has the potential to run 384 reactions at one time.
In conclusion, our method for APOE genotyping is non-invasive, fast, and economical, with the potential for high-throughput application. It is suitable for screening APOE ε4 allele carriers among large samples, which may help elucidate the influence of the APOE ε4 allele in late-onset Alzheimer's disease cases.
Subjects and Methods
Design
A novel test method for clinical research.
Time and setting
Experiments were performed in the Central Laboratory of Peking University Shenzhen Hospital, China from July 2011 to April 2012.
Subjects
A total of 50 late-onset Alzheimer's disease cases (mean age 74.2 ± 5.8 years, 24 males and 26 females), diagnosed according to criteria of the National Institute of Neurological and Communicative Disorders and Stroke, and the Alzheimer's Diseases and Relative Disorders Associations[62], and 57 age-matched healthy controls (mean age 72.98 ± 4.76 years, 29 males and 28 females), were recruited from the in- and out-patient departments of the Center of Health Examination, Peking University Shenzhen Hospital. All participants were from the Han Chinese population. Written informed consent was obtained from subjects or surrogates. The study was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Peking University Shenzhen Hospital, China.
Methods
Preparation of mouth swab specimens
Sterile mouth swabs (Jiangsu KangJian Medical Apparatus, Jiangyan, Jiangsu Province, China) were used to collect cheek epithelial cells from subjects. The inside of both cheeks was rubbed and swabs were swirled five times. Tips of mouth swab specimens were cut into 1.5 mL microcentrifuge tubes and stored at room temperature until DNA extraction.
Human genomic DNA extraction
The Magnetic Nanoparticles DNA Extraction Kit (Wawasye Nanotech, Wuhan, Hubei Province, China), including magnetic nanoparticles, and lysis, binding, washing, and elution buffers, was used. Lysis buffer (200 µL) was added to mouth swab specimens and vortexed for seconds. Tubes were centrifuged briefly at 12,000 × g at room temperature, and then incubated at 80°C for 30 minutes. Tubes were again briefly centrifuged at 12,000 × g and supernatants transferred to clean microcentrifuge tubes. Magnetic nanoparticles (20 µL) were added and mixed by inversion 20 times. Mixtures were then incubated at room temperature for 10 minutes. Tubes were again briefly centrifuged and supernatants discarded. Tubes were placed into a magnetic stand to preserve magnetic nanoparticles when discarding supernatants. Washing buffer (100 µL) was added and mixed by inversion 20 times. Supernatants were discarded using the magnetic stand. The washing procedure was repeated, and magnetic nanoparticles permitted to air dry for five minutes. Following this, elution buffer (50 µL) was added and incubated at 70°C for 10 minutes. After brief centrifugation, supernatants (including genomic DNA) were transferred to clean microcentrifuge tubes and stored at −20°C until use.
APOE genotyping by real-time PCR
Primer and probe sequences for rs429358 genotyping were designed by Shanghai Genecore Biotechnologies (Genecore, Shanghai, China), and were: forward primer 5′-ACC TCG CCG CGG TAC TG-3′, reverse primer 5′-GGG CAC GGC TGT CCA A-3′; TaqMan probe pair 5′-FAM-CGG CCG CAC ACG TCC TCC-BHQ-3′, 5′-HEX-CGG CCG CGC ACG TCC T-BHQ-3′. For rs7412 genotyping, sequences according to Koch et al.[33] were used: forward primer 5′-CGC GGC CCT GTT CCA-3′, reverse primer 5′-CTC CGC GAT GCC GAT G-3′; TaqMan probe pair 5′-FAM-ACT GCC AGG CGC TTC TGC AGG-BHQ-3′, 5′-HEX-CAC TGC CAG GCA CTT CTG CAG GT-BHQ-3′. All primers and probes were synthesized and purified by Genecore.
Real-time PCR amplification reactions for rs429358 and rs7412 genotyping were performed using the Roche LightCycler 480II system (Roche Diagnostics, Basel, Switzerland). Reactions contained 5 µL of 2 × premix Ex Taq (TaKaRa, Dalian, Liaoning Province, China), 2.5 µmol/L of each primer, 1.25 µmol/L of each TaqMan probe, and 2 µL of genomic DNA. Sterile water was added to a final volume of 10 µL. Negative template controls, using sterile water instead of genomic DNA, were included in every amplification. Cycling parameters were: an initial denaturation at 95°C for 30 seconds, followed by 40 cycles of denaturation at 95°C for 5 seconds, annealing and extension at 60°C for 31 seconds, and finally cooling to 4°C. After amplification, genotypes were automatically determined by the Roche LightCycler 480 service software, equipped with the Endpoint Genotyping module.
DNA sequencing
DNA sequencing was performed to validate APOE genotyping obtained by real-time PCR. The primer pair sequences (5′-GGG CAC GGC TGT CCA A-3′ and 5′-CGC GGC CCT GTT CCA-3′) used, produced a 300 bp fragment encompassing rs429358 and rs7412, and were designed and synthesized by Genecore. PCR amplification reactions were performed using the ABI 2720 Thermal Cycler (Applied Biosystems, CA, USA). PCR reaction volumes were 25 µL, and included 12.5 µL of 2 × premix Ex Taq (TaKaRa), 5 µmol/L of each primer, 2 µL of genomic DNA, and 9.5 µL of sterile water. Appropriate negative controls were included in each run. Amplification conditions were: an initial denaturation at 95°C for 30 seconds, followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 18 seconds.
Southern blots were performed to verify amplification of the expected 300 bp APOE products. PCR products (5 µL) were mixed with 6 × loading buffer (1 µL) (TaKaRa) and electrophoresed on 2% agarose gels. Amplicons were measured using the DL1000 molecular mass marker (TaKaRa). PCR products of 300 bp were sent to Beijing Genomics Institute (Shenzhen, Guangdong Province, China) for DNA sequencing.
Statistical analysis
Genotype distributions and allele frequencies of rs429358 and rs7412 were obtained by direct counting. To determine if genotype distributions were in Hardy-Weinberg equilibrium, tests were performed using R[63]. Other analyses were performed using SPSS 13.0 software (SPSS, Chicago, IL, USA). Genotype frequencies between two groups, were compared using chi-square tests and assessment of APOE ε4 allele risk for late-onset Alzheimer's disease determined. P ≤ 0.05 was considered statistically significant.
Footnotes
Funding: This work was supported by two grants from Science, Industry, Trade and Information Technology Commission of Shenzhen Municipality in China, grant No. 201002063, JC20110518075 7A.
Conflicts of interest: None declared.
Peer review: A high-sensitivity, non-invasive, fast and economical method to genotype APOE gene polymorphisms by collecting buccal mucosa epithelial cells from cases with late-onset Alzheimer's disease has been established, which is suitable for clinical research.
Copyedited by Phillips A, Li KY, Wang X, Yu J, Qiu Y, Li CH, Song LP, Liu WJ, Zhao M
References
- [1].Brookmeyer R, Johnson E, Ziegler-Graham K, et al. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- [2].Lauderback CM, Kanski J, Hackett JM, et al. Apolipoprotein E modulates Alzheimer's Abeta(1-42)-induced oxidative damage to synaptosomes in an allele-specific manner. Brain Res. 2002;924(1):90–97. doi: 10.1016/s0006-8993(01)03228-0. [DOI] [PubMed] [Google Scholar]
- [3].Nussbaum RL, Ellis CE. Alzheimer's disease and Parkinson's disease. N Engl J Med. 2003;348(14):1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- [4].Raffai RL, Dong LM, Farese RV, Jr, et al. Introduction of human apolipoprotein E4 “domain interaction” into mouse apolipoprotein E. Proc Natl Acad Sci U S A. 2001;98(20):11587–11591. doi: 10.1073/pnas.201279298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nelson PT, Braak H, Markesbery WR. Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol. 2009;68(1):1–14. doi: 10.1097/NEN.0b013e3181919a48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li L, Thompson PA, Kitchens RL. Infection induces a positive acute phase apolipoprotein E response from a negative acute phase gene: role of hepatic LDL receptors. J Lipid Res. 2008;49(8):1782–1793. doi: 10.1194/jlr.M800172-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Laffont I, Takahashi M, Shibukawa Y, et al. Apolipoprotein E activates Akt pathway in neuro-2a in an isoform-specific manner. Biochem Biophys Res Commun. 2002;292(1):83–87. doi: 10.1006/bbrc.2002.6586. [DOI] [PubMed] [Google Scholar]
- [8].Huang Y. Abeta-independent roles of apolipoprotein E4 in the pathogenesis of Alzheimer's disease. Trends Mol Med. 2010;16(6):287–294. doi: 10.1016/j.molmed.2010.04.004. [DOI] [PubMed] [Google Scholar]
- [9].Luchsinger JA, Mayeux R. Cardiovascular risk factors and Alzheimer's disease. Curr Atheroscler Rep. 2004;6(4):261–266. doi: 10.1007/s11883-004-0056-z. [DOI] [PubMed] [Google Scholar]
- [10].Lahiri DK. Apolipoprotein E as a target for developing new therapeutics for Alzheimer's disease based on studies from protein, RNA, and regulatory region of the gene. J Mol Neurosci. 2004;23(3):225–233. doi: 10.1385/JMN:23:3:225. [DOI] [PubMed] [Google Scholar]
- [11].Kline A. Apolipoprotein E, amyloid-ß clearance and therapeutic opportunities in Alzheimer's disease. Alzheimers Res Ther. 2012;4(4):32. doi: 10.1186/alzrt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ferri CP, Prince M, Brayne C, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366(9503):2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lasser RA, Dukoff R, Levy J, et al. Apolipoprotein E epsilon 4 allele in association with global cognitive performance and CSF markers in Alzheimer's disease. Int J Geriatr Psychiatry. 1998;13(11):767–774. doi: 10.1002/(sici)1099-1166(1998110)13:11<767::aid-gps866>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- [14].Ewers M, Zhong Z, Bürger K, et al. Increased CSF-BACE 1 activity is associated with ApoE-epsilon 4 genotype in subjects with mild cognitive impairment and Alzheimer's disease. Brain. 2008;131(Pt 5):1252–1258. doi: 10.1093/brain/awn034. [DOI] [PubMed] [Google Scholar]
- [15].Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- [16].Hampel H, Bürger K, Teipel SJ, et al. Core candidate neurochemical and imaging biomarkers of Alzheimer's disease. Alzheimers Dement. 2008;4(1):38–48. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]
- [17].Rall SC, Jr, Weisgraber KH, Innerarity TL, et al. Structural basis for receptor binding heterogeneity of apolipoprotein E from type III hyperlipoproteinemic subjects. Proc Natl Acad Sci U S A. 1982;79(15):4696–4700. doi: 10.1073/pnas.79.15.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63(3):287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brown CM, Wright E, Colton CA, et al. Apolipoprotein E isoform mediated regulation of nitric oxide release. Free Radic Biol Med. 2002;32(11):1071–1075. doi: 10.1016/s0891-5849(02)00803-1. [DOI] [PubMed] [Google Scholar]
- [20].Agosta F, Vossel KA, Miller BL, et al. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer's disease and frontotemporal dementia. Proc Natl Acad Sci U S A. 2009;106(6):2018–2022. doi: 10.1073/pnas.0812697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Thal DR, Rüb U, Orantes M, et al. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58(12):1791–1800. doi: 10.1212/wnl.58.12.1791. [DOI] [PubMed] [Google Scholar]
- [22].Drzezga A, Grimmer T, Henriksen G, et al. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology. 2009;72(17):1487–1494. doi: 10.1212/WNL.0b013e3181a2e8d0. [DOI] [PubMed] [Google Scholar]
- [23].Solomon A, Kivipelto M, Wolozin B, et al. Midlife serum cholesterol and increased risk of Alzheimer's and vascular dementia three decades later. Dement Geriatr Cogn Disord. 2009;28(1):75–80. doi: 10.1159/000231980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wolozin B, Manger J, Bryant R, et al. Re-assessing the relationship between cholesterol, statins and Alzheimer's disease. Acta Neurol Scand Suppl. 2006;185:63–70. doi: 10.1111/j.1600-0404.2006.00687.x. [DOI] [PubMed] [Google Scholar]
- [25].Bu G. Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10(5):333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Prince JA, Zetterberg H, Andreasen N, et al. APOE epsilon4 allele is associated with reduced cerebrospinal fluid levels of Abeta42. Neurology. 2004;62(11):2116–2118. doi: 10.1212/01.wnl.0000128088.08695.05. [DOI] [PubMed] [Google Scholar]
- [27].Lahiri DK, Sambamurti K, Bennett DA. Apolipoprotein gene and its interaction with the environmentally driven risk factors: molecular, genetic and epidemiological studies of Alzheimer's disease. Neurobiol Aging. 2004;25(5):651–660. doi: 10.1016/j.neurobiolaging.2003.12.024. [DOI] [PubMed] [Google Scholar]
- [28].Parker GR, Cathcart HM, Huang R, et al. Apolipoprotein gene E4 allele promoter polymorphisms as risk factors for Alzheimer's disease. Psychiatr Genet. 2005;15(4):271–275. doi: 10.1097/00041444-200512000-00009. [DOI] [PubMed] [Google Scholar]
- [29].Mahley RW, Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012;76(5):871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103(15):5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wallis SC, Rogne S, Gill L, et al. The isolation of cDNA clones for human apolipoprotein E and the detection of apoE RNA in hepatic and extra-hepatic tissues. EMBO J. 1983;2(12):2369–2373. doi: 10.1002/j.1460-2075.1983.tb01748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Calero O, Hortigüela R, Bullido MJ, et al. Apolipoprotein E genotyping method by real time PCR, a fast and cost-effective alternative to the TaqMan and FRET assays. J Neurosci Methods. 2009;183(2):238–240. doi: 10.1016/j.jneumeth.2009.06.033. [DOI] [PubMed] [Google Scholar]
- [33].Koch W, Ehrenhaft A, Griesser K, et al. TaqMan systems for genotyping of disease-related polymorphisms present in the gene encoding apolipoprotein E. Clin Chem Lab Med. 2002;40(11):1123–1131. doi: 10.1515/CCLM.2002.197. [DOI] [PubMed] [Google Scholar]
- [34].Lahiri DK, Schnabel B. DNA isolation by a rapid method from human blood samples: effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochem Genet. 1993;31(7-8):321–328. doi: 10.1007/BF02401826. [DOI] [PubMed] [Google Scholar]
- [35].Walker AH, Najarian D, White DL, et al. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect. 1999;107(7):517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rogers NL, Cole SA, Lan HC, et al. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol. 2007;19(3):319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Maloney B, Ray B, Hayden EP, et al. Development and validation of the high-quality ‘rapid method for swab’ to genotype the HTTLPR serotonin transporter (SLC6A4) promoter polymorphism. Psychiatr Genet. 2009;19(2):72–82. doi: 10.1097/YPG.0b013e3283208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].London SJ, Xia J, Lehman TA, et al. Collection of buccal cell DNA in seventh-grade children using water and a toothbrush. Cancer Epidemiol Biomarkers Prev. 2001;10(11):1227–1230. [PubMed] [Google Scholar]
- [39].Ng DP, Koh D, Choo S, et al. Saliva as a viable alternative source of human genomic DNA in genetic epidemiology. Clin Chim Acta. 2006;367(1-2):81–85. doi: 10.1016/j.cca.2005.11.024. [DOI] [PubMed] [Google Scholar]
- [40].Rogers NL, Cole SA, Lan HC, et al. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol. 2007;19(3):319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ilveskoski E, Lehtimäki T, Erkinjuntti T, et al. Rapid apolipoprotein E genotyping from mailed buccal swabs. J Neurosci Methods. 1998;79(1):5–8. doi: 10.1016/s0165-0270(97)00157-x. [DOI] [PubMed] [Google Scholar]
- [42].Wang G, Su X. A novel technology for the detection, enrichment, and separation of trace amounts of target DNA based on amino-modified fluorescent magnetic composite nanoparticles. Anal Bioanal Chem. 2010;397(3):1251–1258. doi: 10.1007/s00216-010-3625-8. [DOI] [PubMed] [Google Scholar]
- [43].Hoe HS, Harris DC, Rebeck GW. Multiple pathways of apolipoprotein E signaling in primary neurons. J Neurochem. 2005;93(1):145–155. doi: 10.1111/j.1471-4159.2004.03007.x. [DOI] [PubMed] [Google Scholar]
- [44].Soininen H, Kosunen O, Helisalmi S, et al. A severe loss of choline acetyltransferase in the frontal cortex of Alzheimer patients carrying apolipoprotein epsilon 4 allele. Neurosci Lett. 1995;187(2):79–82. doi: 10.1016/0304-3940(95)11343-6. [DOI] [PubMed] [Google Scholar]
- [45].Sunderland T, Mirza N, Putnam KT, et al. Cerebrospinal fluid beta-amyloid1-42 and tau in control subjects at risk for Alzheimer's disease: the effect of APOE epsilon4 allele. Biol Psychiatry. 2004;56(9):670–676. doi: 10.1016/j.biopsych.2004.07.021. [DOI] [PubMed] [Google Scholar]
- [46].Cedazo-Mínguez A, Cowburn RF. Apolipoprotein E: a major piece in the Alzheimer's disease puzzle. J Cell Mol Med. 2001;5(3):254–266. doi: 10.1111/j.1582-4934.2001.tb00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Khachaturian AS, Corcoran CD, Mayer LS, et al. Apolipoprotein E epsilon4 count affects age at onset of Alzheimer disease, but not lifetime susceptibility: The Cache County Study. Arch Gen Psychiatry. 2004;61(5):518–524. doi: 10.1001/archpsyc.61.5.518. [DOI] [PubMed] [Google Scholar]
- [48].Berr C, Hauw JJ, Delaère P, et al. Apolipoprotein E allele epsilon 4 is linked to increased deposition of the amyloid beta-peptide (A-beta) in cases with or without Alzheimer's disease. Neurosci Lett. 1994;178(2):221–224. doi: 10.1016/0304-3940(94)90763-3. [DOI] [PubMed] [Google Scholar]
- [49].Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Heun R, Gühne U, Luck T, et al. Apolipoprotein E allele 4 is not a sufficient or a necessary predictor of the development of Mild Cognitive Impairment. Eur Psychiatry. 2010;25(1):15–18. doi: 10.1016/j.eurpsy.2009.02.009. [DOI] [PubMed] [Google Scholar]
- [51].Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009;30(9):1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Laskowitz DT, Thekdi AD, Thekdi SD, et al. Downregulation of microglial activation by apolipoprotein E and apoE-mimetic peptides. Exp Neurol. 2001;167(1):74–85. doi: 10.1006/exnr.2001.7541. [DOI] [PubMed] [Google Scholar]
- [53].Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43(8):1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- [54].Siest G, Pillot T, Régis-Bailly A, et al. Apolipoprotein E: an important gene and protein to follow in laboratory medicine. Clin Chem. 1995;41(8 Pt 1):1068–1086. [PubMed] [Google Scholar]
- [55].Youmans KL, Tai LM, Nwabuisi-Heath E, et al. APOE4-specific changes in Aβ accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 2012;287(50):41774–41786. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Baum L, Chen L, Ng HK, et al. Apolipoprotein E isoforms in Alzheimer's disease pathology and etiology. Microsc Res Tech. 2000;50(4):278–281. doi: 10.1002/1097-0029(20000815)50:4<278::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- [57].Tai LM, Bilousova T, Jungbauer L, et al. Levels of soluble apolipoprotein E/amyloid-β (Aβ) complex are reduced and oligomeric Aβ increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. J Biol Chem. 2013;288(8):5914–5926. doi: 10.1074/jbc.M112.442103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Thomson DM, Brown NN, Clague AE. Routine use of hair root or buccal swab specimens for PCR analysis: advantages over using blood. Clin Chim Acta. 1992;207(3):169–174. doi: 10.1016/0009-8981(92)90116-8. [DOI] [PubMed] [Google Scholar]
- [59].Zhao X, Tapec-Dytioco R, Wang K, et al. Collection of trace amounts of DNA/mRNA molecules using genomagnetic nanocapturers. Anal Chem. 2003;75(14):3476–3483. doi: 10.1021/ac034330o. [DOI] [PubMed] [Google Scholar]
- [60].Zivelin A, Rosenberg N, Peretz H, et al. Improved method for genotyping apolipoprotein E polymorphisms by a PCR-based assay simultaneously utilizing two distinct restriction enzymes. Clin Chem. 1997;43(9):1657–1659. [PubMed] [Google Scholar]
- [61].Nauck M, Hoffmann MM, Wieland H, et al. Evaluation of the apo E genotyping kit on the LightCycler. Clin Chem. 2000;46(5):722–724. [PubMed] [Google Scholar]
- [62].McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- [63].Vienna, Austria: 2012. R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org . [Google Scholar]


