Extract
Progress in developing robust therapies for spinal cord injury (SCI), traumatic brain injury (TBI) and peripheral nerve injury has been slow. A great deal has been learned over the past 30 years regarding both the intrinsic factors and the environmental factors that regulate axon growth, but this large body of information has not yet resulted in clinically available therapeutics. This therapeutic bottleneck has many root causes, but a consensus is emerging that one contributing factor is a lack of standards for experimental design and reporting. The absence of reporting standards, and even of commonly accepted definitions of key words, also make data mining and bioinformatics analysis of neural plasticity and regeneration difficult, if not impossible. This short review will consider relevant background and potential solutions to this problem in the axon regeneration domain.
Lack of reproducibility
In 2003, the U.S. National Institute of Neurological Disorders and Stroke (NINDS) initiated a groundbreaking project to replicate important studies in the SCI field. The “Facilities of Research Excellence—Spinal Cord Injury” (FORE—SCI) project was designed to explore two issues; the failure of basic science breakthroughs to lead to successful therapeutics and the perceived lack of robustness of findings reported in many high impact papers (Steward et al., 2012). Three SCI centers undertook replication studies to validate interesting results from the original literature. Publications from the University of California at Irvine (Sharp et al., 2012; Sharp et al., 2013; Steward et al., 2006; Steward et al., 2012; Steward et al., 2008; Nielson et al., 2011), the University of Miami (Marcillo et al., 2012; Pinzon et al., 2008; Pinzon et al., 2008), and the Ohio State University (Popovich et al., 2012a; Popovich et al., 2012b) revealed a surprisingly high failure rate in confirming the original studies. In twelve replication experiments, six failed to replicate, four gave partial replication, one was inconclusive and one succeeded but only after three attempts. In a summary of the project the team leaders identified a number of potential explanations for the failures (Steward et al., 2012). If we ignore explanations such as the original results being statistical outliers or scientific misconduct, many of the potential explanations concerned variation in animals and animal care (sources, strains, housing, drugs, handling), injuries (different injury devices, different surgical methods), and differences in reagents (different drug lots, different sources for cell therapies). A major conclusion was that method sections in papers in most journals currently are incomplete, making effective replication studies impossible without direct communication with the original authors.
While the FORE-SCI project was remarkable for its vision and comprehensive investigation of issues related to lack of reproducibility in SCI studies, it is not the only project of this type. The drug discovery industry depends on basic science research, especially that done in academic laboratories, for the identification of many of its therapeutic targets. Scientists at Bayer Healthcare routinely validate studies from the original literature prior to launching a drug discovery campaign. They found in 61 internal projects that only 21% of original studies could be replicated (Prinz et al., 2011). Similarly, scientists at Abbot could reproduce only 11% of landmark studies (Begley et al., 2012). Both groups called for better scientific practices in basic research labs.
Investigator bias
While it is usually difficult to prove in individual cases, investigator bias, including inadvertent and unrecognized bias (Ransohoff et al., 2010), is widely believed to contribute to the reproducibility problem. To overcome investigator bias, leaders in neuroscience recently argued for more rigorous standards in study design and reporting (Landis et al., 2012). Key recommendations included randomization of animals to different study groups and in data collection; blinding investigators, especially those doing assessment or data collection and analysis, to treatments; sample-size estimation; and improved data handling. Moreover, these aspects of a study should be addressed in grant proposals and manuscripts submitted for publication.
Reporting standards
Curiously, while translational and clinical researchers have long had specific recommendations and standards for conducting and reporting research with humans, such standards do not exist in most basic science fields. For example NINDS has extensive recommendations/requirements about common data elements (CDEs) used in clinical research for stroke and TBI (http://www.commondataelements.ninds.nih.gov). Worldwide leaders in the SCI field have proposed design and reporting standards for SCI clinical trials (Anderson et al., 2005; Kwon et al., 2011). Neural Regeneration Research has adopted this idea and has well developed guidelines for authors publishing clinical trial data. However, similar reporting guidelines for animal SCI studies do not yet exist.
Basic science reporting standards are often developed by professional organizations. A well-established example is the data file standard for fluorescent activated cell sorting developed by the International Society for Advancement of Cytometry. In other cases, ad hoc working groups develop such standards. A framework to promote this approach was put in place by the Minimal Information for Biological and Biomedical Investigations (MIBBI) project (Taylor et al., 2008). Perhaps the most widely used MIBBI guideline is the Minimum Information About a Microarray Experiment (MIAME), which established reporting standards for microarray experiments. MIAME established recommendations for describing raw data, reporting how the data were normalized, how samples were prepared, and details of the experimental design (Brazma et al., 2001).
An SCI reporting standard
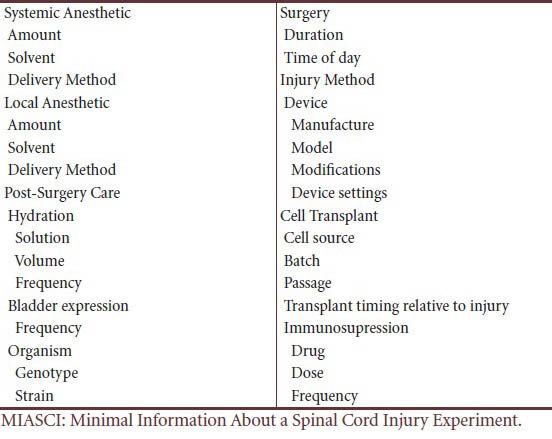
In 2011 a group of scientists in the U.S. and Japan began planning a project to develop reporting standards for animal studies related to SCI. Thirty-five scientists held a 3 day meeting in New Orleans in the Fall of 2012 titled: Growth Cones and Axon Regeneration: Entering The Age of Informatics. There were a number of scientific talks, and breakout sessions were held to develop a proposal for Minimal Information About a Spinal Cord Injury experiment (MIASCI). Because of the very wide-ranging way SCI experiments are done the MIASCI draft includes a relatively large number of metadata elements (>250, examples in Table 1) that could limit acceptance by authors, editors and reviewers. Many of these elements concern things like animal source and strain, housing conditions, and experimental design issues such as whether the investigators were blinded to treatment conditions. The majority covers various kinds of treatments that are unlikely to be used simultaneously in a given study. For example, it is unlikely, at present, that a study involving a cell therapy would also involve screening a siRNA library AND a compound library. Consequently, the number of data elements that need to be annotated for a given SCI study should be in line with other reporting standards. Nonetheless, it is clear that for the MIASCI standard to be widely adopted and used by authors and annotators, a MIASCI reporting tool will need to be developed that provides simple entry using standardized terminologies, provides definitions and eliminates repetitive entry of required information. Output from such a tool should be both human- and machine-readable, and available in a variety of formats.
Table 1.
Representative examples of data elements in the draft MIASCI
Having SCI researchers report the metadata from experiments along with the data will go a long way toward achieving the transparency of research recommended by Landis et al. (2012). As members of the SCI community become accustomed to reporting issues related to study design (blinded assessment of outcome measures, power analysis to determine sample size, exclusion criteria) they will inevitably adopt more rigorous approaches to the research they conduct. An additional major impact on the SCI domain will be to facilitate data mining and bioinformatic analysis. As the amount of SCI-related information published each year becomes more and more overwhelming, this will become increasingly important.
The SCI data flood
PubMed indexes over 20,000 papers on SCI with 1,723 having been published in 2012. Related topics such as neuroprotection, axon regeneration, axon growth and nerve regeneration account for more than 40,000 additional papers. Scientists cannot digest that much information and, at present, computers can’t either.
Computer scientists specializing in artificial intelligence have developed impressive tools for analyzing structured data of the kind found in relational databases and unstructured data such as that found in the corpus of a particular field. Relational databases are extremely efficient at storing and retrieving data but need to be carefully designed at the beginning and are not easily adapted to incorporate new types of data. They also typically require that the data be stored in a single storage system. To overcome these limitations and to enable the use of data spread across the internet the concept of the “semantic web” was developed (Berners-Lee et al., 2001), along with languages such as Extensible Markup Language (XML), Resource Description Framework (RDF), and Web Ontology Language (OWL). For this latter approach to work ontologies need to be developed that identify key concepts in a particular field, provide definitions and relationships among the concepts and give various synonyms for a given concept. With appropriate ontologies in place large text mining projects can be performed to build a knowledge base. If the ontologies are created with expressive descriptive logic to define relationships among concepts, description logic reasoning engines can be used to answer complex queries made against the knowledge base. Finally, semantic web approaches allow queries across dispersed resources such as the Gene Ontology and the Chemical Entities of Biological Interest (ChEBI) provided that the data are annotated with concepts from ontologies (Shah et al., 2009; Callahan et al., 2011).
The development of an ontology for a particular research area, such as SCI, necessarily requires a combination of text mining to identify recurring terms and phrases as well as input from domain experts who can provide definitions for concepts, identify synonyms and establish relationships among these concepts. Having computer scientists involved is also essential as they develop the logical framework and tools to make the ontology and related knowledge base useful. Finally, members of the scientific community need to participate by providing input as to the type of questions they would like to ask (so-called “use cases”) and how they would like output to be visualized.
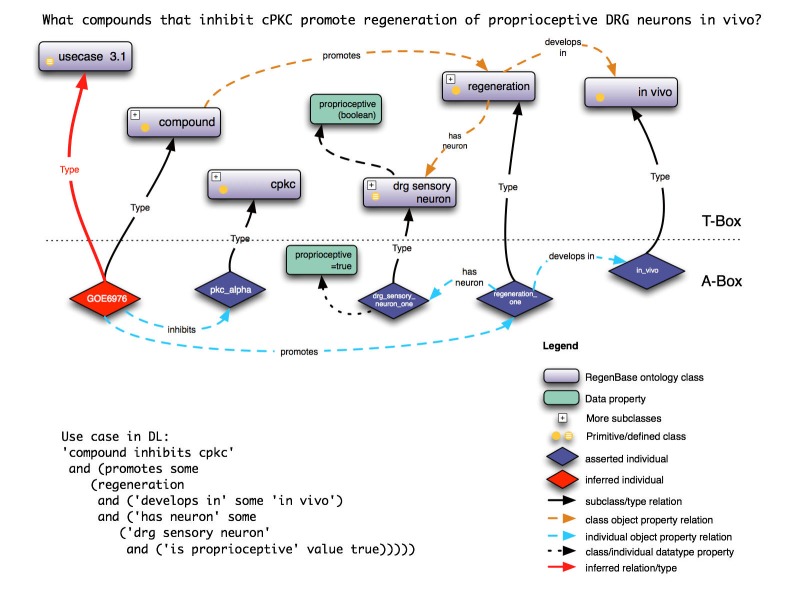
We (V.P.L., J.L.B. and U.V.) are leading a team of neuroscientists, informaticists and computer scientists at the University of Miami and Stanford University to develop an ontology about axon regeneration, called the RegenBase Ontology. The long-term goal of this project is to connect information about SCI experiments to information about biological processes, molecular networks and high-throughput screening data to speed the identification and testing of novel therapeutics. The metadata and data that can be obtained from well-annotated research articles are critical to developing a useful web-based resource for the SCI community. Obtaining input from SCI researchers worldwide regarding MIASCI and use cases for RegenBase is critical to the success of this project. An example of a use case using a draft RegenBase ontology is shown in Figure 1.
Figure 1.
A query example using the RegenBase Ontology.
The T-Box includes concepts, such as “compound” and “regeneration”, and relationships, such as “promotes” and “develops in vivo”. When data from real world experiments are incorporated into the A-Box of the ontology, it is possible to ask questions such as “What compounds that inhibit cPKC promote regeneration of proprioceptive DRG neurons in vivo?”. Using a Descriptive Logic (DL) reasoning engine, we found that a compound, Gö6976, satisfied this logical query.
We were able to successfully query the knowledge base for protein kinase C inhibitors that promote regeneration of sensory neurons. Another interesting question for stakeholders, for example, is “would a Chinese language version of MIASCI or of RegenBase facilitate use and adoption?”. In addition, collaboration with journal editors and publishers needs to be explored. Cooperation of these constituencies is critical for the development of true transparency in SCI research.
Footnotes
Conflicts of interest: None declared.
Funding: Research in the Lemmon/Bixby lab is supported by NIH grants NS080145 and NS059866, and by the Miami Project to Cure Paralysis.
Copyedited by Phillips A, Li KY, Wang X, Yu J, Qiu Y, Li CH, Song LP, Liu WJ, Zhao M
References
- [1].Anderson DK, Beattie M, Blesch A, Bresnahan J, Bunge M, Dietrich D, Dietz V, Dobkin B, Fawcett J, Fehlings M, Fischer I, Grossman R, Guest J, Hagg T, Hall ED, Houle J, Kleitman N, McDonald J, Murray M, Privat A, et al. Recommended guidelines for studies of human subjects with spinal cord injury. Spinal Cord. 2005;43:453–458. doi: 10.1038/sj.sc.3101746. [DOI] [PubMed] [Google Scholar]
- [2].Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- [3].Berners-Lee T, Hendler J. Publishing on the semantic web. Nature. 2001;410:1023–1024. doi: 10.1038/35074206. [DOI] [PubMed] [Google Scholar]
- [4].Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- [5].Callahan A, Dumontier M, Shah NH. HyQue: evaluating hypotheses using Semantic Web technologies. J Biomed Semantics. 2011;2(Suppl 2):S3. doi: 10.1186/2041-1480-2-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kwon BK, Okon EB, Tsai E, Beattie MS, Bresnahan JC, Magnuson DK, Reier PJ, McTigue DM, Popovich PG, Blight AR, Oudega M, Guest JD, Weaver LC, Fehlings MG, Tetzlaff W. A grading system to evaluate objectively the strength of pre-clinical data of acute neuroprotective therapies for clinical translation in spinal cord injury. J Neurotrauma. 2011;28:1525–1543. doi: 10.1089/neu.2010.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, Bradley EW, Crystal RG, Darnell RB, Ferrante RJ, Fillit H, Finkelstein R, Fisher M, Gendelman HE, Golub RM, Goudreau JL, Gross RA, Gubitz AK, Hesterlee SE, Howells DW, Huguenard J, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 2012;490:187–191. doi: 10.1038/nature11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marcillo A, Frydel B, Bramlett HM, Dietrich WD. A reassessment of P2X7 receptor inhibition as a neuroprotective strategy in rat models of contusion injury. Exp Neurol. 2012;233:687–692. doi: 10.1016/j.expneurol.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nielson JL, Strong MK, Steward O. A reassessment of whether cortical motor neurons die following spinal cord injury. J Comp Neurol. 2011;519:2852–2869. doi: 10.1002/cne.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pinzon A, Marcillo A, Pabon D, Bramlett HM, Bunge MB, Dietrich WD. A re-assessment of erythropoietin as a neuroprotective agent following rat spinal cord compression or contusion injury. Exp Neurol. 2008;213:129–136. doi: 10.1016/j.expneurol.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pinzon A, Marcillo A, Quintana A, Stamler S, Bunge MB, Bramlett HM, Dietrich WD. A re-assessment of minocycline as a neuroprotective agent in a rat spinal cord contusion model. Brain Res. 2008;1243:146–151. doi: 10.1016/j.brainres.2008.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Popovich PG, Lemeshow S, Gensel JC, Tovar CA. Independent evaluation of the effects of glibenclamide on reducing progressive hemorrhagic necrosis after cervical spinal cord injury. Exp Neurol. 2012a;233:615–622. doi: 10.1016/j.expneurol.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Popovich PG, Tovar CA, Wei P, Fisher L, Jakeman LB, Basso DM. A reassessment of a classic neuroprotective combination therapy for spinal cord injured rats: LPS/pregnenolone/indomethacin. Exp Neurol. 2012b;233:677–685. doi: 10.1016/j.expneurol.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- [15].Ransohoff DF, Gourlay ML. Sources of bias in specimens for research about molecular markers for cancer. J Clin Oncol. 2010;28:698–704. doi: 10.1200/JCO.2009.25.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shah NH, Jonquet C, Chiang AP, Butte AJ, Chen R, Musen MA. Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinformatics. 2009;10(Suppl 2):S1. doi: 10.1186/1471-2105-10-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sharp K, Yee KM, Steward O. A re-assessment of the effects of treatment with an epidermal growth factor receptor (EGFR) inhibitor on recovery of bladder and locomotor function following thoracic spinal cord injury in rats. Exp Neurol. 2012;233:649–659. doi: 10.1016/j.expneurol.2011.04.013. [DOI] [PubMed] [Google Scholar]
- [18].Sharp KG, Yee KM, Stiles TL, Aguilar RM, Steward O. A re-assessment of the effects of treatment with a non-steroidal anti-inflammatory (ibuprofen) on promoting axon regeneration via RhoA inhibition after spinal cord injury. Exp Neurol. 2013;248:321–337. doi: 10.1016/j.expneurol.2013.06.023. [DOI] [PubMed] [Google Scholar]
- [19].Steward O, Popovich PG, Dietrich WD, Kleitman N. Replication and reproducibility in spinal cord injury research. Exp Neurol. 2012;233:597–605. doi: 10.1016/j.expneurol.2011.06.017. [DOI] [PubMed] [Google Scholar]
- [20].Steward O, Sharp K, Selvan G, Hadden A, Hofstadter M, Au E, Roskams J. A re-assessment of the consequences of delayed transplantation of olfactory lamina propria following complete spinal cord transection in rats. Exp Neurol. 2006;198:483–499. doi: 10.1016/j.expneurol.2005.12.034. [DOI] [PubMed] [Google Scholar]
- [21].Steward O, Sharp K, Yee KM. A re-assessment of the effects of intracortical delivery of inosine on transmidline growth of corticospinal tract axons after unilateral lesions of the medullary pyramid. Exp Neurol. 2012;233:662–673. doi: 10.1016/j.expneurol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Steward O, Sharp K, Yee KM, Hofstadter M. A re-assessment of the effects of a Nogo-66 receptor antagonist on regenerative growth of axons and locomotor recovery after spinal cord injury in mice. Exp Neurol. 2008;209:446–468. doi: 10.1016/j.expneurol.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, Brazma A, Brinkman RR, Michael Clark A, Deutsch EW, Fiehn O, Fostel J, Ghazal P, Gibson F, Gray T, Grimes G, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]