Abstract
Preliminary basic research and clinical findings have demonstrated that electroacupuncture therapy exhibits positive effects in ameliorating depression. However, most studies of the underlying mechanism are at the single gene level; there are few reports regarding the mechanism at the whole-genome level. Using a rat genomic gene-chip, we profiled hippocampal gene expression changes in rats after electroacupuncture therapy. Electroacupuncture therapy alleviated depression-related manifestations in the model rats. Using gene-chip analysis, we demonstrated that electroacupuncture at Baihui (DU20) and Yintang (EX-HN3) regulates the expression of 21 genes. Real-time PCR showed that the genes Vgf, Igf2, Tmp32, Loc500373, Hif1a, Folr1, Nmb, and Rtn were upregulated or downregulated in depression and that their expression tended to normalize after electroacupuncture therapy. These results indicate that electroacupuncture at Baihui and Yintang modulates depression by regulating the expression of particular genes.
Keywords: nerve regeneration, acupuncture, traditional Chinese medicine, depression, gene expression profiling, gene-chip, electroacupuncture, Baihui (DU20), Yintang (EX-HN3), chronic stress, behavior, NSFC grant, neural regeneration
Introduction
Chronic stress presents macroscopic changes in psychoactivity, causing stress-related mood disorders. Microscopically, it shows structural and functional changes in neurites and neural circuits. In essence, chronic stress leads to abnormal expression of stress-induced genes[1,2,3]. There is strong evidence that abnormal gene expression in turn worsens the disease, and this vicious circle makes it difficult to cure conditions such as depression[4,5,6,7]. Recently, a large number of genes, such as the 5-hydroxytryptamine receptor, 5-hydroxytryptamine transporter, and tryptophan hydroxylase, have been found to be related to depression[8,9,10]. Depression is a complex, clinically common mental disorder, which involves various brain nuclei, multi-region neuronal associations, and diverse transmitters[11,12,13,14]. It cannot be fully explained in terms of single-gene disorders. A previous study found evidence for multiple gene imbalances in the brain of patients with depression, and proposed this as a pathological factor. Various nerve cells and multiple brain functions lead to a complex interaction between gene expression and the clinical manifestations of depression[16].
The hippocampus is a brain region closely associated with learning and memory abilities and emotion. It is composed of the hippocampal gyrus and the dentate gyrus. The former consists of CA1−4 regions and is made of pyramidal cells; the latter is composed of granular cells and can produce new neurons throughout life[17,18,19].
Chronic stress can persistently activate the hypothalamic-pituitary-adrenal (HPA) axis, leading to an increase in glucocorticoid levels. The hippocampus regulates this axis and is a major target for glucocorticoids. It is selectively affected by the high levels of glucocorticoids in stress, resulting in decreased hippocampal synaptic plasticity, and neuronal atrophy and loss, which consequently influences learning and memory abilities. Therefore, damage to hippocampal neuronal structure and function plays a key role in the onset of depression[20,21,22,23,24,25,26,27].
There is strong ancient and modern evidence for the utility of electroacupuncture in the treatment of depression. However, most studies report the underlying mechanism at the single gene level[28,29,30,31]. To the best of our knowledge, there are no studies reporting the underlying mechanism at the whole-genome level. This study was the first to use a rat genome gene-chip to investigate changes in hippocampal gene expression after electroacupuncture therapy.
Results
Quantitative analysis of experimental animals
After habituation for 1 week, 45 Wistar rats were randomly and evenly divided into normal control, model, and electroacupuncture groups. A model of chronic unpredictable mild stress-induced depression was established in the latter two groups. Rats in the electroacupuncture group only received electroacupuncture at acupoints Baihui (DU20) and Yintang (EX-HN3). All animals were included in the final analysis.
Electroacupuncture did not influence body weight
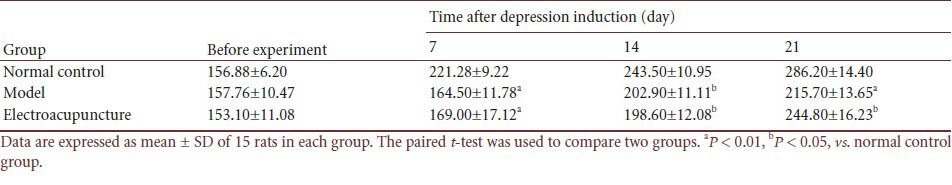
Compared with control rats, the rats that received chronic unpredictable mild stress and the rats that received electroacupuncture both exhibited significantly decreased body weight (P < 0.01 or P < 0.05; Table 1).
Table 1.
Effect of electroacupuncture on body weight (g) in a rat depression model
Electroacupuncture ameliorated the manifestations of depression in the model rats
Electroacupuncture increased open-field test scores
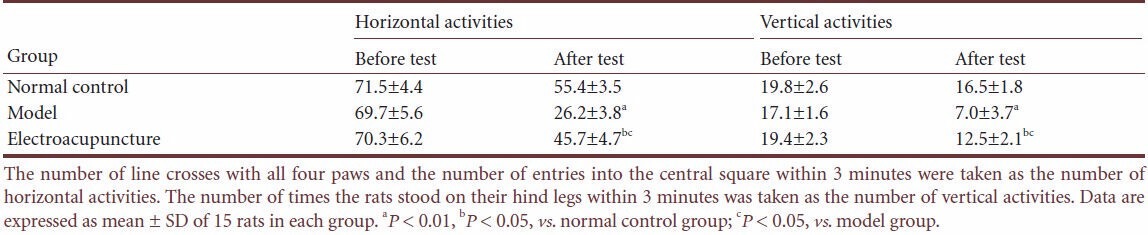
Compared with control rats, the rats that received chronic unpredictable mild stress exhibited significantly decreased scores for both horizontal and vertical activities in the open-field (P < 0.01 or P < 0.05). However, after electroacupuncture, the scores for both were increased (P < 0.05; Table 2).
Table 2.
Effect of electroacupuncture on open-field test scores in a rat depression model
Electroacupuncture increased sucrose consumption in the depression model rats
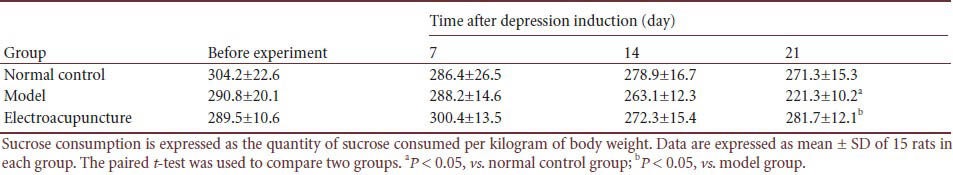
Before and 21 days after chronic unpredictable mild stress, sucrose consumption was significantly lower in the model group than that in the control group (P < 0.05). However, after electroacupuncture, sucrose consumption was significantly increased in the depression model rats (P < 0.05; Table 3).
Table 3.
Effect of electroacupuncture on sucrose consumption (g/kg) in a rat depression model
Electroacupuncture shortened the time spent in an immobile state in the depression model rats
Compared with control rats, the rats that received chronic unpredictable mild stress spent a significantly longer time immobile in the forced swimming test (P < 0.01 or P < 0.05). However, after electroacupuncture, the time spent immobile was shortened (P < 0.05 or P < 0.01; Table 4).
Table 4.
Effect of electroacupuncture on time spent immobile (seconds) in a rat depression model

Electroacupuncture influenced hippocampal depression-related gene expression in the depression model rats
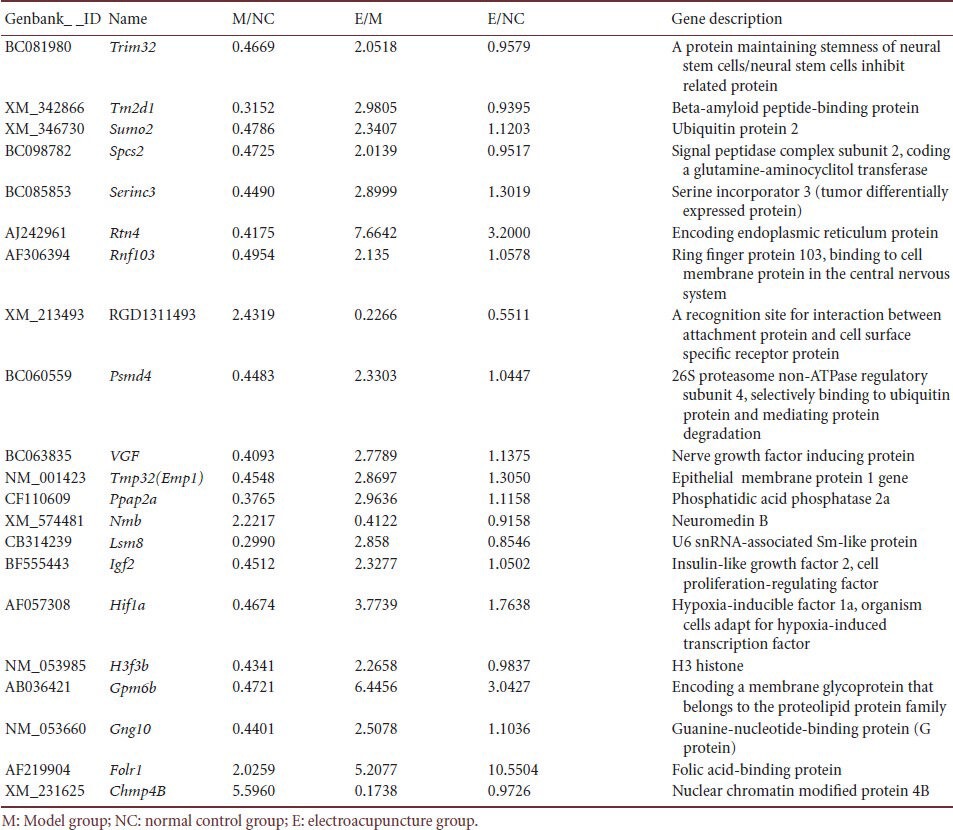
A total of 21 genes (Table 5), Trim32, Tm2d1, Sumo2, Spcs2, Serinc3, Rtn4, Rnf103, Loc500373, Rnf103, RGD1311493, Psmd4, Ppap2a, Vgf, Tmp32, Nmb, Lsm8, Igf2, Hif1a, H3f3b, Gpm6b, Gng10, Folr1, and XM_231625 were identified to be differentially expressed in the hippocampus of the depression model rats compared with control rats. After electroacupuncture, the expression level of these genes was normalized. Among the 21 differentially expressed genes, RGD1311493, Nmb, Folr1, and Chmp4B were upregulated, i.e., gene expression in the model group was more than twice that in the control group. The remaining 17 genes were downregulated, i.e., gene expression in the model group was less than 0.5-times that in the control group. These 21 genes encode stress-response proteins, DNA injury and repair proteins, apoptotic proteins, ion channel receptors, growth factors, and cell cycle-related proteins.
Table 5.
Effect of electroacupuncture on hippocampal depression-related gene expression in a rat depression model
After 21 days of electroacupuncture, the expression of 21 genes (all except Folrl) was closer to a normal level. Moreover, 14 genes (Trim32, Tm2d1, Sumo2, Spcs2, Rnf103, Psmd4, Vgf, Ppap2a, Nmb, Lsm8, Igf2, H3f3b, Gng10, and Chmp4B) showed close-to-normal expression after electroacupuncture (i.e., a ratio of gene expression in the electroacupuncture group to that in the normal control group of 1.0 ± 0.2). The genes Vgf, Igf2, Tmp32, Loc500373, Hif1a, Folr1, Nmb, and Rtn4, were selected for PCR analysis. Amplification bands were obtained for all of them (Figure 1).
Figure 1.
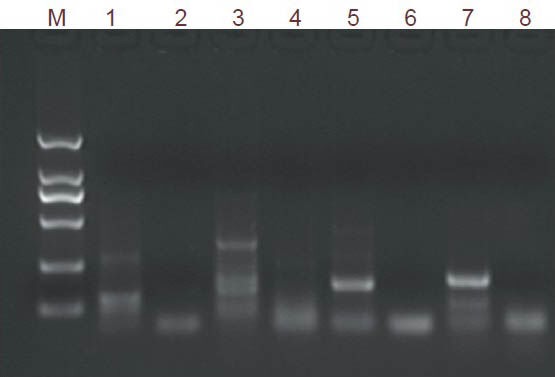
Electrophoresis results of PCR amplification products of genes from rat hippocampal tissue.
The band size in the marker lane, from top to bottom, is 2,000 bp, 1,000 bp, 750 bp, 500 bp, 250 bp, and 100 bp. M: Marker DL2000; 1: Vgf; 2: Igf2; 3: Tmp32; 4: Loc500373; 5: Hif1a; 6: Folr1; 7: Nmb; 8: Rtn4.
Each gene showed good amplification specificity upon examination of the dissociation curve in real-time quantitative PCR. The quantitative PCR findings were consistent with those of the gene chip analysis (data not shown).
Discussion
Our present results suggest that (1) depression is attributable to comprehensive regulation of multiple hippocampal genes that are concerned with metabolism, signal transduction, and cell growth. In the diseased state, genes with altered expression are closely related to the pathological mechanism and are the main factors leading to depression; and (2) electroacupuncture can treat depression by modifying or regulating the expression of various genes. The mechanism underpinning the effect of electroacupuncture on depression is related to gene regulation. The known function of the eight genes selected here is as follows.
(1) Tmp32 and Vgf: Tmp32 and Vgf regulate steroid hormone levels. These genes were significantly downregulated in depression and upregulated after electroacupuncture. Neuroactive steroids are the active steroids in the nervous system. They are endogenous neuromodulators, which can be synthesized in the brain, adrenal gland, ovary, and testis, and mainly comprise progesterone, deoxycortone, dehydroepiandrosterone, testosterone, and the latter's metabolites[23]. By binding to intracellular receptors, neuroactive steroids regulate synaptic inhibitory transmission, inflammation, myelination, central nervous system development and post-injury repair, and the HPA axis and its stress effect[32,33]. Recently, increasing attention has been paid to the role of the stress hypothesis in the onset of depression. According to the stress hypothesis, depression is caused by excessive stress mechanisms in the brain, in which the HPA axis plays a key role. The HPA axis is considered the common pathway of many symptoms and signs of depression, and the stress hypothesis is opening new and exciting avenues for the treatment of this condition[34].
(2) Trim32: Trim32 is a widely expressed ubiquitin ligase of 655 amino acids that is closely involved with cellular apoptosis[35]. In this study, we found that Trim32 expression was downregulated in depression and returned to normal after electroacupuncture. This warrants further study.
(3) Nmb: Nmb encodes a 117-amino-acid protein that functions in: (i) estrogen release and regulation during individual maturation; (ii) energy metabolism in adipocytes; (iii) thyroxine release; and (vi) neural C-fos gene expression and nerve cell growth[36]. Energy metabolism and nerve cell growth are both closely associated with depression, suggesting that Nmb is a depression-related gene that deserves further study.
(4) Igf2: Igf2 expression is likely to be related to the Haxis and the activity of signal transduction pathways. Here, Igf2 gene expression decreased in the brain of depression model rats and its expression returned to normal after electroacupuncture. This might relate to normalization of the endocrine system by electroacupuncture, and of the HPA axis in particular. In addition, Igf2 can stimulate the phosphatidylinositol 3-kinase/protein kinase B pathway, and the products downstream of this signal transduction pathway overlap considerably with those of 5-hydroxytryptamine metabolism. Igf2 promotes nerve cell proliferation, increased transmitter levels between synapses, and synaptic plasticity[37].
(5) Cytochrome c oxidase: Cytochrome c oxidase is located at the terminus of the cytochrome system of cellular respiration: it transmits electrons onto oxygen molecules to bind to protons to form water molecules. Cytochrome c oxidase carries four protons simultaneously to form a difference in transmembrane chemical potential energy, which contributes to ATP formation. The active cytochrome c oxidase molecule is an assembly of many subunits and cofactors[38]. We found that expression of the related gene Loc500373 was decreased in the brain tissue of depression model rats. This suggests that energy metabolism is inhibited and ATP formation is reduced in this rat model. However, after electroacupuncture therapy, the increased Loc500373 gene expression would promote ATP formation, thus benefiting cell function.
(6) Ribosomal protein: Ribosomal proteins are an important component of the ribosome that plays an essential role in protein synthesis. We demonstrated that Rtn4 expression was decreased in the brain tissue of depression model rats, indicating a deficit in protein biosynthesis. After electroacupuncture, Rtn4 expression was increased, which would facilitate protein biosynthesis[39].
(7) Hif1a: Hif1a is a receptor that mainly functions in cellular apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). After specifically binding to TRAIL, it stimulates and transmits a death signal via its death domain, and activates the caspase cascade that leads to cellular apoptosis. Hif1a causes tumor cell death and is also related to thymocyte and nerve cell apoptosis[40]. We found that Hif1a expression was downregulated in depression model rats, which would suggest a reduction in hippocampal nerve cell apoptosis. Hif1a expression recovered after electroacupuncture, which suggests that it is a depression-related gene that warrants further study.
In the model group, we demonstrated decreased expression of genes related to transcription/translation, neurotransmission/signal transduction, inflammation/the immune system, metabolism, enzymatic reactions, metabolism, and protein biosynthesis. Together, this would be predicted to lead to impaired hippocampal structure and function, which might even result in cell death. We believe that these gene changes underlie the manifestations of depression in this model. The upregulated genes in the model group/normal control group and the downregulated genes in the electroacupuncture group/model group are related to inflammation/immunity and neurotransmission/signal transduction. The downregulated genes in the model group/normal control group and the upregulated genes in the electroacupuncture group/model group are related to cell cycle/cell structure, neurotransmission/signal transduction, and transcription.
In the model group, the upregulated genes are related to inflammation/immunity and oxidative stress, and therefore the model rats would be expected to exhibit increased inflammation/immunity, oxidative stress, injury responses, and clotting. The downregulated genes are related to the cell cycle, signal transduction/neurotransmission, and metabolism (in particular protein metabolism). After electroacupuncture, the expression of these genes tended to return to normal. This is highly important for maintaining tissue structure, restoring cell function, and alleviating the symptoms of depression. The mechanism underlying the effect of electroacupuncture on depression relates to the regulation of multiple genes. This adds experimental evidence to the observed clinical effect of electroacupuncture on depression. We validated eight genes by real-time PCR, and the results were consistent with the gene-chip findings. This indicates that the data determined by gene-chip technology are reliable.
It should be emphasized that knowledge of gene function is still limited. The post-genomic era has only just begun. Many genes have multiple effects and their functions interact. The function of these and other depression-related genes should be studied further.
Materials and Methods
Design
A cytological gene level-based contrast observation animal study with bioinformatics analysis of high-throughput gene-chip data.
Time and setting
This study was performed in the Laboratory Animals Facility, Chinese PLA General Hospital, China, between September 2011 and September 2012.
Materials
Forty-five adult healthy male Wistar rats of specific pathogen-free grade, aged 5–6 weeks, weighing 160–180 g, were provided by SPF (Beijing) Laboratory Animals Science & Technology Co., Ltd., China (license No. SCXK (Jing) 2011-0001). Rats were raised in an artificially illuminated environment, with a 12-hour day-night cycle and a controlled temperature of 22°C. The animal experiment protocols were in strict accordance with the Guidance Suggestions for the Care and Use of Laboratory Animals issued by the Ministry of Science and Technology of China[41].
Methods
Establishment of a depression model in rats using chronic unpredictable mild stress
We established a rat model of chronic unpredictable mild stress-induced depression using a slight modification of the method of Willner[42,43]. In summary, after a 7-day habituation, rats received 21 days of unpredictable stimuli, including movement restriction, illumination, day-night reversal over a 24-hour period, electrical stimulation on the sole of the foot (1.0 mA, 10-second stimulation once every other minute, a total of 30 times), swimming in iced water (4°C, 5 minutes), thermal stress (45°C, 5 minutes), vibration (once per second for 15 minutes), tail clamping (1 minute), water deprivation (24 hours), and food deprivation (48 hours). One of the above-mentioned stimuli was performed every day. Each stimulus was performed four times on average. No stimulus was performed on any two or more successive days, so that the rats could not predict which stimulus would be given. The depression model rats were raised separately, one rat per cage. The control group rats were housed five to a cage and did not receive any stimulus.
Electrical stimulation
In accordance with The Rat Brain in Stereotaxic Coordinates[44], electroacupuncture at rat acupoints Baihui (the center of parietal bone) and Yintang (the middle point of the line between the brow bones) was performed from the day of establishing the depression model to day 21 of treatment. The acupuncture needle (HWATO Brand, No. 30, 0.5 cun; Suzhou Medical Instrument Factory, Suzhou, Jiangsu Province, China) was pricked into the skin at Baihui and Yintang at a depth of 2 mm. A forward oblique needling was required for Baihui and an upward oblique needling for Yintang. Electroacupuncture was carried out with a cluster-shaped wave, a frequency of 2 Hz and an intensity of 1 mA, once a day for 20 minutes. The level was such that the animals did not struggle or vocalize.
Body weight measurement
Body weight was measured 1 day before and on days 7, 14, and 21 of the treatment.
Open-field test
One day before the induction of depression and on day 21 of the treatment, rats were placed into the central square of an open-field apparatus. The number of line crosses with all four paws, number of entries into the central square, and the frequency with which the rats stood on their hind legs were counted during a 3-minute trial[45].
Sucrose consumption test
The sucrose consumption test was performed 1 day before the induction of depression and on days 7, 14, and 21 of the treatment. Following water deprivation for 24 hours, rats were given 1% (w/v) sucrose solution and were simultaneously food deprived. Twenty-four hours later, the sucrose solution-containing bottle was weighed and the sucrose consumption was calculated. The data were expressed as sucrose solution mass per kilogram of body weight (g/kg)[46].
Forced swimming test
Rats were placed into a plastic cylinder (100 cm high, 40 cm diameter) filled with 25 ± 1°C water at a depth of 75 cm. After two minutes, the rats were observed for a further 5 minutes, and the time that the rats spent struggling or maintaining an immobile state with their head above water was recorded[47]. The forced swimming test was performed 1 day before the induction of depression and on days 7, 14, and 21 of the treatment. In the electroacupuncture group, the forced swimming test was performed 12 hours after electroacupuncture. This was to avoid false positives, i.e., reduced immobility because of transient increases in mobility after stimulation of the cortical motor area.
Measurement of rat hippocampal gene expression by gene-chip analysis
The rats were decapitated on day 21 after the induction of depression. Following washes with pre-cooled 4°C physiological saline, a cruciform incision was made at the base of the skull. The cranial bone was separated using blood vessel forceps and the entire brain was harvested. The brain tissue and fascia on the surface of the hippocampus[44] was stripped using ophthalmic surgical forceps. Then the entire hippocampus was gently harvested and its wet weight recorded. The hippocampus was placed immediately into a pre-tagged 1.5-mL freezing tube. The entire procedure of sample harvesting was performed in an ice bath. The freezing tube was stored in liquid nitrogen. For RNA extraction, the hippocampus was cut into small blocks and ground in a pre-cooled mortar. After the addition of 2–3 mL Trizol reagent (Invitrogen, Gaithersburg, MD, USA), total RNA was extracted by the one-step method. Total RNA was precipitated from the aqueous phase by isopropanol and then further purified with a Nucleospin® RNA Clean-up kit (Macherey-Nagel, Düren, Germany). The RNA was quantified using a spectrophotometer (Beijing Huashengpuxin Instrument Co., Ltd., Beijing, China).
The RNA sample was then fluorescently labeled using a Crystal Core® cRNA amplification labeling kit (CapitalBio Corporation, Beijing, China). The labeled sample was dissolved in 80 µL hybridization solution (3 × SSC, 0.2% SDS, 5 × Denhardt's solution, 25% fluoroamine) and hybridized to a 27K Rat Genome Array gene-chip (CapitalBio Corporation; Rat Genome Version 3.0.5) at 42°C overnight. This chip contains 26,962 70-mer oligo DNAs representing approximately 22,012 genes and 27,044 transcripts. After hybridization, the sample was washed for 5 minutes in a 42°C solution containing 0.2% SDS, 2 × SSC, for 5 further minutes in 0.2 × SSC, and then dried. The chip images were scanned using a double-channel laser scanner (CapitalBio Corporation). The typing was determined according to the difference in mass spectrum molecular weight.
Validation of differentially expressed genes
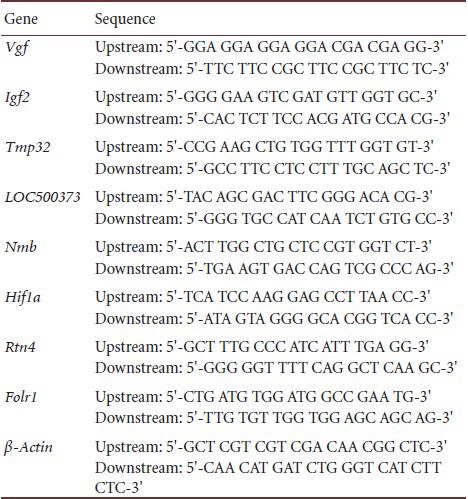
The candidate genes that appeared repeatedly in the experiment were selected for further analysis. The candidate genes were selected depending on their high frequency, their extent of up-regulation or down-regulation, and their expression ratio relative to the normal control group or the model group[47]. Differentially expressed genes were validated by real-time quantitative PCR. The genes chosen for validation and their corresponding primer sequences are in Table 6.
Table 6.
Prime sequences of the chosen genes
For real-time PCR, 2 µg RNA sample was mixed with 5 µg diethyl pyrocarbonate-treated RNase-free water, then incubated at 70°C for 10 minutes and at 4°C for 5 minutes in a PCR instrument (Hangzhou Jingle Scientific Instrument Co., Ltd., Hangzhou, Zhejiang Provcince, China). The sample was then reverse transcribed in a 20-µL volume containing 4 µL moloney murine leukemia virus 5 × buffer, 2 µL 10 mmol/L dNTPs, 0.5 µL recombinant RNase inhibitor, 1 µL moloney murine leukemia virus reverse transcriptase (CapitalBio Corporation; 200 units), and 0.5 µg primer at 25°C for 10 minutes, 37°C for 60–120 minutes, 99°C for 5 minutes, and 4°C for 5 minutes, before being stored at -20°C for later use.
A fragment of the rat β-actin gene was PCR amplified from each reverse-transcribed template. The PCR products were separated on a 1% agarose gel. Eight genes with high frequency were selected for PCR analysis. Amplification was performed at a temperature 3–5°C lower than primer annealing temperature, for 40 cycles. The PCR reagent mix included SYBR Green dye (CapitalBio Corporation). An IQ5PCR instrument (Bio-Rad, Hercules, CA, USA) was used.
Statistical analysis
SPSS 16.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. All data were expressed as mean ± SD. The paired t-test was used for comparison between groups and the F test for comparison among groups. A level of P < 0.05 was considered statistically significant.
Footnotes
Funding: This study was supported by the National Natural Science Foundation of China, No. 81273847.
Conflicts of interest: None declared.
Copyedited by Devon R, Raye W, Liu SG, Sui HB, Yu J, Li CH, Song LP, Liu WJ, Zhao M
References
- [1].Revollo HW, Qureshi A, Collazos F, et al. Acculturative stress as a risk factor of depression and anxiety in the Latin American immigrant population. Int Rev Psychiatry. 2011;23(1):84–92. doi: 10.3109/09540261.2010.545988. [DOI] [PubMed] [Google Scholar]
- [2].Rogóz Z, Skuza G, Legutko B. Repeated treatment with mirtazepine induces brain-derived neurotrophic factor gene expression in rats. J Physiol Pharmacol. 2005;56(4):661–671. [PubMed] [Google Scholar]
- [3].Dwivedi Y. Brain-derived neurotrophic factor: role in depression and suicide. Neuropsychiatr Dis Treat. 2009;5:433–449. doi: 10.2147/ndt.s5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Urbach A, Bruehl C, Witte OW. Microarray-based long-term detection of genes differentially expressed after cortical spreading depression. Eur J Neurosci. 2006;24(3):841–856. doi: 10.1111/j.1460-9568.2006.04862.x. [DOI] [PubMed] [Google Scholar]
- [5].Kohli MA, Salyakina D, Pfennig A, et al. Association of genetic variants in the neurotrophic receptor-encoding gene NTRK2 and a lifetime history of suicide attempts in depressed patients. Arch Gen Psychiatry. 2010;67(4):348–359. doi: 10.1001/archgenpsychiatry.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Perroud N, Uher R, Ng MY, et al. Genome-wide association study of increasing suicidal ideation during antidepressant treatment in the GENDEP project. Pharmacogenomics J. 2012;2(1):68–77. doi: 10.1038/tpj.2010.70. [DOI] [PubMed] [Google Scholar]
- [7].Schosser A, Butler AW, Ising M, et al. Genomewide association scan of suicidal thoughts and behaviour in major depression. PLoS One. 2011;6(7):e20690. doi: 10.1371/journal.pone.0020690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kanellopoulos D, Gunning FM, Morimoto SS, et al. Hippocampal volumes and the brain-derived neurotrophic factor val66met polymorphism in geriatric major depression. Am J Geriatr Psychiatry. 2011;19(1):13–22. doi: 10.1097/jgp.0b013e3181f61d62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cole J, Weinberger DR, Mattay VS, et al. No effect of 5HTTLPR or BDNF Val66Met polymorphism on hippocampal morphology in major depression. Genes Brain Behav. 2011;10(7):756–764. doi: 10.1111/j.1601-183X.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Benjamin S, McQuoid DR, Potter GG, et al. The brain-derived neurotrophic factor Val66Met polymorphism, hippocampal volume, and cognitive function in geriatric depression. Am J Geriatr Psychiatry. 2010;18(4):323–331. doi: 10.1097/JGP.0b013e3181cabd2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Steffens DC, McQuoid DR, Payne ME, et al. Change in hippocampal volume on magnetic resonance imaging and cognitive decline among older depressed and nondepressed subjects in the neurocognitive outcomes of depression in the elderly study. Am J Geriatr Psychiatry. 2011;19(1):4–12. doi: 10.1097/JGP.0b013e3181d6c245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zeman M, Jáchymová M, Jirák R, et al. Polymorphisms of genes for brain-derived neurotrophic factor, methylenetetrahydrofolate reductase, tyrosine hydroxylase, and endothelial nitric oxide synthase in depression and metabolic syndrome. Folia Biol (Praha) 2010;56(1):19–26. [PubMed] [Google Scholar]
- [13].Kishida I, Aklillu E, Kawanishi C, et al. Monoamine metabolites level in CSF is related to the 5-HTT gene polymorphism in treatment-resistant depression. Neuropsychopharmacology. 2007;32(10):2143–2151. doi: 10.1038/sj.npp.1301336. [DOI] [PubMed] [Google Scholar]
- [14].Yokota C, Kuge Y, Inoue H, et al. Bilateral induction of the S-100A9 gene in response to spreading depression is modulated by the cyclooxygenase-2 activity. J Neurol Sci. 2005;234(1-2):11–16. doi: 10.1016/j.jns.2005.02.008. [DOI] [PubMed] [Google Scholar]
- [15].Cui Y, Kataoka Y, Inui T, et al. Up-regulated neuronal COX-2 expression after cortical spreading depression is involved in non-REM sleep induction in rats. J Neurosci Res. 2008;86(4):929–936. doi: 10.1002/jnr.21531. [DOI] [PubMed] [Google Scholar]
- [16].Porter RJ, Mulder RT, Joyce PR, et al. Tryptophan hydroxylase gene (TPH1) and peripheral tryptophan levels in depression. J Affect Disord. 2008;109(1-2):209–212. doi: 10.1016/j.jad.2007.11.010. [DOI] [PubMed] [Google Scholar]
- [17].Dulawa SC, Holick KA, Gundersen B, et al. Effects of chronic fluoxetine in animal models of anxiety and depression. Neuropsychopharmacology. 2004;29(7):1321–1330. doi: 10.1038/sj.npp.1300433. [DOI] [PubMed] [Google Scholar]
- [18].Navailles S, Zimnisky R, Schmauss C. Expression of glucocorticoid receptor and early growth response gene 1 during postnatal development of two inbred strains of mice exposed to early life stress. Dev Neurosci. 2010;32(2):139–148. doi: 10.1159/000293989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Helmreich DL, Parfitt DB, Lu XY, et al. Relation between the hypothalamic-pituitary-thyroid (HPT) axis and the hypothalamic-pituitary-adrenal (HPA) axis during repeated stress. Neuroendocrinology. 2005;81(3):183–192. doi: 10.1159/000087001. [DOI] [PubMed] [Google Scholar]
- [20].Jarrell H, Hoffman JB, Kaplan JR, et al. Polymorphisms in the serotonin reuptake transporter gene modify the consequences of social status on metabolic health in female rhesus monkeys. Physiol Behav. 2008;93(4-5):807–819. doi: 10.1016/j.physbeh.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Morag A, Pasmanik-Chor M, Oron-Karni V, et al. Genome-wide expression profiling of human lymphoblastoid cell lines identifies CHL1 as a putative SSRI antidepressant response biomarker. Pharmacogenomics. 2011;12(2):171–184. doi: 10.2217/pgs.10.185. [DOI] [PubMed] [Google Scholar]
- [22].Oved K, Morag A, Pasmanik-Chor M, et al. Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics. 2012;13(10):1129–1139. doi: 10.2217/pgs.12.93. [DOI] [PubMed] [Google Scholar]
- [23].Cattaneo A, Sesta A, Calabrese F, et al. The expression of VGF is reduced in leukocytes of depressed patients and it is restored by effective antidepressant treatment. Neuropsychopharmacology. 2010;35(7):1423–1428. doi: 10.1038/npp.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Molteni R, Calabrese F, Cattaneo A, et al. Acute stress responsiveness of the neurotrophin BDNF in the rat hippocampus is modulated by chronic treatment with the antidepressant duloxetine. Neuropsychopharmacology. 2009;34(6):1523–1532. doi: 10.1038/npp.2008.208. [DOI] [PubMed] [Google Scholar]
- [25].Parihar VK, Hattiangady B, Kuruba R, et al. Predictable chronic mild stress improves mood, hippocampal neurogenesis and memory. Mol Psychiatry. 2011;16(2):171–183. doi: 10.1038/mp.2009.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kálmán J, Palotás A, Juhász A, et al. Impact of venlafaxine on gene expression profile in lymphocytes of the elderly with major depression--evolution of antidepressants and the role of the “neuro-immune” system. Neurochem Res. 2005;30(11):1429–1138. doi: 10.1007/s11064-005-8513-9. [DOI] [PubMed] [Google Scholar]
- [27].Lee JH, Ko E, Kim YE, et al. Gene expression profile analysis of genes in rat hippocampus from antidepressant treated rats using DNA microarray. BMC Neurosci. 2010;11:152. doi: 10.1186/1471-2202-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yoon HK, Kim YK. Association between serotonin-related gene polymorphisms and suicidal behavior in depressive patients. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(5):1293–1297. doi: 10.1016/j.pnpbp.2008.04.004. [DOI] [PubMed] [Google Scholar]
- [29].Jacobsen JP, Medvedev IO, Caron MG. The 5-HT deficiency theory of depression: perspectives from a naturalistic 5-HT deficiency model, the tryptophan hydroxylase 2Arg439His knockin mouse. Philos Trans R Soc Lond B Biol Sci. 2012;367(1601):2444–2459. doi: 10.1098/rstb.2012.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cervo L, Canetta A, Calcagno E, et al. Genotype-dependent activity of tryptophan hydroxylase-2 determines the response to citalopram in a mouse model of depression. J Neurosci. 2005;25(36):8165–8172. doi: 10.1523/JNEUROSCI.1816-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Savignac HM, Dinan TG, Cryan JF. Resistance to early-life stress in mice: effects of genetic background and stress duration. Front Behav Neurosci. 2011;5:13. doi: 10.3389/fnbeh.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Larsen MH, Hay-Schmidt A, Rønn LC, et al. Temporal expression of brain-derived neurotrophic factor (BDNF) mRNA in the rat hippocampus after treatment with selective and mixed monoaminergic antidepressants. Eur J Pharmacol. 2008;578(2-3):114–122. doi: 10.1016/j.ejphar.2007.08.050. [DOI] [PubMed] [Google Scholar]
- [33].Park SH, Choi SH, Lee J, et al. Effects of repeated citalopram treatments on chronic mild stress-induced growth associated protein-43 mRNA expression in rat hippocampus. Korean J Physiol Pharmacol. 2008;12(3):117–123. doi: 10.4196/kjpp.2008.12.3.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wong ML, Dong C, Maestre-Mesa J, et al. Polymorphisms in inflammation-related genes are associated with susceptibility to major depression and antidepressant response. Mol Psychiatry. 2008;13(8):800–812. doi: 10.1038/mp.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mandelli L, Serretti A, Marino E, et al. Interaction between serotonin transporter gene, catechol-O-methyltransferase gene and stressful life events in mood disorders. Int J Neuropsychopharmacol. 2007;10(4):437–447. doi: 10.1017/S1461145706006882. [DOI] [PubMed] [Google Scholar]
- [36].Lotrich FE. Gene-environment interactions in geriatric depression. Psychiatr Clin North Am. 2011;34(2):357–376. doi: 10.1016/j.psc.2011.02.003. viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ramanathan M, Kumar SN, Suresh B. Evaluation of cognitive function of fluoxetine, sertraline and tianeptine in isolation and chronic unpredictable mild stress-induced depressive Wistar rats. Indian J Exp Biol. 2003;41(11):1269–1272. [PubMed] [Google Scholar]
- [38].Heine VM, Maslam S, Zareno J, et al. Suppressed proliferation and apoptotic changes in the rat dentate gyrus after acute and chronic stress are reversible. Eur J Neurosci. 2004;19(1):131–144. doi: 10.1046/j.1460-9568.2003.03100.x. [DOI] [PubMed] [Google Scholar]
- [39].Veena J, Rao BS, Srikumar BN. Regulation of adult neurogenesis in the hippocampus by stress, acetylcholine and dopamine. J Nat Sci Biol Med. 2011;2(1):26–37. doi: 10.4103/0976-9668.82312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Frye CA, Walf AA. Changes in progesterone metabolites in the hippocampus can modulate open field and forced swim test behavior of proestrous rats. Horm Behav. 2002;41(3):306–315. doi: 10.1006/hbeh.2002.1763. [DOI] [PubMed] [Google Scholar]
- [41].The Ministry of Science and Technology of the People's Republic of China. Guidance Suggestions for the Care and Use of Laboratory Animals. 2006-09-30 [Google Scholar]
- [42].Dziedzicka-Wasylewska M, Willner P, Papp M. Changes in dopamine receptor mRNA expression following chronic mild stress and chronic antidepressant treatment. Behav Pharmacol. 1997;8(6-7):607–618. doi: 10.1097/00008877-199711000-00017. [DOI] [PubMed] [Google Scholar]
- [43].Ye Y, Wang G, Wang H, et al. Brain-derived neurotrophic factor (BDNF) infusion restored astrocytic plasticity in the hippocampus of a rat model of depression. Neurosci Lett. 2011;503(1):15–19. doi: 10.1016/j.neulet.2011.07.055. [DOI] [PubMed] [Google Scholar]
- [44].Zhuge QC. 3rd ed. Beijing: People's Medical Pubishing House; 2005. The Rat Brain in Stereotaxic Coordinates. [Google Scholar]
- [45].Yu H, Wang DD, Wang Y, et al. Variant brain-derived neurotrophic factor Val66Met polymorphism alters vulnerability to stress and response to antidepressants. J Neurosci. 2012;32(12):4092–4101. doi: 10.1523/JNEUROSCI.5048-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].South T, Westbrook F, Morris MJ. Neurological and stress related effects of shifting obese rats from a palatable diet to chow and lean rats from chow to a palatable diet. Physiol Behav. 2012;105(4):1052–1057. doi: 10.1016/j.physbeh.2011.11.019. [DOI] [PubMed] [Google Scholar]
- [47].Spasojevic N, Jovanovic P, Dronjak S. Chronic fluoxetine treatment affects gene expression of catecholamine enzymes in the heart of depression model rats. Indian J Exp Biol. 2012;50(11):771–775. [PubMed] [Google Scholar]


