Abstract
Heat shock protein 90α (Hsp90α)/p23 and Hsp90β/p23 interactions are crucial for proper folding of proteins involved in cancer and neurodegenerative diseases. Small molecule Hsp90 inhibitors block Hsp90α/p23 and Hsp90β/p23 interactions in part by preventing ATP binding to Hsp90. The importance of isoform-selective Hsp90α/p23 and Hsp90β/p23 interactions in determining the sensitivity to Hsp90 was examined using 293T human kidney cancer cells stably expressing split Renilla luciferase (RL) reporters. Interactions between Hsp90α/p23 and Hsp90β/p23 in the split RL reporters led to complementation of RL activity, which was determined by bioluminescence imaging of intact cells in cell culture and living mice using a cooled charge-coupled device camera. The three geldanamycin-based and seven purine-scaffold Hsp90 inhibitors led to different levels of inhibition of complemented RL activities (10–70%). However, there was no isoform selectivity to both classes of Hsp90 inhibitors in cell culture conditions. The most potent Hsp90 inhibitor, PU-H71, however, led to a 60% and 30% decrease in RL activity (14 hr) in 293T xenografts expressing Hsp90α/p23 and Hsp90β/p23 split reporters respectively, relative to carrier control–treated mice. Molecular imaging of isoform-specific Hsp90α/p23 and Hsp90β/p23 interactions and efficacy of different classes of Hsp90 inhibitors in living subjects have been achieved with a novel genetically encoded reporter gene strategy that should help in accelerating development of potent and isoform-selective Hsp90 inhibitors.
Introduction
Protein-protein interactions play very important roles in different biological processes, including cell proliferation, differentiation, and death (1). However, proteins must be properly folded before they can interact with each other. In mammalian cells, protein folding is mediated by the heat shock protein 90 (Hsp90) chaperone system, including Hsp90 and the cochaperones p23, Hip, Hop, and Hsp70 (2, 3). Overexpression of Hsp90 in human cancers correlates with poor prognosis (4, 5). Hsp90 interacts strongly with the cochaperones as a fully active multichaperone complex (6), and its ATP affinity and ATPase activity in cancer cells are higher than in normal cells (4, 6). The most important interaction within the Hsp90 chaperone system is between Hsp90(α/β) and p23, which occurs only when Hsp90 is in the ATP bound form. Hsp90/p23 interactions are important for assembly of functional Hsp90/client proteins complex, release of client proteins, and disassembly of transcriptionally active complex (2, 3).
Small molecule inhibitors have been developed to inhibit Hsp90 ATPase activity by targeting its ATP-binding pocket (2, 7–9). These inhibitors have higher binding affinities to Hsp90 in cancer compared with that of normal cells (6). They preclude p23 binding to Hsp90 and lead to misfolding, partly by competitively blocking ATP binding to Hsp90. The subsequent degradation of the misfolded proteins leads to simultaneous inhibition of multiple signal transduction pathways and cell growth arrest. The two classes of Hsp90 inhibitors that are in advanced developmental stages are the geldanamycin-based and the purine-scaffold Hsp90 inhibitors. Two of the geldanamycin-based Hsp90 inhibitors, 17-allylamino-17-demethoxygeldanamycin (17-AAG) and 17-(dime-thylaminoethylamino)-17-demethoxygeldanamycin (17-DMAG), are now in phase I/phase II clinical trials for patients with advanced malignancies (10–12). Likewise, purine-scaffold Hsp90 inhibitors have also been rationally designed and are in both advanced preclinical and phase I clinical evaluation (12, 9).
The investigation of Hsp90/p23 interactions has been limited to in vitro analyses, such as binding assays and coimmunoprecipitation (13–15). These methods are sensitive to the ionic strength of the detergents/buffers and may not accurately reflect the nature of interactions between Hsp90/p23 in intact living cells. Phenotypic assays have also been developed to examine the downstream effects of the inhibition of Hsp90/p23 interactions (i.e., degradation of Hsp90 client proteins) by different Hsp90 inhibitors (7, 16–19). Some of these Hsp90 inhibitors have shown efficacy for growth inhibition in cell culture and xenograft models in living mice (9, 19). Recently, a noninvasive imaging method was developed to monitor Her2 degradation by Hsp90 inhibitors in a breast cancer xenograft model (20). In spite of these advances, longitudinal studies for monitoring the efficacies of Hsp90 inhibitors cannot be achieved noninvasively without sacrificing the mice at each time point before excision of tumors for in vitro analyses. Furthermore, because both Hsp90 isoforms (α and β) are expressed in cancer cells, it is not possible to decipher the individual contribution of each isoform in determining the sensitivity of Hsp90 inhibitors.
To examine the contribution of isoform-selective Hsp90α/p23 and Hsp90β/p23 interactions in tumor responsiveness to Hsp90 inhibitors noninvasively in cell culture and living mice, we used genetically encoded reporters that are based on split Renilla luciferase (RL) protein fragment–assisted complementation (SRL-PFAC) developed by our laboratory (21–23). This system is based on the complementation of two inactive halves of the full-length RL mediated by the interaction between two positively interacting proteins. SRL-PFAC was previously used to monitor heterodimerization between MyoD/Id (24), homodimerization of herpes simplex virus thymidine kinase (23), and rapamycin-mediated mTOR/FKBP12 interactions (22), both in cell culture and living mice by optical bioluminescence imaging.
The SRL-PFAC system is advantageous because (a) the interaction of each Hsp90 isoform (α/β) with p23 can be individually monitored; (b) signal amplification is achieved through the enzymatic properties of complemented RL activity; (c) RL exhibits flash kinetics (25) and, thus, provides a real-time monitoring of Hsp90α/p23 and Hsp90β/p23 interactions; (d) SRL-PFAC has a high signal to background ratio and potentially less steric hindrance due to the small size of RL fragments (NRL, 23 kDa; CRL, 13 kDa); (e) RL does not require ATP for its activity, thus important for screening inhibitors that target the ATP-binding pocket of Hsp90; (f) the fusion constructs can be optimized and validated in cell culture before generation of stable cells; and (g) repetitive imaging of the same mouse allows dynamic indirect monitoring of Hsp90α/p23 and Hsp90β/p23 interactions in response to treatment with Hsp90 inhibitors, wherein the pharmacokinetic properties of each Hsp90 inhibitor influence efficacy.
In the current work, we implemented the SRL-PFAC system to indirectly examine the kinetics of Hsp90α/p23 and Hsp90β/p23 interactions and showed its feasibility in monitoring both in cell culture and in small living subjects the efficacy and isoform selectivity of Hsp90 inhibitors.
Materials and Methods
Chemicals, enzymes, and reagents
The pET3a vectors encoding the cDNA for human Hsp90α, Hsp90β, p23, and a p23 (F103A) mutant were gifts from Dr. David Toft (Mayo Clinic). The plasmids pCMV-hRL encoding the full-length synthetic RL and pCMV-FL encoding the full-length firefly luciferase (FL), LARII substrate for FL assay, and 5× passive lysis buffer, EnduRen Live Cell Substrate, were purchased from Promega. Restriction enzymes, modification enzymes, and ligase were purchased from New England Biolabs. TripleMaster TaqDNA polymerase for PCR amplification was purchased from Brinkmann Eppendorf. Site-directed mutagenesis was performed using the Stratagene kit. Amplicillin and kanamycin for bacterial culture, DMSO were purchased from Sigma. Plasmid extraction kits and DNA gel extraction kits were purchased from Qiagen. Coelenterazine was purchased from Nanolight Technology. All animal cell culture media, fetal bovine serum (FBS), antibiotics streptomycin and penicillin, and plastic wares for cell cultures, bacterial culture media, Lipofecta-mine 2000 transfection reagents, and 4% to 12% gradient SDS-PAGE gels were purchased from Invitrogen. Geldanamycin, 17-AAG, 17-DMAG, and puromycin hydrochloride (100 mg/mL) were purchased from Invivogen. Geldanamycin, 17-AAG, and 17-DMAG were dissolved in DMSO as stock solutions (1 mg/mL), aliquoted, and stored at −20°C. Purine-scaffold Hsp90 inhibitors were synthesized as previously reported (16, 26, 27), dissolved as 100 mmol/L DMSO stock, and stored at −20°C.
Construction of plasmids
The NH2 terminal portion (NRL, amino acids 1–229) and COOH terminal portion (CRL, amino acids 230–311) of the hRluc gene was PCR amplified as described before (21–25). The full-length human Hsp90α and a full-length p23 (F103A) mutant and Hsp90β fragments (corresponding to amino acids 1–486 and full length) were amplified using the forward primers designed with Nhe1 or BamH1 and the corresponding reverse primers designed with BamH1 or XhoI with a stop codon and were subcloned upstream or downstream of NRL or CRL using corresponding restriction enzymes. Mutageneses of Hsp90β were performed using the Stratagene mutagenesis kit and confirmed by direct sequencing. To attach a Myc-tag to the COOH terminus of Hsp90β2.2-CRL, the c-myc peptide sequences (EQKLISEEDL) were incorporated into the XhoI reverse primer to use in conjunction with the Hsp90β Nhe1 forward primer for PCR cloning.
Cell culture
All cell lines used in this study were purchased from American Type Culture Collection. Human 293T embryonic kidney cancer cells were grown in MEM supplemented with 10% FBS and 1% penicillin/ streptomycin solutions.
Cell transfection, FL, and RL assays
Transfections were performed in 80% confluent 24-h-old cultures of 293T. For transfection, 250 ng/well of pcDNA molar equivalent of each split RL reporters, full-length RL, were used in 12-well tissue culture plates, and 5 AL Lipofectamine 2000 was used per transfection. pcDNA3.1(+) vector (5 ng) expressing full-length FL was cotransfected per well to normalize for transfection efficiency. The cells were assayed after 24-h incubation at 37°C at 5% CO2. The luminometer assays for FL and RL activity were performed as previously described (21, 22, 24). Protein concentrations in cell lysates were determined by Bradford assay (Bio-Rad). RL activities were normalized for protein content and transfection efficiency using FL activity and expressed as relative light units per microgram protein per minute of counting (RLU/μg protein/min).
Selection of 293T cells stably expressing NRL(M185V)-p23/Hsp90α2.2-CRL and NRL(M185V)-p23/Hsp90β2.2-CRL and evaluation of the efficacy of Hsp90 inhibitors in cell culture
Eighty percent confluent 293T cells were cotransfected with 5 μg each of pcDNA3.1+ plasmids expressing NRL(M185V)-p23 with Hsp90α2.2-CRL or NRL(M185V)-p23 with Hsp90β2.2-CRL for 24 h, before replating in 1.5 μg/mL of puromycin hydrochloride to allow formation of individual colonies. To screen for 293T cells expressing both Hsp90α/p23 and Hsp90β/p23 split RL reporters, cell culture medium was aspirated, 3 μg coelenterazine diluted in 3 mL of PBS were added, and bioluminescence imaging was performed using the IVIS 50 imaging system (Xenogen Corp.) with an acquisition time of 3 min. Positive colonies with the highest RL activities were trypsinized and transferred to individual wells in a 24-well plate and grown in MEM containing 1.5 μg/mL of puromycin.
To determine the effect of Hsp90 inhibitors on complementation of Hsp90α/p23 and Hsp90β/p23 in intact cells, 3.5 × 104 293T cells stably transfected with split RL reporters [NRL(M185V)-p23/Hsp90α-CRL and NRL(M185V)-p23/Hsp90β-CRL] were plated in each well in the 96-well black well plate (Costar) and allowed to attach for 24 h before treatment with different concentrations of the geldanamycin-based and purine-scaffold Hsp90 inhibitors for 24 h. EnduRen live cell substrate (10 μg/mL; diluted in 50 μL of medium) was added to each well for 1.5 h, and RL activities were determined by bioluminescence imaging. Cell number in each well was determined by sulforhodamine B assay as previously described (28). Complemented RL activities were normalized for cell number and to that of cells treated with carrier controls (0.12% DMSO for geldanamycin, 17-AAG, 17-DMAG, PU3, PU24FCL, PU-H58, PU-L66, and PU-DZ7; 0.012% DMSO for PU-H71 and PU-DZ8).
Western blotting and coimmunoprecipitation
Coimmunoprecipitation of NRL(M185V)-p23/Hsp90β2.2-CRL-Myc was performed with the agarose-conjugated goat anti-myc antibody (Bethyl Laboratories). Agaroseconjugated goat IgG was used as a negative control. 293T cells were transiently cotransfected with 5 μg each of NRL(M185V)-p23 and Hsp90β2.2-CRL-Myc and treated with different concentrations of PU-H71 for 24 h before lysis with 0.1% NP40 buffer [20 mmol/L Tris-HCl (pH 7.4), 25 mmol/L NaCl, 2 mmol/L DTT, 20 mmol/L Na2MoO4, and 0.1% NP40] for 2 h at 4°C on a rotating platform. Cell debris was removed by centrifugation at 1.3 × 104 × g for 15 min at 4°C. The RL activity for each sample was determined by luminometer assay, whereas protein concentrations were determined by Bradford assay. RL activities in PU-H71–treated cells were normalized to that of carrier control–treated cells (as 100%). Three hundred micrograms of the total cell lysates were incubated with 20 μL of agaroseconjugated goat IgG or goat anti-myc antibody for 3 h on a rotating platform at 4°C. The beads were washed four times with ice-cold lysis buffer, and immunoprecipitated proteins were eluted in 20 μL of 2 × Tris (2-carboxy-ethyl)-phosphin-HCl (TCEP) buffer [156 mmol/L Tris-HCl, 50 mmol/L Tris base (Sigma), 12.5 mmol/L TCEP HCl (Pierce), 0.006% bromophenol blue (w/v), 24% glycerol (v/v), and 5% SDS (w/v)] at room temperature for 15 min. Eluted proteins were resolved on a 4% to 12% SDS-PAGE gradient gel and electroblotted to a nitrocellulose membrane (Schleicher & Schuell). The expression of split RL fusion constructs and Hsp70 was determined by Western blotting using 1 of 10 of the input for coimmunoprecipitation (30 μg of total protein). The membranes were blocked with 5% nonfat dry milk in TBS containing 0.01% Tween 20 for 1 h and probed overnight at 4°C on a rotating platform with mouse monoclonal antibody against RL (1:1,000 dilution; Chemicon) or Myc-Tag (Clone 9B11), rabbit polyclonal antibodies against human Hsp70 (both 1:1,000 dilution; Cell Signaling), and mouse monoclonal antibody against human α-tubulin (1:5,000 dilution, Clone B-5-1-2, Sigma). Secondary antibodies were peroxidase-conjugated goat anti-mouse IgG or anti-rabbit IgG (1:3,000 dilution for both; Cell Signaling). Immunoblots were developed using the LumiGlo enhanced chemiluminescence method (Cell Signaling) using blue films (Midwest Scientific). The quantitation of coimmunoprecipitation was performed as follows: the band intensities of Hsp90β2.2-CRLMyc and NRL(M185V)-p23 from the Myc-Ab beads lanes (treated with carrier control or PU-H71) were quantitated using NIH imaging J. The ratio of the intensities [Hsp90β2.2-CRL-Myc/NRL(M185V)-p23] was normalized to that of the carrier control–treated cells (as 100%).
Optical charge-coupled device imaging in living mice
Animal handling was performed in accordance with Stanford University Animal Research Committee guidelines. Mice were gas anesthetized using isofluorane (2% isoflurane in 100% oxygen, 1 L/min) during all injection and imaging procedures. Mice were imaged using a cooled charge-coupled device (CCD) camera (Xenogen IVIS29; Xenogen Corp.). 293T cells (5 × 106) stably cotransfected with NRL(M185V)-p23/Hsp90α2.2-CRL or NRL(M185V)-p23/Hsp90β2.2-CRL were implanted s.c. in the bottom left and right flanks of each female nude mouse of 7 weeks old (nu/nu, Charles River), respectively, for 14 days for tumor establishment. To determine RL activity in the implanted tumors in living mice, 3 μg of coelenterazine in 5% ethanol:95% PBS (final volume, 100 μL) were injected via tail vein, and the mice were immediately imaged with the IVIS system with an acquisition time of 3 min. The animals were placed prone in a light-tight chamber, and a gray scale reference image was obtained under low-level illumination. Photons emitted from cells implanted in the mice were collected and integrated for 3 min. Images were obtained using Living Image Software (Xenogen Corp.) and Igor Image Analysis Software (Wavemetrics). To quantify the measured light, the maximum photons per second per square centimeter per steradian were obtained over regions of implanted cells as validated previously. One of the sets were i.p. injected with 200 μL of 75 mg/kg of PU-H71 dissolved in 6.7% DMSO/PBS or 6.7% DMSO/PBS as carrier control. At 6 and 14 h later, 30 μg of coelenterazine were injected in the tail vein for follow-up imaging of the effects of the Hsp90 inhibitors on complemented RL activity, as described above. Max photons at 6 and 14 h were normalized to that of at 0 h for each individual mouse and expressed as average max photons ± SE.
Data analysis
Each experiment was repeated at least thrice, and results were expressed as mean F SE. Statistical differences were determined by Student's t test using P < 0.05 as cutoff point.
Results SRL-PFAC system was specific for indirect monitoring of Hsp90β/p23 interactions
We tested the hypothesis that SRLPFAC can be used to indirectly monitor interactions between p23 and each of the Hsp90 isoforms (Fig. 1A, top). In the presence of ATP, interactions between Hsp90 and p23 bring the two inactive halves of full-length RL (NRL and CRL) in close proximity, leading to complementation of RL activity and photon production in the presence of the RL substrate coelenterazine (Fig. 1A, middle). To determine if inhibition of Hsp90α/p23 and Hsp90β/p23 interactions would lead to reduction in complemented RL activity, we made use of geldanamycin, two geldanamycin derivatives (17-AAG and 17-DMAG), and seven purine-scaffold Hsp90 inhibitors (Fig. 1B). Binding of geldanamycin-based and purine-scaffold Hsp90 inhibitors to Hsp90 leads to conformation changes in Hsp90 that prevents ATP binding, thus precludes Hsp90 from interacting with p23 and reduces complementation of NRL/CRL and RL activity (Fig. 1A, bottom). To create fusion constructs expressing p23 and Hsp90, full-length human p23 and the human 1.4 kb Hsp90β fragment (29) were subcloned upstream or downstream of NRL and CRL fragments. These constructs were transiently cotransfected in pairs into 293T human embryonic kidney cancer cells to determine the orientation that led to the highest complementation of RL activity, relative to pcDNA vector control and full-length RL-transfected cells. Because the substrates of RL (coelenterazine) and FL (d-luciferin) do not cross-react (30), a pcDNA vector expressing FL was cotransfected to normalize for transfection efficiency.
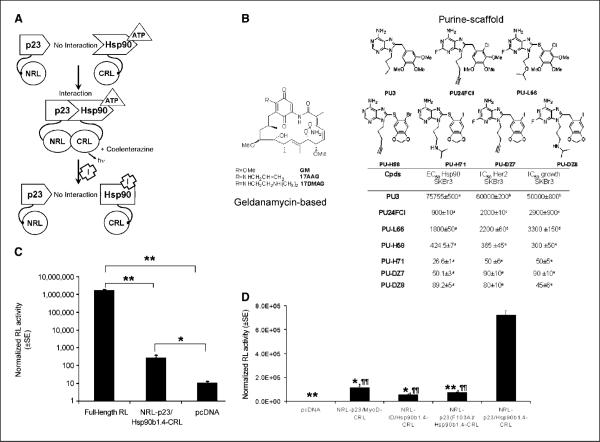
Figure 1.
SRL-PFAC was sensitive and specific for indirect monitoring of Hsp90/p23 interactions. A, schematic diagram for monitoring Hsp90/p23 interaction using the SRL-PFAC. The two interacting proteins p23 and Hsp90 were fused to the NRL (amino acids 1–229) and CRL (amino acids 230–311) portion of the RL through an EF and (G4S)2 peptide linker, respectively (top). In the presence of ATP, p23/Hsp90 interactions brought NRL and CRL in close proximity and led to complementation of RL enzyme activity and photon production in the presence of the substrate coelenterazine. Binding of Hsp90 inhibitors (I) to Hsp90-CRL leads to a conformation change and prevents ATP from binding, thus diminishing the interaction between NRL-p23 and Hsp90-CRL and complementation of RL activity. B, structure of geldanamycin-based Hsp90 inhibitors used in this study (left). Structure (top right) and Hsp90-binding affinity, SKBr3 cell growth inhibition, and IC50 values for Her2 degradation (bottom right) of purine-scaffold Hsp90 inhibitors used in this study. a–e, determined as described in refs. 15, 19, 45–47, respectively. C, complementation of Hsp90/p23 split RL reporter was orientation specific. 293T cells were transiently cotransfected with plasmids expressing the Hsp90β1.4 and p23 fused to NRL and CRL in eight possible orientations. FL was transfected to control for transfection efficiency. RL and FL activities were determined by luminometer assays 24 h posttransfection and normalized for protein content. Cotransfection of NRL-p23/Hsp90β1.4-CRL led to RL activity that was significantly higher than that of pcDNA vector control–transfected cells (P < 0.05). 293T cells transfected with full-length RL were used as a positive control. Columns, mean; bars, SE. *, P < 0.05; **, P < 0.005. D, specificity of SRL-PFAC for monitoring Hsp90β1.4/p23 interaction was determined using noninteracting protein partners (NRL-p23/MyoD-CRL and NRL-Id/Hsp90β1.4-CRL) and a p23 (F103A) mutant that does not interact with Hsp90 [NRL-p23(F103A)/Hsp90β1.4-CRL]. RL and FL activities were determined as described in B. Columns, mean; bars, SE. *, P < 0.05; **, P < 0.005 relative to 293T cells transiently cotransfected with NRL-p23/Hsp90β1.4-CRL (normalized to 100%). ¶, P < 0.005 relative to 293T cells transiently transfected with pcDNA.
Among the eight possible orientations of fusion constructs, cotransfection of NRL-p23/Hsp90β1.4-CRL led to the highest RL activity (Fig. 1C) compared with cells transfected with pcDNA vector control (P < 0.05). Similar results were observed in MCF-7 human breast cancer cells, HT1080 human fibrosarcoma cells, and Chinese hamster ovary cells (data not shown). To determine the specificity of NRL-p23/Hsp901.4h-CRL interactions, 293T cells were transiently cotransfected with constructs expressing noninteracting protein partners of NRL-p23/MyoD-CRL or NRL-Id/Hsp90β1.4-CRL. Furthermore, a human p23(F103A) mutant that does not interact with chicken Hsp90α (31) was incorporated into NRL as NRL-p23(F103A) and transiently cotransfected with Hsp90β1.4-CRL into 293T cells. Figure 1D shows that RL activity in 293T cells transiently cotransfected with NRL-p23/Hsp90β1.4-CRL was significantly higher than that of cells cotransfected with noninteracting protein partners of NRL-p23/MyoD-CRL, NRL-Id/Hsp90β-CRL (P < 0.05 in both cases), or NRL-p23(F103A)/Hsp90β1.4-CRL (P < 0.005). All together, our data support that SRL-PFAC was specific for indirect monitoring of Hsp90β/p23 interactions.
Full-length Hsp90α and Hsp90β in CRL and M185V mutation in NRL further increased sensitivity of SRL-PFACfor monitoring Hsp90/p23 interactions
Constitutive dimerization in the COOH terminus of purified full-length human Hsp90α was shown to enhance p23 binding (32). To determine if higher levels of complemented RL activity can be achieved using full-length Hsp90β and Hsp90α (both 2.2 kb), Hsp90α2.2-CRL and Hsp90β2.2-CRL fusion constructs were transiently cotransfected with NRL-p23. Figure 2A shows that cotransfection of NRL-p23 with Hsp90α2.2-CRL or Hsp90β2.2-CRL led to a 2-fold and 3- fold increase in complemented RL activities, respectively, compared with that of NRL-p23/Hsp90β1.4-CRL. Specificity of NRL-p23/ Hsp90α2.2-CRL and NRL-p23/Hsp90β2.2-CRL complemented RL activity was confirmed using the NRL-p23 (F103A) mutant control (P < 0.001). Transient transfection of Hsp90α2.2-CRL and Hsp90β2.2-CRL alone led to similar background RL activity compared with that of Hsp90β1.4-CRL (P > 0.05).
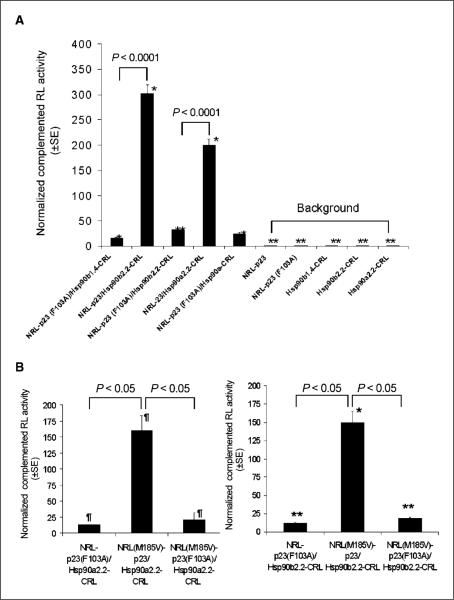
Figure 2.
Sensitivity of SRL-PFAC for indirect monitoring of Hsp90/p23 interactions was increased by using full-length Hsp90 (α and β) and incorporation of the M185V mutation in NRL. A, effect of incorporating full-length Hsp90α and Hsp90β into CRL. Full-length Hsp90αand Hsp90β (both 2.2 kb) were subcloned upstream of CRL as the Hsp90α2.2-CRL and Hsp90β2.2-CRL fusion constructs, respectively, and transiently cotransfected with NRL-p23 in 293T cells. FL was used to control for transfection efficiency. Specificity of interactions was determined using an NRL-p23(F103A) mutant control as described in Fig. 1D and expressed as percentage of RL activity of 293T cells transiently cotransfected with NRL-p23/ Hsp90β1.4-CRL (normalized to 100%). *, P < 0.05; **, P < 0.005 relative to NRL-p23/Hsp90β1.4-CRL transfected cells. B, effect of M185V mutation in NRL on complemented RL activity. 293T cells were transiently cotransfected with NRL(WT)-p23 or NRL(M185V)-p23 and Hsp90α2.2-CRL or Hsp90β2.2-CRL. FL was used to control for transfection efficiency. Specificity of interactions was determined using the NRL(WT)-p23(F103A) or NRL(M185V)-p23(F103A) mutant controls and expressed as percentage of RL activity of 293T cells transiently cotransfected with NRL(WT)-p23/Hsp90α2.2-CRL or NRL(WT)-p23/ Hsp90β2.2-CRL (normalized to 100%). ¶, P < 0.05 relative to NRL(WT)-p23/ Hsp90α2.2-CRL. *, P < 0.05; **, P < 0.01 relative to NRL(WT)-p23/Hsp90β2.2-CRL.
The M185V mutation in RL was shown to increase photon production by ~3-fold, without significant changes in serum stability (25). To determine if incorporation of the M185V mutation into NRL-p23 fusion construct would lead to higher complemented RL activity, NRL(M185V)-p23 was transiently cotransfected with Hsp90α2.2-CRL or Hsp90β2.2-CRL. Figure 2B shows that transient cotransfection of Hsp90α2.2-CRL (left) or Hsp90β2.2-CRL (right) with NRL(M185V)-p23 led to a 1.5-fold increase in complemented RL activity compared with that of NRL(WT)-p23/Hsp90α2.2-CRL or NRL(WT)-p23/Hsp90β2.2-CRL (P < 0.05 in both cases). The specificity of RL complementation was also shown using the NRL(M185V)-p23(F103A) mutant construct. Compared with pcDNA vector control–transfected cells, NRL(M185V)-p23/ Hsp90α2.2-CRL–transfected and NRL(M185V)-p23/Hsp90β2.2-CRL–transfected cells led to signal to background ratios of f12 and 14, respectively (data not shown).
SRL-PFAC was specific for indirect monitoring of disruption of Hsp90β/p23 interactions by Hsp90 inhibitors
To determine if SRL-PFAC can be used to indirectly monitor disruption of Hsp90/ p23 interactions by Hsp90 inhibitors, 293T cells transiently cotransfected with NRL(M185V)-p23/Hsp90β2.2-CRL were treated with the purine-scaffold Hsp90 inhibitor PU-DZ7 for 24 h before analysis of RL activity. The specificity of using NRL(M185V)-p23/ Hsp90β-CRL was confirmed using Hsp90β2.2(K107A)-CRL, Hsp90β2.2(K111A)-CRL, and Hsp90β2.2(K107AK111A)-CRL mutant fusion constructs. These mutations are analogous to the K111A mutation in chicken Hsp90α that reduces binding to geldanamycin (29). Figure 3A shows that PU-DZ7 lead to greater reduction in complemented RL activity of 293T cells cotransfected with NRL(M185V)-p23/Hsp90β2.2-CRL compared with that of NRL(M185V)-p23/Hsp90β2.2(K107A)-CRL or NRL(M185V)-p23/ Hsp90β2.2(K107AK111A)-CRL (P < 0.05 at 100 nmol/L and 200 nmol/L of PU-DZ7). Furthermore, complemented RL activity in 293T cells transiently transfected with NRL(M185V)-p23/ Hsp90β2.2(K111A)-CRL was insensitive to the effect of PU-DZ7 (P < 0.05 at all PU-DZ7 concentrations).
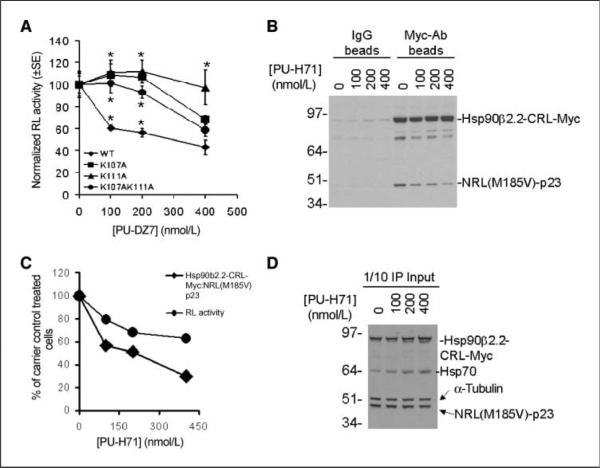
Figure 3.
SRL-PFAC was specific for evaluation of the efficacy of Hsp90 inhibitors in disruption of NRL-p23/Hsp90β-CRL interactions. A, specificity of Hsp90 inhibitors–mediated disruption of Hsp90/p23 interactions in cell culture was determined by Hsp90β mutants that do not bind to Hsp90 inhibitors. 293T cells transiently transfected with the indicated constructs for 24 h were treated with the Hsp90 inhibitor PU-DZ7 for 12 h before analysis of RL activity as described in Fig. 1C. RL activity was normalized for transfection efficiency and protein content and then to carrier control–treated cells. Points, mean; bars, SE. *, P < 0.05 relative to NRL(WT)-p23/Hsp90β2.2-CRL. B, coimmunoprecipitation of split RL reporters. 293T cells were transiently cotransfected with NRL(M185V)-p23/Hsp90β2.2-CRL-Myc and treated with the Hsp90 inhibitor PU-H71 for 24 h. Interactions between NRL(M185V)-p23/Hsp90β2.2-CRL-Myc in the absence or presence of PU-H71 were determined by immunoprecipitation using goat IgG control antibodies (left) or goat polyclonal anti-myc antibodies (right). The association with NRL(M185V)-p23 was detected using an RL antibody. C, quantitation of expression of NRL(M185V)-p23 and Hsp90β2.2-CRL-Myc (B) shows that the decrease in RL activity correlates with decrease association between NRL(M185V)-p23/Hsp90β2.2-CRL. D, the expression of NRL(M185V)-p23, Hsp90β2.2-CRL-Myc, and Hsp70 in transfected cells was determined by Western blotting using 1 of 10 of the cell lysates used for coimmunoprecipitation (IP).
To validate SRL-PFAC for indirectly monitoring Hsp90α/p23 and Hsp90β/p23 interactions, a myc-tag was appended to Hsp90β2.2-CRL as Hsp90β2.2-CRL-Myc to facilitate its detection by coimmunoprecipitation and western blotting. 293T cells transiently cotransfected with NRL(M185V)-p23/Hsp90β2.2-CRL-Myc were treated with the Hsp90 inhibitor PU-H71 (Fig. 1B) for 24 h before coimmunoprecipitation of Hsp90β2.2-CRL-Myc with Myc polyclonal antibodies. Treatment with PU-H71 decreased association between NRL(M185V)-p23/Hsp90β2.2-CRL-Myc compared with carrier control–treated cells (Fig. 3B), which also correspond to decrease in complemented RL activity (Fig. 3C). Inhibition of cellular Hsp90 by PU-H71 was confirmed by an increase in the expression of Hsp70, which is a feed-back mechanism characteristic of Hsp90 inactivation (33). The activity of PU-H71 was confirmed by the increased expression of Hsp70 in PU-H71–treated cells compared with that of carrier control cells. Furthermore, the protein expression of both NRL(M185V)-p23 and Hsp90β2.2-CRL-Myc was not significantly affected by PU-H71 (Fig. 3D), suggesting that the decrease in complemented RL activity was due to inhibition of NRL(M185V)-p23/Hsp90β2.2-CRL-Myc interactions per se instead of decreased expression of either split RL reporter. Thus, SRL-PFAC was specific for indirect monitoring of disruption of Hsp90β/p23 interactions by Hsp90 inhibitors. Because the ATP-binding domains of Hsp90α and Hsp90β are highly conserved (34), we therefore proceeded with NRL(M185V)-p23/Hsp90α2.2-CRL and NRL(M185V)-p23/Hsp90β2.2-CRL for subsequent experiments.
SRL-PFAC reveals differential sensitivities of Hsp90α/p23 and Hsp90β/p23 interactions to geldanamycin-based and purine-scaffold Hsp90 inhibitors
To circumvent the variability in expression of the Hsp90/p23 split reporters in transient transfection studies (Figs. 1–3), 293T cells were stably transfected with the constructs expressing NRL(M185V)-p23/Hsp90α2.2-CRL or NRL(M185V)-p23/Hsp90β2.2-CRL. To evaluate the efficacy of geldanamycin-based and purine-scaffold Hsp90 inhibitors in intact cells in 96-well format, these stable cells were treated with different concentrations of the inhibitors or carrier controls for 24 h. RL activities were determined by optical bioluminescence imaging of intact cells using a cooled CCD camera 1.5 h after addition of the RL substrate EnduRen, followed by normalization of cell number by sulforhodamine B cell proliferation assay (28).
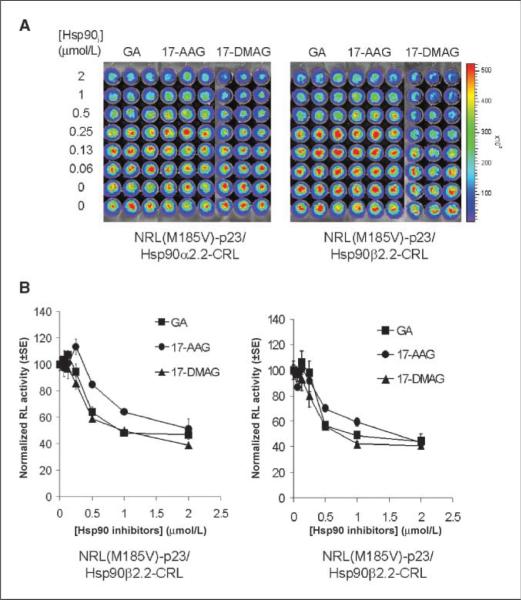
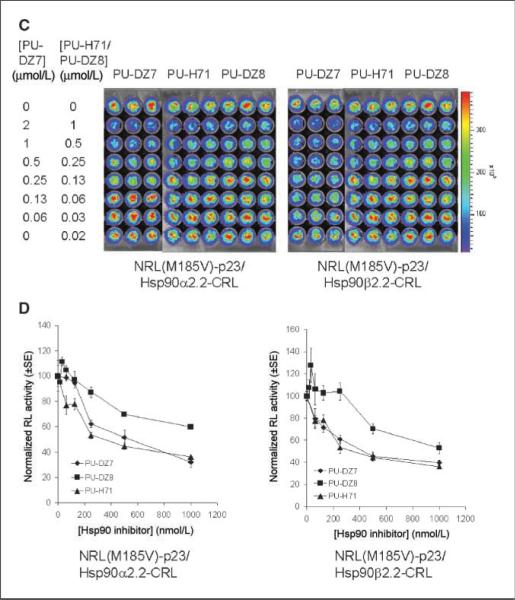
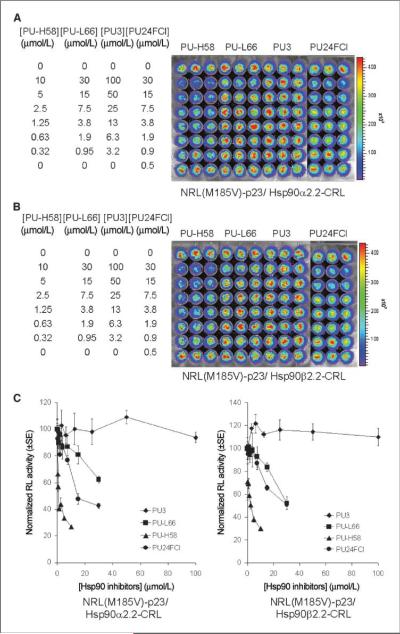
The geldanamycin-based inhibitors (Fig. 4A and B) led to similar inhibition of both NRL(M185V)-p23/Hsp90α-CRL and NRL(M185V)-p23/Hsp90β-CRL interactions, with a maximum inhibition of 53% to 55%, 49% to 57%, and 60% to 61% observed for geldanamycin, 17-AAG, and 17-DMAG, respectively, at 2 μmol/L. In the purine-scaffold series, the ability of these seven compounds to disrupt Hsp90α/p23 and Hsp90β/p23 interactions (Figs. 4C, D and 5) correlated with their previously reported affinity for Hsp90 from tumor lysates (Fig. 1B; refs. 13, 17, 20, 24). Maximum inhibition of 64% to 68% and 60% to 64% was observed at 1 μmol/L for both PU-H71 and PU-DZ7, respectively (Fig. 4D). Overall, there was no significant difference between the two Hsp90 isoforms (α and β) in determining the sensitivity to both geldanamycin-based and purine-scaffold Hsp90 inhibitors. There was no significant differences in the expression of MDR1 or the efflux of rhodamine 123 in PU-H71 or carrier control–treated 293T stable cells (data not shown); thus, the decrease in complemented RL activity was not due to up-regulation/activation of MDR1 that reduces accumulation of coelenterazine (35).
Figure 4.
Geldanamycin-based and purine-scaffold Hsp90 inhibitors led to different levels of inhibition of NRL(M185V)-p23/Hsp90-CRL complementation. A, effect of geldanamycin-based Hsp90 inhibitors (Hsp90i). 293T cells stably transfected with NRL(M185V)-p23/Hsp90α2.2-CRL (left) or NRL(M185V)-p23/Hsp90β2.2-CRL (right) were treated with geldanamycin (GA), 17-AAG, or 17-DMAG for 24 h before determination of complemented RL activity by bioluminescence imaging of intact cells. B, complemented RL activities in cells treated with Hsp90 inhibitors were normalized for cell number and then to carrier control–treated cells. Points, mean of the triplicates; bars, SE. C, effect of purine-scaffold Hsp90 inhibitors PU-DZ7, PU-H71, and PU-DZ8 (24 h) on complemented RL activities. RL activities were determined as in A. D, quantitation of complemented RL activity as in B.
Figure 5.
Purine-scaffold Hsp90 inhibitors led to different levels of inhibition of NRL(M185V)-p23/Hsp90-CRL complementation. Effect of purine-scaffold Hsp90 inhibitors (PU-H58, PU-H66, PU3, and PU24FCl) on complemented RL activities in 293T cells stably transfected with NRL(M185V)-p23/Hsp90β2.2-CRL (A) or NRL(M185V)-p23/Hsp90β2.2-CRL (B). RL activities were determined at 24 h posttreatment as described in Fig. 4A. C, quantitation of complemented RL activity.
The purine-scaffold Hsp90 inhibitor PU-H71 was more effective in disruption of Hsp90α/p23 interactions in living mice
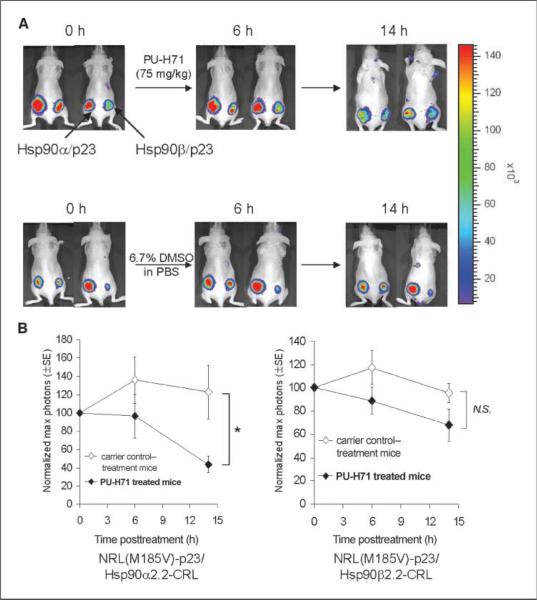
Low micromolar concentrations of purine-scaffold Hsp90 inhibitors were previously reported to be achieved intratumorally in s.c. xenografted mice after one administered dose of these inhibitors (36). PU-H71 is a highly water-soluble inhibitor which can be given to mice formulated in PBS (compared with that of PU-DZ7); it was, therefore chosen to validate the mechanisms of purine-scaffold Hsp90 inhibitors in living subjects. To determine the efficacy of PU-H71 in disruption of Hsp90α/p23 and Hsp90β/ p23 interactions in living mice, 293T cells stably transfected with the Hsp90α/p23 and Hsp90β/p23 split RL reporters used in the above cell culture studies (Figs. 4 and 5) were implanted in the lower left and right flank of nude mice (n = 10), respectively, for establishment of tumor xenografts. Bioluminescence imaging was performed immediately after tail vein injection of the RL substrate coelenterazine (30 μg) using a cooled CCD camera as previously described (21, 22, 24). After baseline scan, the mice were randomized (n = 5 per group) and treated with 75 mg/kg of PUH71 or equal volume of carrier control (6.7% DMSO in PBS) i.p. and reimaged at 6 and 14 h (Fig. 6A).
Figure 6.
Disruption of Hsp90α/p23 and Hsp90β/p23 interactions by Hsp90 inhibitors in living mice. 5 × 106 293T cells stably transfected with NRL(M185V)-p23/ Hsp90α2.2-CRL (left) or NRL(M185V)-p23/ Hsp90β2.2-CRL (right) were implanted s.c. in the lower flanks of each female nude mouse to allow xenograft establishment for 2 wk, respectively. A, mice (n = 10) were imaged at 0 h to determine the RL activities in the implanted cells by optical bioluminescence imaging using a cooled CCD camera with an acquisition time of 3 min immediately after tail vein injection of 30 μg of coelenterazine. Mice (n = 5) were subsequently i.p. injected with 75 mg/kg of purine-scaffold Hsp90 inhibitor PU-H71 (n = 5) diluted in 6.7% DMSO in PBS (carrier control) i.p. in a final volume of 200 μL. Control mice (n = 5) were treated i.p. with equal volume of carrier control. RL activity was determined at 6 and 14 h posttreatment. Representative images from two mice from each treatment group at each time point are shown with the optical bioluminescence image superimposed on the visible light image. Representative images from two mice were shown here in the PU-H71 and carrier control–treated group. B, quantitation of the maximum photons from site implanted with NRL(M185V)-p23/Hsp90α2.2-CRL (left) or NRL(M185V)-p23/Hsp90β2.2-CRL (right). Points, average of normalized maximum photons at 0 h; bars, SE. *, P < 0.05 relative to carrier control– treated mice; N.S., P > 0.05.
At 6 h post–PU-H71 administration, the average max photons decreased by 4% and 11% for xenograft expressing NRL(M185V)-p23/Hsp90α2.2-CRL and NRL(M185V)-p23/Hsp90β2.2-CRL compared with 36% and 18% increase in complemented RL activity in carrier control–treated mice, respectively (P > 0.05 in both cases). At 14 h, 56% and 32% decrease in maximum photons for xenograft expressing NRL(M185V)-p23/Hsp90α2.2-CRL (P < 0.05 relative to carrier control–treated mice at 0 h) and NRL(M185V)-p23/ Hsp90β2.2-CRL (P > 0.05 relative to carrier control–treated mice at 0 h) was observed, compared with 23% increase and 4% decrease in maximum photons in carrier control–treated mice, respectively (Fig. 6B). Therefore, PU-H71 was more selective in inhibition of Hsp90α/p23 interactions in living mice compared with that of Hsp90β/p23.
Discussion
The up-regulation of the Hsp90 chaperone system is thought to allow cancer cells to sustain expression of client proteins that are overexpressed or mutated in cancer (6, 37, 38). Different classes of small molecule inhibitors have been developed to inhibit Hsp90 (α and β), but the mechanisms by which these inhibitors disrupt Hsp90/p23 interactions, as well as the role of each Hsp90 isoform (α and β) in determining sensitivity to these inhibitors, have not been noninvasively and longitudinally examined in living subjects. Because both Hsp90α and Hsp90β are expressed in cancer cells and their expression was compensatory (38), the individual role(s) of Hsp90α/p23 and Hsp90β/p23 in determining sensitivity and selectivity to Hsp90 inhibitors cannot be easily deciphered.
In the current study, the nature of Hsp90α/p23 and Hsp90β/p23 interactions and the efficacy of geldanamycin-based and purine- scaffold Hsp90 inhibitors were first indirectly monitored in intact cells in cell culture using genetically encoded SRL-PFAC reporters. The efficacy of the lead compound (PU-H71) was subsequently evaluated in living mice through repetitive, noninvasive imaging and was found to be more selective for disruption of Hsp90α/p23 interactions compared with that of Hsp90β/p23 interactions.
Direct comparison of the efficacy of geldanamycin-based and purine-based Hsp90 inhibitors in disruption of Hsp90α/p23 and Hsp90β/p23 interactions
The efficacies of geldanamycin-based and purine-scaffold Hsp90 inhibitors in disruption of Hsp90α/p23 and Hsp90β/p23 interactions were directly compared using SRL-PFAC (Figs. 4 and 5). In agreement with their identical Hsp90-binding moieties (16, 26, 27, 36), geldanamycin-based Hsp90 inhibitors led to similar maximal inhibition of RL activity relative to carrier control–treated cells. On the other hand, purine-scaffold Hsp90 inhibitors that differ in both their adenine moieties for Hsp90 binding and aryl functional groups that alter conformation of Hsp90 (Fig. 1B) led to different levels of inhibition in RL activity. Their levels of inhibition of RL activity also positively correlated with their affinity for Hsp90 from tumor cell lysates (Figs. 1B, 4C, D, and 5). However, geldanamycin-based and purine-scaffold Hsp90 inhibitors tested were not isoform selective in their inhibition of Hsp90α/p23 or Hsp90β/p23 interactions under cell culture conditions. We have also extended our study using MCF-7 human breast cancer cells stably expressing the Hsp90α/p23 and Hsp90β/ p23 split reporters. Both PU-H71 and 17-DMAG led to dose-dependent decreases in complemented RL activities in cell culture studies, as in the case for 293T cells (Supplementary Fig. S1).
Selectivity of PU-H71 in disruption of Hsp90α/p23 inter-actions in living mice
The mechanism and kinetics of PU-H71 in disruption of Hsp90α/p23 and Hsp90β/p23 interactions in living mice was further validated in living mice by noninvasive, repetitive imaging of 293T tumor xenografts expressing the Hsp90α/p23 and Hsp90β/p23 split reporters. Under cell culture conditions, PU-H71 was equally effective in disruption of both Hsp90α/p23 and Hsp90β/p23 interactions (Figs. 4B,C and 5). In living mice, however, PU-H71 was more effective in disruption of Hsp90α/p23 interactions compared with that of Hsp90β/p23 interactions (Fig. 6B). The data suggest the nature of Hsp90β/p23 interactions was stronger than that of Hsp90α/p23 under the influence of tumor microenvironment. Thus, development of isoform-selective Hsp90 inhibitors is very important in cancer treatment because Hsp90α and Hsp90β play distinct roles in determination of drug responses and resistance (39). In addition to validating the mechanism of Hsp90 inhibitors in intact cells in cell culture studies, the efficacy of Hsp90 inhibitors in living mice should also be monitored because (a) the immediate effects of Hsp90 inhibitors on disruption of Hsp90/p23 interactions can be indirectly monitored, (b) pharmacokinetics and delivery in living mice can be directly optimized, (c) repeated monitoring of efficacy can be accomplished with molecular imaging, and (d) individual variations can be studied much better because each mouse serves as its own control. We have also obtained preliminary data on comparing the efficacy of 17-AAG and PU-H71 in disruption of Hsp90α/p23 and Hsp90β/p23 interactions in living mice. At the same concentration (75 mg/kg) and route of injection (i.p.), PU-H71 was more effective in disruption of split Hsp90α/p23 and Hsp90β/p23 reporter interactions compared with that of 17-AAG.7
Comparison of SRL-PFAC with other strategies for monitoring protein-protein interactions in living subjects
To our knowledge, this is the first demonstration of indirect monitoring of isoform-specific Hsp90α/p23 and Hsp90β/p23 interactions and evaluation of the efficacy of different classes of Hsp90 inhibitors in cell culture conditions. The same genetically encoded reporter system was then used to validate the lead compound from the cell culture screen (PU-H71) for its ability to disrupt Hsp90α/p23 and Hsp90β/ p23 interactions in living mice. In addition to SRL-PFAC, split FL protein fragment–assisted complementation assays (SFL-PFAC) have also been used to monitor other protein-protein interactions in living mice (40–42). The smaller fragment sizes for the split RL strategy (23 kDa for NRL and 13 kDa for CRL) relative to split FL fragments (~32 kDa) may minimize steric hindrance for Hsp90α/ p23 and Hsp90β/p23 interactions in the split reporters. Because FL requires ATP for its enzymatic activity (43), SFL-PFAC may not be suitable for evaluating the efficacy of Hsp90 inhibitors (and other ATP inhibitors per se) that may inhibit complemented split FL activities.
Feasibility of using SRL-PFAC for development of isoform-selective Hsp90 inhibitors
Given (a) the Hsp90 chaperone system is highly up-regulated in multiple cancers (37), (b) both Hsp90α and Hsp90β are expressed in cancer cells and may play distinct roles in determining drug responses, and (c) the expression of Hsp90α and Hsp90β can be compensatory (38), Hsp90 inhibitors aimed at disruption of Hsp90α/p23 and Hsp90β/p23 interactions specifically or both interactions are being developed through indirect imaging of Hsp90α/p23 and Hsp90β/p23 interactions in conjunction with high-throughput screening. The lead compounds from the cell culture screen will be validated in living mice using xenografts, orthotopic and transgenic cancer models that express the split reporters. Furthermore, SRL-PFAC can be extended to indirectly monitor the efficacy of disruptors for Hsp90α and Hsp90β homodimerization (44) and Hsp90α/Hsp90β heterodimerization, both in cell culture and in living subjects.
Molecular imaging of Hsp90α/p23 and Hsp90β/p23 interactions in living subjects with the most potent Hsp90 inhibitor evaluated in cell culture has been accomplished using the SRL-PFAC. This will allow the screening and subsequent validation of the next generation of Hsp90 inhibitors with improved pharmacodynamics and pharmacokinetic properties. The SRL-PFAC also serves as a gene- ralizable platform for development and validation of other pharmacologic modulators aimed at disrupting specific protein-protein interactions pertaining to cancer and other human diseases in living subjects.
Supplementary Material
Acknowledgments
Grant support: NIH RO1 CA082214, National Cancer Institute NCI ICMIC P50 at Stanford University School of Medicine (S.S. Gambhir), and in part by a postdoctoral fellowship award PDF0023706 from Susan G. Komen for the Cure (C.T. Chan).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Manish Patel for flow cytometry analysis and William Sullivan and Dr. David Toft from Mayo Clinic for providing us with the Hsp90 and p23 expression plasmids, as well as helpful discussion.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Unpublished results.
References
- 1.Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Burrows F. Targeting multiple signal transduction pathways through inhibition of Hsp90. J Mol Med. 2004;82:488–99. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 3.Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–24. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Yano M, Naito Z, Yokoyama M, et al. Expression of hsp90 and cyclin D1 in human breast cancer. Cancer Lett. 1999;137:45–51. doi: 10.1016/s0304-3835(98)00338-3. [DOI] [PubMed] [Google Scholar]
- 5.Myung J-K, Afjehi-Sadat L, Felizardo-Cabatic M, Slavc I, Lubec G. Expressional patterns of chaperones in ten human tumor cell lines. Proteome Sci. 2004;2:8. doi: 10.1186/1477-5956-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamal A, Thao L, Sensintaffar J, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors [see comment]. Nature. 2003;425:407–10. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 7.Soga S, Shiotsu Y, Akinaga S, Sharma SV. Development of radicicol analogues. Curr Cancer Drug Targets. 2003;3:359–69. doi: 10.2174/1568009033481859. [DOI] [PubMed] [Google Scholar]
- 8.Le Brazidec JY, Kamal A, Busch D, et al. Synthesis and biological evaluation of a new class of geldanamycin derivatives as potent inhibitors of Hsp90. J Med Chem. 2004;47:3865–73. doi: 10.1021/jm0306125. [DOI] [PubMed] [Google Scholar]
- 9.Chiosis G. Discovery and development of purine-scaffold Hsp90 inhibitors. Curr Top Med Chem. 2006;6:1183–91. doi: 10.2174/156802606777812013. [DOI] [PubMed] [Google Scholar]
- 10.Banerji U, O'Donnell A, Scurr M, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23:4152–61. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 11.Goetz MP, Toft D, Reid J, et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J Clin Oncol. 2005;23:1078–87. doi: 10.1200/JCO.2005.09.119. [DOI] [PubMed] [Google Scholar]
- 12.Chiosis G, Caldas Lopes E, Solit D. Heat shock protein-90 inhibitors: a chronicle from geldanamycin to today's agents. Curr Opin Investig Drugs. 2006;7:534–41. [PubMed] [Google Scholar]
- 13.Rowlands MG, Newbatt YM, Prodromou C, Pearl LH, Workman P, Aherne W. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal Biochem. 2004;327:176–83. doi: 10.1016/j.ab.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 14.Soti C, Vermes A, Haystead TA, Csermely P. Comparative analysis of the ATP-binding sites of Hsp90 by nucleotide affinity cleavage: a distinct nucleotide specificity of the C-terminal ATP-binding site. Eur J Biochem. 2003;270:2421–8. doi: 10.1046/j.1432-1033.2003.03610.x. [DOI] [PubMed] [Google Scholar]
- 15.He H, Zatorska D, Kim J, et al. Identification of Potent Water Soluble Purine-Scaffold Inhibitors of the Heat Shock Protein 90. J Med Chem. 2006;49:381–90. doi: 10.1021/jm0508078. [DOI] [PubMed] [Google Scholar]
- 16.Chiosis G, Lucas B, Shtil A, Huezo H, Rosen N. Development of a purine-scaffold novel class of Hsp90 binders that inhibit the proliferation of cancer cells and induce the degradation of Her2 tyrosine kinase. Bioorg Med Chem. 2002;10:3555–64. doi: 10.1016/s0968-0896(02)00253-5. [DOI] [PubMed] [Google Scholar]
- 17.Chiosis G, Lucas B, Huezo H, Solit D, Basso A, Rosen N. Development of purine-scaffold small molecule inhibitors of Hsp90. Curr Cancer Drug Targets. 2003;3:371–6. doi: 10.2174/1568009033481778. [DOI] [PubMed] [Google Scholar]
- 18.Munster PN, Srethapakdi M, Moasser MM, Rosen N. Inhibition of heat shock protein 90 function by ansamycins causes the morphological and functional differentiation of breast cancer cells. Cancer Res. 2001;61:2945–52. [PubMed] [Google Scholar]
- 19.Vilenchik M, Solit D, Basso A, et al. Targeting wide-range oncogenic transformation via PU24FCl, a specific inhibitor of tumor Hsp90. Chem Biol. 2004;11:787–97. doi: 10.1016/j.chembiol.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Smith-Jones PM, Solit DB, Akhurst T, Afroze F, Rosen N, Larson SM. Imaging the pharmacodynamics of HER2 degradation in response to Hsp90 inhibitors. Nat Biotechnol. 2004;22:701–6. doi: 10.1038/nbt968. [DOI] [PubMed] [Google Scholar]
- 21.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic Renilla lucif-erase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–9. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulmurugan R, Massoud TF, Huang J, Gambhir SS. Molecular imaging of drug-modulated protein-protein interactions in living subjects. Cancer Res. 2004;64:2113–9. doi: 10.1158/0008-5472.can-03-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massoud TF, Paulmurugan R, Gambhir SS. Molecular imaging of homodimeric protein-protein interactions in living subjects. FASEB J. 2004;18:1105–7. doi: 10.1096/fj.03-1128fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies.[see comment]. Proc Natl Acad Sci U S A. 2002;99:15608–13. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 26.He H, Zatorska D, Kim J, et al. Identification of Potent Water Soluble Purine-Scaffold Inhibitors of the Heat Shock Protein 90. 2006:381–90. doi: 10.1021/jm0508078. [DOI] [PubMed] [Google Scholar]
- 27.Zhou V, Han S, Brinker A, Klock H, Caldwell J, Gu X-J. A time-resolved fluorescence resonance energy transfer-based HTS assay and a surface plasmon resonance-based binding assay for heat shock protein 90 inhibitors. Anal Biochem. 2004;331:349–57. doi: 10.1016/j.ab.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 29.Chadli A, Bouhouche I, Sullivan W, et al. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. PNAS. 2000;97:12524–9. doi: 10.1073/pnas.220430297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002;99:377–82. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wochnik GM, Young JC, Schmidt U, Holsboer F, Hartl FU, Rein T. Inhibition of GR-mediated transcription by p23 requires interaction with Hsp90. FEBS Lett. 2004;560:35–8. doi: 10.1016/S0014-5793(04)00066-3. [DOI] [PubMed] [Google Scholar]
- 32.Nemoto T, Ohara-Nemoto Y, Ota M, Takagi T, Yokoyama K. Mechanism of dimer formation of the 90-kDa heat-shock protein. Eur J Biochem. 1995;233:1–8. doi: 10.1111/j.1432-1033.1995.001_1.x. [DOI] [PubMed] [Google Scholar]
- 33.Workman P, Powers MV. Chaperoning cell death: a critical dual role for Hsp90 in small-cell lung cancer. Nat Chem Biol. 2007;3:455–7. doi: 10.1038/nchembio0807-455. [DOI] [PubMed] [Google Scholar]
- 34.Chen B, Piel WH, Gui L, Bruford E, Monteiro A. The HSP90 family of genes in the human genome: insights into their divergence and evolution. Genomics. 2005;86:627–37. doi: 10.1016/j.ygeno.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Pichler A, Prior JL, Piwnica-Worms D. Imaging reversal of multidrug resistance in living mice with bioluminescence: MDR1 P-glycoprotein transports coelenterazine. Proc Natl Acad Sci U S A. 2004;101:1702–7. doi: 10.1073/pnas.0304326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solit DB, Rosen N. Hsp90: a novel target for cancer therapy. Curr Top Med Chem. 2006;6:1205–14. doi: 10.2174/156802606777812068. [DOI] [PubMed] [Google Scholar]
- 37.Kamal A, Boehm MF, Burrows FJ. Therapeutic and diagnostic implications of Hsp90 activation. Trends Mol Med. 2004;10:283–90. doi: 10.1016/j.molmed.2004.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterjee M, Jain S, Stuhmer T, et al. STAT3 and MAPK signaling maintains overexpression of the heat shock proteins 90{α} and {β} in multiple myeloma cells, which critically contribute to tumor cell survival. Blood. 2006 doi: 10.1182/blood-2006-05-024372. blood-2006-05-024372. [DOI] [PubMed] [Google Scholar]
- 39.Subbarao Sreedhar A, Kalmar E, Csermely P, Shen Y-F. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–5. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 40.Luker GD, Sharma V, Pica CM, et al. Noninvasive imaging of protein-protein interactions in living animals. Proc Natl Acad Sci U S A. 2002;99:6961–6. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luker GD, Sharma V, Piwnica-Worms D. Visualizing protein-protein interactions in living animals. Methods. 2003;29:110–22. doi: 10.1016/s1046-2023(02)00285-2. [DOI] [PubMed] [Google Scholar]
- 42.Luker KE, Piwnica-Worms D. Optimizing luciferase protein fragment complementation for bioluminescent imaging of protein-protein interactions in live cells and animals. Methods Enzymol. 2004;385:349–60. doi: 10.1016/S0076-6879(04)85019-5. [DOI] [PubMed] [Google Scholar]
- 43.Fraga H, Fernades D, Novotny J, et al. Firefly luciferase produces hydrogen peroxide as a coproduct in dehydroluciferyl adenylate formation. Chem Biochem. 2006;7:929–35. doi: 10.1002/cbic.200500443. [DOI] [PubMed] [Google Scholar]
- 44.Chiosis G, Aguirre J, Nicchitta CV. Synthesis of Hsp90 dimerization modulators. Bioorg Med Chem Lett. 2006;16:3529–32. doi: 10.1016/j.bmcl.2006.03.092. [DOI] [PubMed] [Google Scholar]
- 45.Moulick K, Clement CC, Aguirre J, et al. Synthesis of a red-shifted fluorescence polarization probe for Hsp90. Bioorg Med Chem Lett. 2006;16:4515–8. doi: 10.1016/j.bmcl.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Chiosis G, Timaul MN, Lucas B, et al. A small molecule designed to bind to the adenine nucleotide pocket of Hsp90 causes Her2 degradation and the growth arrest and differentiation of breast cancer cells. Chem Biol. 2001;8:289–99. doi: 10.1016/s1074-5521(01)00015-1. [DOI] [PubMed] [Google Scholar]
- 47.Llauger L, He H, Kim J, et al. Evaluation of 8-arylsulfanyl, 8-arylsulfoxyl, and 8-arylsulfonyl adenine derivatives as inhibitors of the heat shock protein 90. J Med Chem. 2005;48:2892–905. doi: 10.1021/jm049012b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.