Abstract
The underlying pathogenesis of type-II diabetes mellitus is in the dysfunction and selective loss of pancreatic islet β-cells, which ultimately leads to underproduction of endogenous insulin. Amylin, a 37-amino-acid human hormone that is cosecreted with insulin, helps regulate gastric emptying and maintain blood glucose homeostasis through improved postprandial satiety. It is hypothesized that amylin protofibrils cause selective loss of pancreatic β-cells in a manner similar to amyloid β aggregation in Alzheimer's disease. β-Cell death occurs in vitro when isolated human or rodent β-cells are exposed to micromolar concentrations of amylin, but the exact mechanism of selective β-cell loss in vivo remains unknown. Therefore, pursuing small-molecule drug discovery for chemoprotectants of amylin-induced β-cell toxicity is a viable phenotypic target that can lead to potential pharmacotherapies for the preservation of β-cell mass, delaying insulin dependence and allowing additional opportunities for lifestyle intervention. Additionally, chronic endoplasmic reticulum (ER) stress induced by chronic hyperglycemia and hyperlipidemia is a potentiating factor of amylin-induced β-cell loss. Herein, we describe a high-content/high-throughput screening (HTS) assay for the discovery of small molecules that are chemoprotective of amylin-induced, ER-stress-potentiated β-cell loss. We also put forth a general method for construction of a robust well-level multivariate scoring system using partial least squares regression analysis to improve high-content assay performance and to streamline the association of complex high-content data into HTS activity databases where univariate responses are typical.
Introduction
Diabetes affects ∼8% of the U.S. population totaling $178 billion annually in federal healthcare costs. The current prediction is that one-third of the U.S. population will develop type 2 diabetes (T2D) in their lifetime.1,2 Diabetes also causes debilitating and costly complications, including neuropathy and nephropathy, and increases risks for limb amputation and other diseases, including cardiovascular disease.
The β-Cell and Diabetes
β-Cells of the pancreatic islets secrete insulin in response to glucose and other nutrients. During T2D, the pancreas is unable to produce sufficient amounts of insulin to maintain blood glucose homeostasis. Insulin deficiency in both type 1 diabetes (T1D) and T2D is characterized by a significant reduction in β-cell mass. The underlying pathogenesis of T2D is in the dysfunction and selective loss of pancreatic islet β-cells, which ultimately leads to underproduction of endogenous insulin. While T1D results from an autoimmune attack on the β-cells,3 in T2D, numerous factors, such as obesity, insulin resistance, hyperinsulinemia, hyperglycemia, and hyperlipidemia, can lead to cellular stress or toxic cellular environments that result in a decrease in β-cell number and function.4 The combination of hyperglycemia and hyperlipidemia is referred to as glucolipotoxicity, and it is most common in patients with T2D. The exact mechanism(s) of glucolipotoxicity in human islets is unclear,5–7 but it involves induction of endoplasmic reticulum (ER) stress, increased apoptosis, and decreased islet function.5,6 Loss of the pancreatic β-cells is the underlying cause of all diabetes, resulting in a loss or considerable reduction in insulin production.3,4 Innovative and efficacious therapies to prevent β-cell loss are urgently needed to prevent the progression to insulin-dependent type-II diabetes mellitus and to allow additional time for lifestyle intervention.
Current Therapies
The main pharmacological approach for treating T1D is insulin-replacement therapy via multiple injections daily or by insulin pump. For T2D, there is primarily a focus on the use of insulin sensitizers (e.g., metformin and thiazolidinediones [TZDs]), incretin mimetics (e.g., GLP1R agonists such as Exenatide), DPP-IV inhibitors (to raise endogenous levels of active GLP1), various insulin secretagogues, and insulin injections. Despite these different classes of drugs for the treatment of T2D, only ∼36% of diabetic patients achieve the A1c goal of ≤7%8; unresponsiveness to treatment hastens the progression to insulin dependence through β-cell loss. Thus, current therapies do not effectively target the underlying loss of β-cell mass. There is significant need to discover therapies that can (1) prevent β-cell loss, and (2) increase β-cell mass via stimulation of β-cell/islet growth.
Amylin Connection to Diabetes
Amylin is cosecreted with insulin and helps regulate gastric emptying and maintain blood glucose homeostasis through improved postprandial satiety.9,10 Depending on concentration, amylin can form small β-cell-toxic aggregates called protofibrils. These fibrils can form either intracellularly or extracellularly with intracellular toxic concentrations several orders of magnitude less than extracellular.11 It has been suggested that irregularities arising during the synthesis, folding, and processing of amylin cause amyloid formation.12 Interestingly, amylin's propensity to form fibrils and amyloids is linked to its amino acid sequence. While human amylin can be toxic, amylin isolated from rodents is nontoxic and does not form fibrils, nor amyloids. Westermark et al. published that the amino acids in the positions 25–29 are responsible for amylin amyloidogenic nature.13 This region is highly variant between humans and rodents; a proline substitution at serine 28 is responsible for nontoxic nature of rodent amylin.13,14 However, transgenic rodents with human amylin have been shown to undergo spontaneous diabetes and show β-cell loss.14 β-Cell death also occurs in vitro when isolated human or rodent β-cells are exposed to micromolar concentrations of amylin, but the exact mechanism of selective β-cell loss remains unknown. Therefore, pursuing small-molecule drug discovery for chemoprotectants of amylin-induced β-cell toxicity is a viable phenotypic target that can lead to potential pharmacotherapies for the preservation of β-cell mass, delaying insulin dependence and allowing additional opportunities for lifestyle intervention. Additionally, chronic ER stress induced by chronic hyperglycemia and hyperlipidemia is a potentiating factor of amylin-induced β-cell loss.15,16 Herein, we describe a high-content/high-throughput screening (HTS) assay for the discovery of small molecules that are chemoprotective of amylin-induced, ER-stress-potentiated β-cell loss.
Increasing High-Content Information in HTS
The multivariate nature of high-content screening (HCS) endpoints poses a significant challenge for screening applications where a single parameter/endpoint (herein the percentage of dead cells) is typically used to select positive compounds for further analysis. In many instances, an ideal univariate response can adequately detect meaningful biological conditions. However, if there is no ideal/robust single parameter or we do not know how to extract it, then additional high-content features can be utilized in a multivariate scoring system to improve assay performance.
A recent analysis of 118 published high-content screens by Singh et al. showed that 60%–80% of the studies utilized only one or two measured features of the cells.17 Underutilization of accessible information is often due to the fitting of HCS into the HTS data infrastructure; extracting “hits” from highly dimensional data can be daunting in the screening process because a single microtiter plate can generate over a billion data points. Principal component analysis (PCA), a data-reduction technique that is often used in early assay development, can help guide the screener to the most important feature(s) by assessing where the most variance in the data set occurs. These highly variant features are often the most relevant but they can also reflect irrelevant biological variability, especially when the signal-to-noise ratio is low. After discovering the most important feature or features, screeners typically only focus on those and ignore the vast number of other parameters in order to facilitate the merging of biological data with chemical information in conventional HTS software. Therefore, there is a dramatic need to increase the information content in high-content/high-throughput screens while working within the confines of our HTS data infrastructure where registration of hundreds of features per cell is prohibitive. Different stages of screening and different assay types require appropriate levels of access to high-content data spanning from a single-well-averaged parameter to multivariate scoring of subpopulations at the single-cell level. Large-scale rapid/single-concentration screening necessitates a more simplistic scoring system that can guide screeners to interesting compounds/conditions for more detailed analysis. Herein we describe, in a simplistic, high-content cytotoxicity assay, a general method for generating a well-averaged multivariate parameter that can dramatically improve assay performance using partial least squares (PLS) regression. This technique can help bridge the univariate-to-multivariate gap in a conventional HTS data infrastructure while allowing the screener appropriate access to underlying detail.
Materials and Methods
Cells and Reagents
Rat insulinoma (INS-1) cells were graciously donated by Christopher Newgard at Duke University. INS-1 cells were cultured according to ATCC guidelines with RPMI-1640 growth medium (Sigma No. r8758), supplemented with 10% FBS, 1% P/S, 1% HEPES, and 1% INS supplements: 10 mM HEPES, 2 mM glutamine, 1 mM sodium pyruvate, and 0.05 mM 2-mercaptoethanol (all concentrations are final). This INS-1 stable cell line was expanded to generate a master cell bank for HCS. Cryogenically preserved stocks were made at the same passage number with ∼2 × 106 cells per 1-mL tube in cryopreservation media consisting of growth media supplemented with 5% dimethyl sulfoxide (DMSO).
A 10 mg/mL stock of tunicamycin was prepared by dissolving 10 mg of tunicamycin in 1 mL of DMSO (tunicamycin, Enzo Life Sciences Cat No. BML-CC104-0010). A 4 mg/mL solution of amylin was prepared fresh in autoclaved ddH2O (Bachem, H-7905).
NuPage Novex 12% Bis-Tris gradient gels were used (Life Technologies, Grand Island, NY) for SDS-PAGE analysis and multiplexed western blotting was performed using a LiCor Odyssey imaging platform. The rabbit anti-BiP monoclonal antibody (C50B12) was purchased from Cell Signaling Technology (Danvers, MA) and used at 1:1,000 dilution. The mouse anti-β-tubulin monoclonal antibody (T5201) was purchased from Sigma-Aldrich (St. Louis, MO) and used at 1:15,000 dilution. Secondary antibodies were purchased from LiCor and used at the manufacturer-recommended dilutions.
High-Content Viability Assay Overview
The amylin-induced toxicity assay was developed in 384-well format with a final assay volume of 56 μL. Cellular health/viability was assessed using a cell-permeant nucleic acid stain to label all cells (Hoechst 33342), and a cell-impermeant nucleic acid stain (YoYo-1) to label dead/membrane-permeable cells. This staining was followed by fixation to preserve the viability signal. The goal of this assay is to detect compounds that can be protective/overcome amylin/ER stress toxicity, to provide chemical probes to further the understanding of islet-specific amylin toxicity, and to discover compounds with clinical translational potential for preserving β-cell mass in the progression of T2D. As a measure of cellular toxicity, total cell counts, live/dead ratio, DNA content, and nuclear morphology were all assessed. Positive-compound effects manifest in high cell counts, high live/dead ratio, normal DNA content (i.e., not sub-G0), and smooth nuclear morphology. Assay timing is as follows. Day 1: cells are plated and allowed to attach overnight. Day 2: a cocktail of amylin/tunicamycin is added and immediately followed by screening-compound addition using a 50-nL pintool array; the cell plate was then allowed to incubate for 24-h. Day 3: live-cell plates were stained with Hoechst-33342 and YoYo-1 and allowed to incubate for 45 min prior to fixation and imaging (Table 1).
Table 1.
High-Content Assay Protocol Table
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate cells | 30 μL | 2,500 INS-1 cells/well |
| 2 | Attachment time | 24 h | Plated cells incubated at 37°C, 5% CO2 |
| 3 | Amylin/tunicamycin | 6 μL | Amylin/tunicamycin mixture added to columns 1–22 |
| 4 | Controls | 6 μL | DMSO control added to control wells in columns 23 and 24 |
| 5 | Library compounds | 50 nL | Compounds added for a direct 1,000-fold dilution |
| 6 | Incubation | 24 h | Cells incubated with compounds at 37°C, 5% CO2 |
| 7 | Staining agents | 20 μL | YoYo-1 and Hoechst added to each well |
| 8 | Incubation time | 45 min | Cells incubated at 37°C, 5% CO2 during staining |
| 9 | Fixing agent | 20 μL | Cells fixed with 4% formaldehyde |
Step Notes
1. Plated in 384-well NUNC poly-D-lysine-coated, black/clear bottom tissue-culture-treated plates.
2. Cells were allowed to attach overnight.
3. Columns 1–2 were controls for max. dead cell signal; columns 3–21 were the experimental wells.
4. Columns 1–2 were controls for max. dead cell signal; columns 22–24 were control wells for min. dead signal.
5. Prestwick Chemical Library and Enzo Life Sciences natural product library were tested. The Biomek NX workstation was used to transfer the compounds to each plate using pintool array.
Final DMSO concentration was 0.1%.
6. The cells were incubated with the tested compounds overnight.
7. The Biomek NX workstation was used to remove media and add the staining agents.
Pipette tip Z-coordinate was −0.35 mm relative to liquid height during aspiration (tips go down as liquid goes down) until 10 μL liquid volume remained. Pipette tips were offset from center to upper left corner for all transfers.
8. The cells were incubated to allow for dye penetration for 45 min.
9. The staining agents were removed and replaced with fixing agent. The fixing agent was incubated with the cells for 20 min before being replaced with PBS prior to imaging.
Cell Plating
The 384-well poly-D-lysine-coated, NUNC black/clear bottom tissue-culture-treated imaging plates (NUNC No. 152029) were used for the assay. Cells were thawed from freezer-stocks, counted using a Vi-Cell XR (Beckman-Coulter, Brea, CA), and diluted to a final concentration of 0.083 × 106 viable cells/mL. Twenty microliters of prewarmed growth medium was added to each well using a Mutidrop-384 (Thermo/Fisher) with a sterile head before depositing 30 μL of cell suspension (2,500 viable cells/well) into each well. The cell plates were then allowed to attach overnight in the incubator.
Amylin and Tunicamycin Addition
To stimulate physiologically relevant ER stress potentiation of amylin toxicity, a combination of tunicamycin and amylin peptide was used to achieve ∼80% cell death. In positive-control wells in the assay plates, the amylin/tunicamycin solution was omitted and cells were highly viable. Tunicamycin was diluted to a 100 μg/mL stock in DMSO. About 2.5 μL of this tunicamycin stock was added to each well using a repeating pipette. Amylin was diluted to a 256 μM stock in autoclaved water and added to each well via a multichannel pipettor. The final concentration of amylin in each well was 20 μM. The final assay volume was 56 μL. These plates were incubated at 37°C for 24 h. The tunicamycin/amylin solution was added to all wells in columns 1–22 and was omitted in positive-control wells in columns 23 and 24.
Compound Libraries and Delivery
Two highly annotated small-molecule libraries were used for pilot screening purposes, including an 1120-molecule, FDA-approved drug set (Prestwick Chemical Library®) and 502 purified natural products (Enzo Life Sciences). Compounds were stored in 5-μL aliquots in 384-well Axygen rigid PCR plates at 1000× concentration for both single-point/rapid screening and dose–response confirmation. Compounds were applied to cell plates with a Biomek NX workstation (Beckman-Coulter, Brea, CA) equipped with a 50-nL pintool array (V&P Scientific, San Diego, CA) for an approximate 1,000-fold dilution, thereby avoiding an intermediate dilution step. The final concentrations of Prestwick library and Enzo natural product library compounds were 1 and 5 μM, respectively, and final DMSO concentration was 0.1%.
Staining and Fixation
Approximately 45 μL of media with compound was removed from each well using the Biomek NX workstation leaving enough liquid behind to wet cells. A solution of 10 μg/mL Hoechst-33342 and 50 nM YoYo-1 (Invitrogen, Carlsbad, CA) was prepared in growth media (omitting serum and antibiotics) and 30 μL was added to each well and then incubated for 45 min at 37°C. Thirty microliters of media was aspirated to remove excess dye and was replaced with 30 μL of fresh 4% formaldehyde. The cell plates were incubated in fixative for 45 min at 37°C. The cells were then washed once with PBS using the Biomek-NX at the slowest dispense speed, sealed with Thermo-ABgene plate sealer, and imaged on a BD Pathway 855 bioimager (Becton Dickenson, San Jose, CA).
Fluorescent Imaging
The BD Pathway 855 was used to acquire multiplexed epifluorescent images in 384-well format with a 10 × /0.40 NA Olympus UPlanSApo objective lens using the BD Attovision software. The YoYo-1 channel (dead cells) was imaged with a 488/10-nm band-pass excitation filter, Fura/FITC epifluorescence dichroic, and a 515-nm long-pass emission filter. The Hoechst-33342 channel (all cells) was imaged with a 380/10-nm band-pass excitation filter, a 400-nm dichroic long-pass, and a 435-nm long-pass emission filter. Laser-based autofocus was performed in each well prior to the collection of YoYo-1 and Hoechst images. Batches of six plates were processed and imaged at a time. Exposure times for both the YoYo-1 and Hoechst channels were adjusted prior to each batch to ensure optimal signal-to-noise ratio. The entire well was imaged using the BD Pathway 855 using a 4 × 5 montage with camera binning=2.
Cell Detection, Segmentation, and Quantitation of Nuclear Morphology
The open-source CellProfiler software was used for cell segmentation, feature extraction, quantitation of signal intensities, and tabulation of cell-level results.18 About 4 × 5 image montages for the Hoechst and YoYo-1 channels were demontaged into 20 separate images and background illumination correction was performed using a 25-pixel block-size Gaussian filter on each image. Demontaged images were coded with parental metadata (plateID and wellID) and also with row/column metadata. Nuclei were identified in the Hoechst/DNA image as primary objects after automatic thresholding using the mixture of Gaussian method. Both shape and intensity measurements were used to segment closely spaced cells in the illumination-corrected demontaged images. Nuclear intensities and shape/morphological properties were tabulated for the Hoechst and YoYo-1 channels. A single-measurement statistical classifier was then used to classify cells as live or dead based on the mean YoYo-1 intensity inside the nuclear mask, with high intensities indicative of cell death/permeabilization. All cellular measurements were exported to a comma-delimited spreadsheet for data analysis. Numerous YoYo-1 and Hoechst intensity and distribution features were tabulated per-cell to achieve a comprehensive phenotypic nuclear description to be used in the multivariate scoring system.
Multivariate Scoring System Overview
The vast majority of high-content assay endpoints when used for HTS are often severely truncated to the most relevant one or two factors to facilitate the merging of biological data with chemical data in HTS database applications. This simple approach, if the univariate parameter is not ideal, can lead to a reduction in screening efficiency and results in false positives/negatives. However, with over 106 cells observed per 384-well plate × 200 primary features extracted for each cell, the incorporation of 109 data points per 384-well plate screened into a screening database is onerous in commercially available HTS software and the majority of observed features are unimportant. We therefore needed a reliable data-reduction technique to capture a reliable and informative well-averaged result for selection of interesting conditions warranting deeper phenotypic analysis. Toward this goal, we developed a multivariate scoring system based on well-averaged cellular features that delivers a single score that is comprised of ∼10–20 of the most important observed features. This approach eliminates the unimportant variables and delivers a single score per well that only includes variables that respond upon treatment in a linear combination with scaling factors that indicate importance.
PLS Regression Analysis
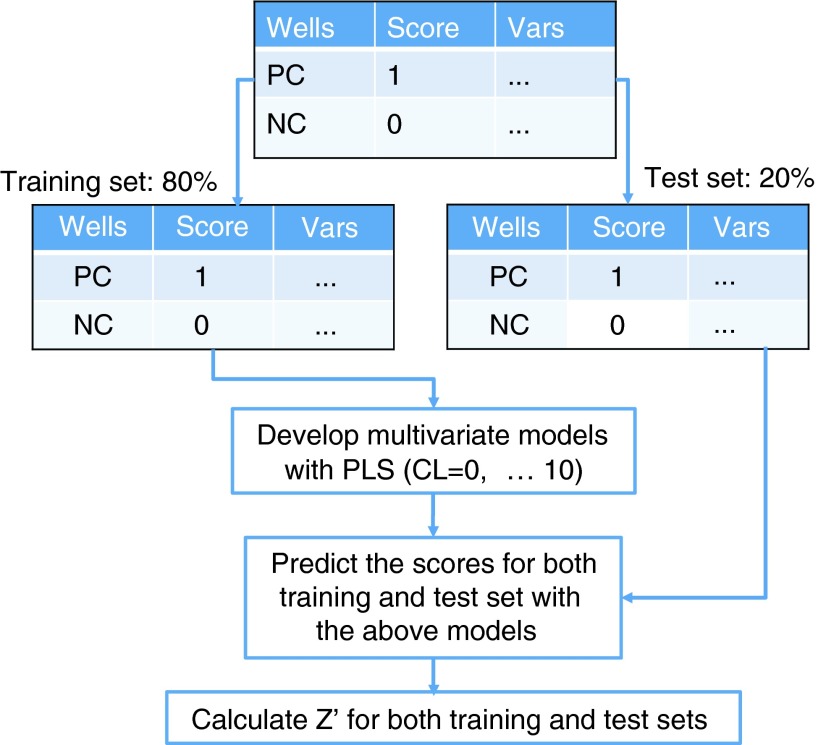
PLS regression, similar to PCA, is a multivariate regression technique that can be used to relate a response (Y) variable to several explanatory (X) variables. It can deal efficiently with high-content data sets where many variables are highly correlated and substantial random noise is involved. PLS was developed in the 1960s by Herman Wold as an econometric technique and was later made popular by Svante Wold in chemometric research.19,20 The Molecular Operating Environment (MOE software; Chemical Computing Group, Montreal, Canada) and JMP software (SAS, Cary, NC) were used to perform PLS regression analysis to determine a linear combination of observed variables that showed the strongest correlation with the positive- and negative-control conditions. Our general workflow for PLC scoring system development is shown in Figure 1.
Fig. 1.
Workflow to develop multivariate models from min-max assay development experiments. PC: the positive controls are assigned to have scores of 1. NC: the negative controls are assigned to be 0. The original data table is split into two sets: 80% of the wells are placed into the training set and the remaining 20% are placed into the test set. Z-prime values are computed per plate with each model generated.
Tabulated features for negative- and positive-control wells were assigned a categorical predicted score of 0 or 1, respectively. A table for regression analysis was constructed using 384-well-averaged features (observed variables) and predicted scores (NC=0, PC=1). The data set was randomly split 80% as a training set for model development and 20% as a test set for model evaluation. Four separate random 80/20 splits were carried through all PLS modeling to ensure that each random selection was a representative subset. Iterative PLS was performed using the 80% training set and the resulting models were evaluated on the 20% test set.
Eleven models for each data split were created where each model included a different number of principal components. Briefly, a principal component is derived from a mathematical procedure that reduced the dimensionality of the data. The PLS procedure takes all the cellular descriptors and generates a set of uncorrelated values, each of which is called a principal component. We varied the number of principal components in the model from PC=0 to PC=10, where PC=0 is the inclusion of all principal components (equal to the total number of cellular features/descriptors). Z-prime values were calculated from the predicted scores for each model (ranging from 0% to 100% effect) and were compared with the univariate PctDead Z-prime values.
Utilizing the INS-1 rodent pancreatic β-cell line, we observed a marked negative synergy between amylin exposure and tunicamycin-induced ER stress, indicating that they are mechanistically linked. Typically, HTS assays select a single primary or derived parameter to report for the selection of interesting compounds. However, this method of selection may exclude other unanticipated phenotypic effects exhibited during screening, thus verifying the need for a multivariate approach to capture this diversity. An improved general method for multivariate scoring is presented using a PLS method for hit determination in a cellular toxicity rescue assay. This assay utilized nuclear count, cell morphology, and plasma membrane permeability as endpoints and the improved scoring system significantly impacted the robustness and quality of screening data.
High-Throughput Screening Data Analysis
ScreenAble HTS software (ScreenAble Solutions, Chapel Hill, NC) was used in the analysis of HTS results in conjunction with Jchem chemical database and Jchem Oracle cartridge (ChemAxon, Budapest, Hungary) for chemical management and integration of biological data with chemical structures. Multiple-well-averaged features were reported back to the ScreenAble database along with classified images (see Fig. 3D) stored as a BLOB data type within the database allowing for registration of multiple features and rapid access to classified images for quality control purposes and visual hit validation.
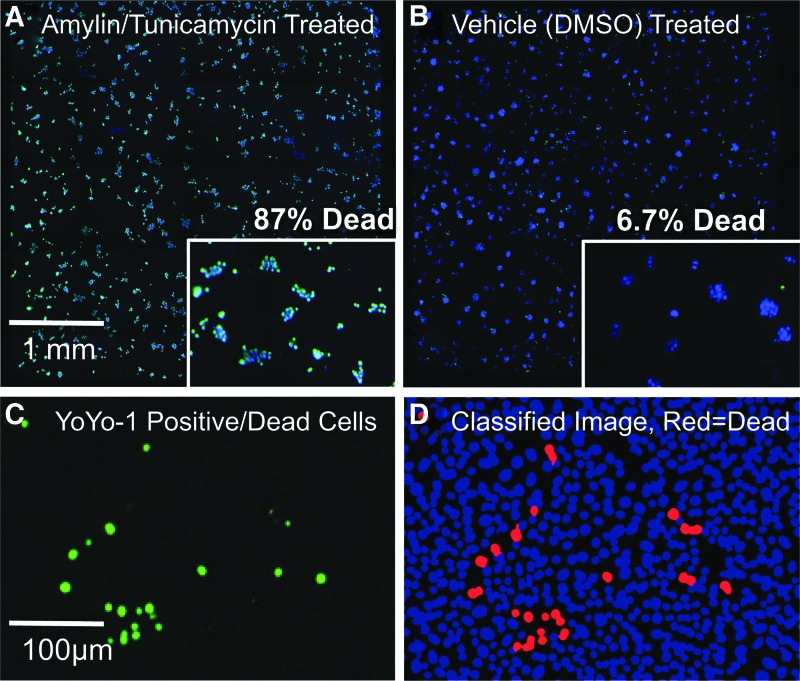
Fig. 3.
Pipeline images used for identifying and categorizing HCS data at the cell level with live nuclei stained with cell-permeant Hoechst-33342 (blue) and dead nuclei stained with both Hoechst-33342 and YoYo-1 (green). (A) Amylin/tunicamycin-treated entire-well image showing mostly double-positive blue/green nuclei indicating dead cells. (B) Vehicle (DMSO)–treated well image showing mostly Hoechst-positive nuclei indicating live cells. (C) YoYo-1-positive nuclei image showing dead cells. (D) Classified nuclei image: live nuclei (blue) and dead nuclei (red).
Results
Amylin-Induced Cytotoxicity
Toward the development of a 384-well high-throughput assay to ameliorate amylin-induced cytotoxicity, the dose-dependent increase in cell death was quantitated with the maximum effect of 30% cell death at 20 μM amylin concentration (Fig. 2), which is consistent with previously published reports of in vitro amylin toxicity wherein an exponential increase in amylin-induced cytotoxicity occurs after reaching a critical concentration for fibrillar aggregate formation, causing membrane blebbing and chromatin condensation.9 Interestingly, there is a narrow concentration window for amylin cytoxicity with the onset at 5 μM in INS-1 cells increasing until a maximum of ∼30% cell death at 20 μM, where below and above this concentration range does not favor the formation of the toxic fibrillar aggregate.
Fig. 2.
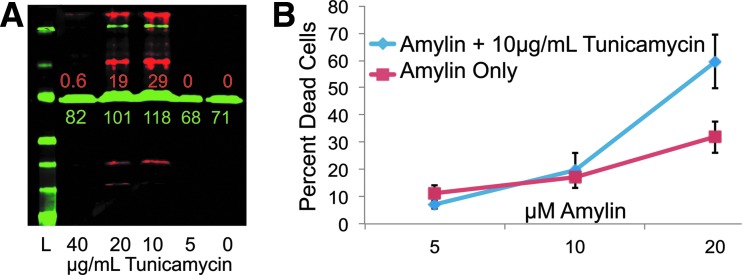
ER-stress-potentiated amylin cytotoxicity. (A) Western blot shows induction of BiP (red) at 10 and 20 μg/mL indicating ER stress response and tubulin (green) as a protein loading control. Densitometry is color coded for red and green bands in relative fluorescent units (RFU). L denotes protein ladder, and numbers correspond to μg/mL concentrations of tunicamycin. (B) INS-1 cells were incubated for 24 h with increasing concentrations of amylin peptide±tunicamycin. The percentage of dead cells was calculated by dividing the number of YoYo1-positive cells by the total number of cells observed. Error bars represent standard error of n=6 values.
To investigate the role of clinically relevant ER stress in the context of amylin-induced cytotoxicity, tunicamycin was added during amylin exposure to induce ER stress through inhibition of protein glycosylation. It has been previously described that ER stress is a mechanism by which amylin induces apoptosis.21 The addition of subtoxic concentrations of tunicamycin (10–20 μM) induces the unfolded protein response (UPR) as detected by induction of Bip/Grp78 expression (Fig. 2A, western blot relative to zero tunicamycin/DMSO control). The western blot in Fig. 2A shows BiP expression (red bands) in response to increased tunicamycin concentration with maximal induction of UPR/ER stress at 10–20 μg/mL tunicamycin. Tunicamycin concentrations above 20 μg/mL result in cell loss/toxicity and fail to induce the UPR likely due to transcriptional shutdown.
To recapitulate the type-II diabetic condition where amylin overexpression accompanies insulin overexpression in the β-cell causing ER stress, we chose the combination of 20 μM amylin and 10 μM tunicamycin as the in vitro disease model for ER-stress-mediated amylin-induced cell death. Figure 2B shows that the addition of subtoxic doses of tunicamycin significantly increases the live/dead ratio from a maximum of ∼30% to over 60% dead, indicative of a negative synergism (i.e., greater than additive cell death caused by each treatment. Note: final optimized negative-control condition resulted in 80%–90% dead cells.).
Assay Validation
With the selection of optimal concentrations and ratio of amylin and tunicamycin, assay validation was performed to determine suitability for HTS purposes. Initial validation attempts in 384-well plates consisted of min-max conditions, where half the plate (192 wells for each condition) was negative control (INS-1 cells treated with amylin/tunicamycin resulting in cell death) and the other half was positive control (treated with vehicle only), signifying rescue from the amylin/tunicamycin insult. These validation experiments assessed the assay window and variability in a univariate readout; percent dead cells—where the dead cell count (YoYo-1 positive) was divided by the total cell count.
Figure 3 shows typical whole-384-well images, which are seamless 4 × 5 image montages, for the negative control (Fig. 3A) where amylin/tunicamycin have been added to induce cytotoxicity, and the positive control (Fig. 3B) where amylin/tunicamycin have been omitted to represent 100% effect in chemoprotection. Figure 3C shows the YoYo-1 image from a single field where green cells are dead/membrane permeabilized, and Figure 3D shows the results of the statistical classifier for the same area where red cells are identified as dead and blue cells are identified as live. In the CellProfiler software using the “Classify Objects” module, the statistical classifier used a single parameter, YoYo-1 average intensity to classify cells as live or dead by setting the threshold approximately halfway between the average intensity for live cells and that of dead cells. Due to the large difference between live and dead YoYo-1 average intensities, this threshold was not modified plate-to-plate or day-to-day.
PLS Scoring System
A PLS model was developed to reduce the 70 primary cellular features, from both fluorescent channels, at the well-averaged level into a single response variable composed only of the most important features. The high-content feature extraction in CellProfiler measured 70 phenotypic parameters in this relatively simple assay and our PLS analysis selected and weighted 19 parameters for computing the well-average “cell health” score for each well. Reducing the 70 primary phenotypic parameters using PLS to a single score allows for the sensitive detection of effects that can manifest in changes in any of the selected 19 extracted features and makes the assay substantially more tolerant to outliers yielding significant improvements in Z-prime/statistical robustness. These parameters were selected automatically with the MOE software with the criteria that they maximize the separation in score between positive and negative controls.
Well-averaged data from the min-max assay validation runs were compiled into a spreadsheet and manually scored as 0.0 for the negative control wells and 1.0 for the positive control wells as the response variable for regression analysis. The data set was split randomly into two sets: 80% of the data as the training set, and 20% of the data as the test set to evaluate the unbiased performance of the model against data not used in the creation of the model. Five separate 80/20 random splits were created and regression analysis was performed on all training sets to avoid any subsampling bias. PLS regression analysis was performed using the manual NC=0.0, PC=1.0 score as the regression variable and incorporation of different numbers of principal components was used to determine the optimal number of principal components for inclusion into the final scoring system. Z-primes were calculated for the entire data set using the univariate response variable (percent dead cells), and for each model scoring system that was generated. Table 2 shows the result of the regression analysis for PctDead, PC1—where the maximum number of principal components was used (equal to the number of primary features ∼70), PC5—where the first five principal components were included, and so on. At PC5 and above, the model reached convergence at Z-prime=0.8, representing a significant improvement over the PctDead parameter.
Table 2.
Partial Least Squares Z-Prime Training Results
| Data Seta | PctDead | PC-1b | PC-5c | PC-10 | PC-70 |
|---|---|---|---|---|---|
| Training | 0.71 | 0.55 | 0.84 | 0.83 | 0.83 |
| Test | 0.55 | 0.80 | 0.80 | 0.80 |
Training data set was used for scoring system development; test data set was used for external validation.
PC-X denotes the inclusion of X number of principal components into the scoring system.
PC-5 was selected as the final model for the highest performance. See Table 3 for final scoring system.
Table 3 shows the final scoring system, including 19 primary features with the associated positive or negative scaling factor. Parameters with constants below 0.0001 were truncated due to low contribution and for ease of implementation. The scoring system is applied using the following linear combination (equation 1):
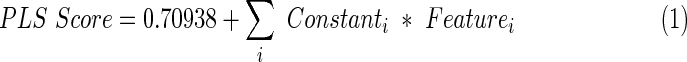 |
Table 3.
Partial Least Squares Scoring Parameters
| Constanta | Feature |
|---|---|
| 0.70938 | Constant |
| −0.0368 | Dead cell percentage |
| −0.0356 | % dead (dead/total) |
| −0.002 | Mean nuclei center X |
| −0.0004 | Mean nuclei classify dead |
| −0.0003 | Mean nuclei orientation |
| −0.0002 | Mean nuclei minor axis length |
| −0.0002 | Threshold weighted variance nuclei |
| −0.0001 | Mean nuclei integrated intensity dead cells |
| −0.0001 | Mean nuclei mass displacement dead cells |
| −0.0001 | Threshold sum of entropies nuclei |
| 0.0001 | Mean nuclei eccentricity |
| 0.0001 | Mean nuclei integrated intensity edge CorrDNA |
| 0.0004 | Mean nuclei classify live |
| 0.0004 | Mean nuclei integrated intensity CorrDNA |
| 0.0005 | Mean nuclei major axis length |
| 0.001 | Mean nuclei center Y |
| 0.001 | Mean nuclei perimeter |
| 0.0013 | Mean nuclei area |
| 0.0368 | Live cell percentage |
Features with constants below 0.0001 were omitted.
Min-Max Scatter: Percent Dead Versus PLS Scoring System
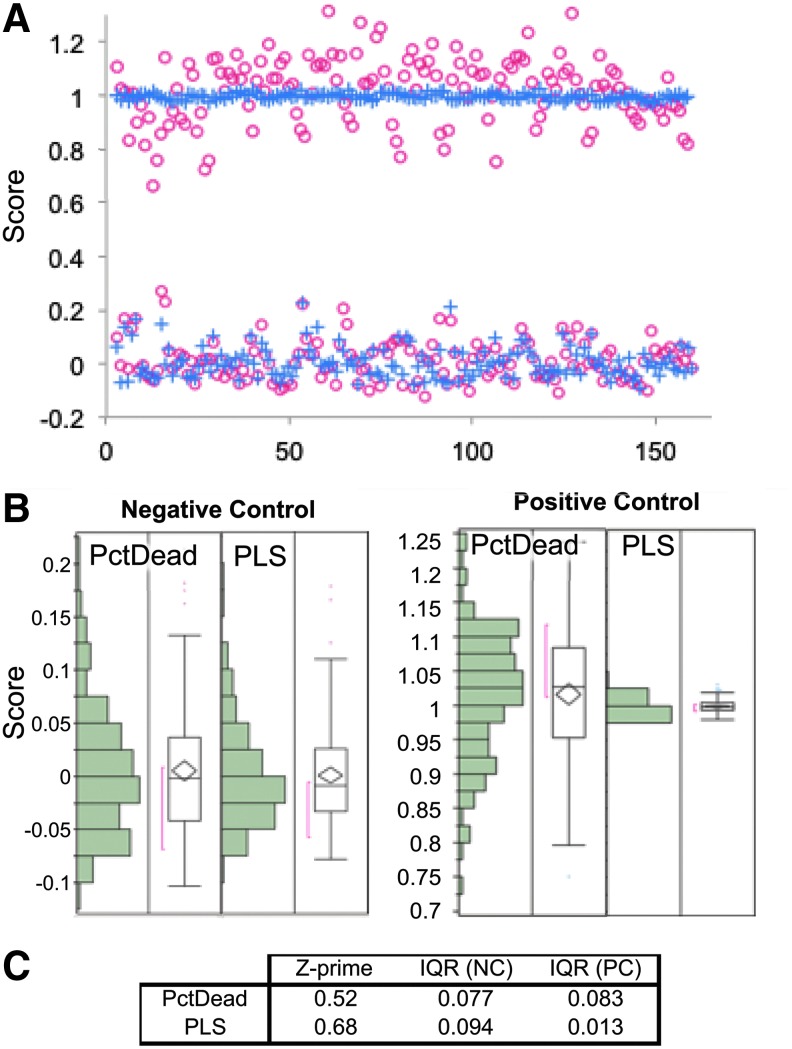
Figure 4 shows the comparison of the PctDead univariate parameter versus the PLS scoring system. The Figure 4A shows a plate-normalized scatter plot for a 384-well min/max plate where red circles indicate PctDead and blue crosses indicate the PLS score. Histograms and boxplots for positive- and negative-control wells are shown in the Figure 4B. The main difference observed is in the positive-control condition (highly viable cells), centered at 1.0, where the standard deviation collapses and dramatically improves the signal-to-noise ratio of the assay. Figure 4C shows tabulated Z-primes for this 384-well min/max plate and interquartile ranges for the negative and positive controls.
Fig. 4.
(A) Scatter plot of the positive and negative controls overlaid comparing the PctDead parameter against the PLS scoring system. The percent dead (PctDead) data are shown as red circles (○) and PLS data as blue crosses (+). (B) Box plots showing distributions of the positive and negative controls for PctDead and PLS scoring system showing a large reduction in variance in the positive-control scatter. (C) Z-primes and interquartile ranges for negative control and positive control distributions.
Pilot Screening
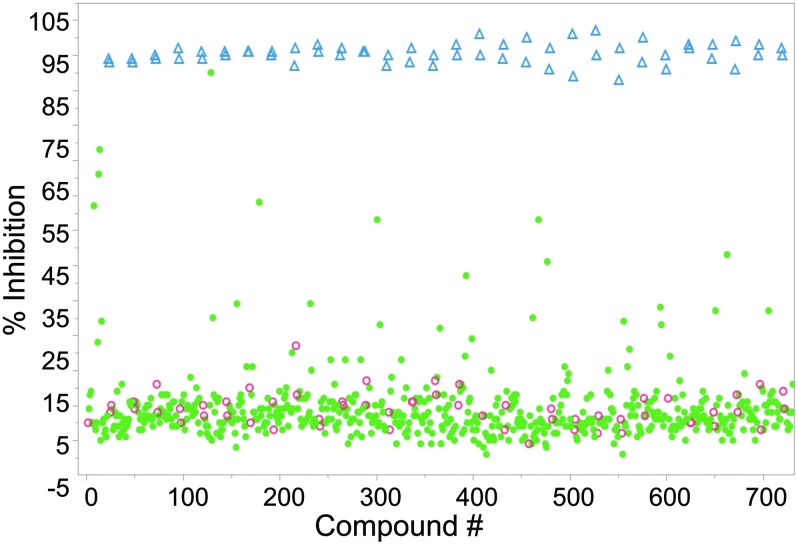
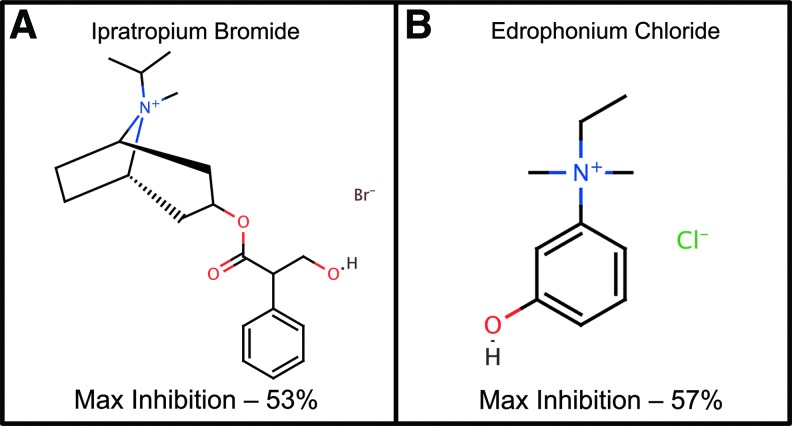
Two highly annotated chemical libraries containing FDA-approved drugs and purified natural products were screened for chemoprotection from ER-stress-mediated amylin-induced cell death. Figure 5 shows a scatter plot of a representative screening set with positive controls (blue triangles), negative controls (red circles), and compounds tested (solid green circles). The average Z-prime for the entire screen was 0.7 and plate-to-plate and day-to-day coefficient of variation was <5%. A total of 18 hits were discovered in primary screening with a hit rate of 1.1%. Of the compounds identified, two muscarinic-receptor-related compounds, ipratropium bromide and edrophonium chloride, were identified (Fig. 6) as chemoprotective in this assay with maximal inhibition of 53% and 57%, respectively. Compounds were considered active in single-point screening if they achieved >50% inhibition.
Fig. 5.
Representative screening data from the Prestwick/Enzo libraries using the PLS scoring system. Positive controls are blue triangles (Δ), negative controls are red circles (○), and test compounds are solid green circles (•). The average Z-prime for the screen was 0.7 for ∼1,600 compounds.
Fig. 6.
Two muscarinic-receptor-modulating compounds identified in pilot screening: (A) ipratropium bromide—a muscarinic receptor antagonist used to treat bronchial spasms, and (B) edrophonium chloride—an acetylcholinesterase inhibitor shown to also interact with the human M1 muscarinic receptor used to treat myasthenia gravis. Maximal percent inhibition shown relative to DMSO controls.
Discussion
The amylin connection to the selective loss of pancreatic β-cells is well established; however, the underlying mechanism and potentiating factors remain unclear. The discovery of small molecules that can prevent/inhibit amylin-induced β-cell loss can lead to the discovery of valuable chemical probes to dissect the underlying molecular mechanisms. Thus, chemoprotection of amylin-induced cytotoxicity is a validated phenotypic target for HCS. Additionally, it is widely recognized that ER stress plays a role in the pathogenesis of diabetes.16 Herein, we describe a high-throughput/high-content assay for the discovery of small molecules that can overcome toxic concentrations of amylin peptide in the presence of ER stress, which mimics the clinical condition of selective pancreatic β-cell loss and the accumulation of amylin plaques in their place.16 It is assumed that the mechanism of amylin-induced cytotoxicity is largely mechanical; that is, amylin protofibrils penetrate the plasma membrane. However, the mechanism for β-cell selectivity is unclear. In T2D patients with substantial insulin resistance, the overproduction of insulin leads to a proportional increase in amylin secretion that should be generally toxic.10 In this state, the remaining β-cells are under stress to produce large quantities of insulin in an attempt to maintain blood glucose homeostasis and ER stress then occurs as part of the UPR due to high translational burden by the cell.22 In an effort to create a clinically relevant disease model for amylin-mediated β-cell loss, we chose to chemically induce ER stress using tunicamycin—an inhibitor of protein glycosylation that induces ER stress/UPR. The addition of tunicamycin had the additional benefit of widening the assay window due to large negative synergy with amylin toxicity that increased the percentage of dead cells observed (up to 90% dead cells) as compared with the relatively nontoxic dosage of tunicamycin (10%–20% dead cells, data not shown).
Endpoint assays for quantifying eukaryotic cell membrane permeability using a combination of fluorescent cell-permeant and -impermeant dyes have long been used to assess cytotoxicity/viability. Multiple assay technologies have been used successfully including high-content analysis, well-averaged fluorescent intensity ratios, and absorbance-based/chromogenic dye exclusion. In this study, HCS was used in conjunction with a multivariate scoring system to determine the benefit of the imaging platform over a conventional univariate/well-averaged readout in 384-well format. Several key factors discovered in assay development were identified and led to a robust assay, including (1) the source of amylin: synthetic, purified amylin produced the highest level of cytotoxicity as compared with bacterially expressed amylin, and (2) entire well imaging using multiple fields at 10×to ensure adequate cell counts to develop statistical significance.
After initial assay validation attempts that consistently yielded Z-prime values above 0.5 for the univariate PctDead response variable, it was noted that this parameter was susceptible to false positive events where the total cell count was extremely low and the few remaining cells in a well appeared to be viable. This result, while clearly the result of compound toxicity, reported a low percent-dead score and was a false positive. With this straightforward, easily understandable high-content assay that was developed against a univariate readout (PctDead), we sought to implement a well-averaged multivariate scoring system as a test case where the resulting scoring system could be easily interpreted. This was done in an effort to leverage the numerous features extracted in this high-content data set to improve the screening outcomes by avoiding false positives/negatives. Development of a well-averaged multivariate scoring system also streamlines the integration of high-content data into HTS screening databases without sacrificing the feature-rich underlying data. This approach in early assay development rapidly directs the screener to the most important features, allowing the rapid detection of hits/interesting conditions in screening. When interesting conditions are discovered, a detailed full analysis of responses can be performed on fewer conditions, thereby reducing the burden of data analysis substantially.
During this process, the scoring system incorporated the anticipated features; dead cell percentage and live cell percentage have equal but opposite constants that contribute to ∼70% of the score. Some interesting unanticipated features were discovered that led to improvements in the robustness of the assay. One example is the “Mean Nuclei Center X” and “Mean Nuclei Center Y”—the average position of a cell in the image that is equivalent to center of mass of cells in the image. When toxicity manifests, cells often detach leaving irregular clusters/patches of cells behind. This effect causes a migration of the center of mass from the center of the image that can be used to buttress other measures of cytotoxicity.
In a thorough analysis of PctDead vs. the PLS score (as shown in Fig. 4 scatter plot), it is apparent that the PLS scoring system dramatically reduced the standard deviation of positive control (i.e., the highly viable/healthy cells). Indeed, the noise relating to the PctDead parameter is largely due to the variable cell counts per well and incomplete cell segmentation of closely packed cell clusters where the YoYo-1 intensity for mixed live/dead cells gets averaged to values less than a single dead cell. The application of the PLS scoring system did not have a large impact on the means of the controls and mainly improved the robustness of the assay through reductions in noise, leading to higher screening efficiency and avoidance of false positives/negatives. This PLS scoring technique is broadly applicable to HCS in the context of HTS for linear responses. In this study we chose a simplistic assay for the sake of illustration to demonstrate that, even when a univariate response is easily understood and robust, the PLS scoring system can still offer substantial benefits. This technique can easily be expanded to more detailed high-content cell death assays at higher magnification to include features like nuclear texture, mitochondrial membrane potential, and micronuclei/nuclear fragmentation. To capture nonlinear responses, artificial neural networks or support vector machines can be used in lieu of PLS. Training a scoring system based on the effects of a positive-control condition or compound can substantially bias the scoring system toward that particular effect but is particularly useful in assays where the positive outcome is straightforward, like cell health/viability. When more nuanced phenotypes are expected or multimodal responses are anticipated, performing PCA on data sets from pilot screening can be highly informative. Seeking to understand the first three principal components in a data set can often lead to the discovery of different effect modalities that can be used as the basis of PLS scoring system development.
Compounds discovered in pilot screening exhibited a potentially shared mechanism through the cholinesterase pathway. Ipratropium bromide (trade name Atrovent) is an anticholinergic drug used in the treatment of bronchial spasms and is a direct antagonist of muscarinic acetylcholine receptors.23 Edrophonium chloride is a reversible acetylcholinesterase inhibitor that prolongs the presence of acetylcholine in the synaptic cleft that has been recently shown to have agonist activity against the human M1 muscarinic receptor (Pubchem Assay AID 588816 and 602250).24 This result demonstrates a potential connection between muscarinic receptor modulation and β-cell survival, which has been previously described and is potentially related to muscarinic receptor inhibition of cytokine (IL-1β and IFNγ)–mediated pancreatic β-cell apoptosis.25–27
In conclusion, this high-content HTS assay platform is a viable approach for phenotypic drug discovery for identifying chemoprotectants against a clinically relevant amylin-induced β-cell death. High-content automated imaging in conjunction with automated feature extraction yields a data-rich phenotypic fingerprint for compound effects that can ultimately lead to new molecular targets, pathways, and compounds with therapeutic effects. Even in a simple live/dead assay as described herein, a PLS scoring system can significantly improve assay performance, reduce the number of false positives/negatives, and help streamline integration of high-content data into conventional HTS data infrastructures. Additionally, two potential chemical probes were discovered in this study that can be used to further mechanistic insight into amylin-induced β-cell loss.
Abbreviations Used
- A1c
glycated hemoglobin or hemoglobin A1c
- DMSO
dimethyl sulfoxide
- DPP-IV
dipeptidyl peptidase-4
- ER
endoplasmic reticulum
- GLP1
glucagon-like peptide-1
- GLP1R
glucagon-like peptide-1 receptor
- HCS
high-content screening
- HTS
high-throughput screening
- INS-1
insulinoma cells
- PCA
principal component analysis
- PctDead
percent dead cells
- PLS
partial least squares
- RFU
relative fluorescent unit
- T1D
type I diabetes
- T2D
type II diabetes
- TZDs
thiazolidinediones
- UPR
unfolded protein response
Acknowledgments
The research reported in this article was partially supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award number R01DK088248 to J.Z.S. and was partially supported by the North Carolina Biotechnology Center Biotechnology Research Grant (BRG) program grant number 2011-BRG-1212 to J.Z.S. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health or the North Carolina Biotechnology Center Biotechnology. The authors would also like to thank Tomas Ding for thoughtful input on experimental design and Kelly B. Sexton for reviewing this article.
Disclosure Statement
No conflict of interest for all listed authors.
References
- 1.Centers for Disease Control and Prevention: 2007National Diabetes Fact Sheet. 2007. Available at http://www.cdc.gov/Features/dsDiabetes/
- 2.Centers for Disease Control and Prevention: 2011 National Diabetes Fact Sheet. 2011. Available at http://www.cdc.gov/diabetes/pubs/factsheet11.htm
- 3.Kloppel G, Lohr M, Habich K, Oberholzer M, Heitz PU: Islet pathology and the pathogenesis of type 1 and type 2 diabetes mellitus revisited. Surv Synth Pathol Res 1985;4:110–125 [DOI] [PubMed] [Google Scholar]
- 4.Lupi R, Del Prato S: Beta-cell apoptosis in type 2 diabetes: quantitative and functional consequences. Diabetes Metab 2008;34Suppl 2:S56–S64 [DOI] [PubMed] [Google Scholar]
- 5.Cunha DA, Hekerman P, Ladriere L, et al. : Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci 2008;121:2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poitout V, Robertson RP: Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocr Rev 2008;29:351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sargsyan E, Bergsten P: Lipotoxicity is glucose-dependent in INS-1E cells but not in human islets and MIN6 cells. Lipids Health Dis 2011;10:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meece J: Dispelling myths and removing barriers about insulin in type 2 diabetes. Diabetes Educ 2006;32:9S–18S [DOI] [PubMed] [Google Scholar]
- 9.Lorenzo A, Razzaboni B, Weir GC, Yankner BA: Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature 1994;368:756–760 [DOI] [PubMed] [Google Scholar]
- 10.O'Brien TD, Butler PC, Westermark P, Johnson KH: Islet amyloid polypeptide: a review of its biology and potential roles in the pathogenesis of diabetes mellitus. Vet Pathol 1993;30:317–332 [DOI] [PubMed] [Google Scholar]
- 11.Hiddinga HJ, Eberhardt NL: Intracellular amyloidogenesis by human islet amyloid polypeptide induces apoptosis in COS-1 cells. Am J Pathol 1999;154:1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter KJ, Scrocchi LA, Warnock GL, et al. : Amyloid inhibitors enhance survival of cultured human islets. Biochim Biophys Acta 2009;1790:566–574 [DOI] [PubMed] [Google Scholar]
- 13.Westermark P, Engström U, Johnson KH, Westermark GT, Betsholtz C: Islet amyloid polypeptide: pinpointing amino acid residues linked to amyloid fibril formation. Proc Natl Acad Sci U S A 1990;87:5036–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zraika S, Hull RL, Verchere CB, et al. : Toxic oligomers and islet beta cell death: guilty by association or convicted by circumstantial evidence? Diabetologia 2010;53:1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pétremand J, Puyal J, Chatton J-Y, et al. : HDLs protect pancreatic β-cells against ER stress by restoring protein folding and trafficking. Diabetes 2012 2012;61:1100–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eizirik DL, Alexssandra K, Cnop M: The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 2008;29:42–61 [DOI] [PubMed] [Google Scholar]
- 17.Singh S, Carpenter AE, Genovesio A: Increasing the content of high-content screening: an overview. J Biomol Screen 2014;19:640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carpenter AE, Jones TR, Lamprecht MR, et al. : CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006;7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wold S, Sjostrom M, Eriksson L: PLS-regression: a basic tool of chemometrics. Chemometrics Intelligent Lab Syst 2001;58:109–130 [Google Scholar]
- 20.Wold S, Trygg J, Berglund A, Antti H: Some recent developments in PLS modeling. Chemometrics Intelligent Lab Syst 2001;58:131–150 [Google Scholar]
- 21.Huang CJ, Lin CY, Haataja L, et al. : High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress mediated beta-cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes 2007;56:2016–2027 [DOI] [PubMed] [Google Scholar]
- 22.Laybutt DR, Preston AM, Akerfeldt MC, et al. : Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 2007;50:752–763 [DOI] [PubMed] [Google Scholar]
- 23.Baigelman W, Chodosh S: Bronchodilator action of the anticholinergic drug, ipratropium bromide (Sch 1000), as an aerosol in chronic bronchitis and asthma. Chest 1977;71:324–328 [DOI] [PubMed] [Google Scholar]
- 24.Fluorescence-based cell-based primary high throughput screening assay to identify antagonists of the human M1 muscarinic receptor (CHRM1). 2011. Avaliable at https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=588852 (last accessed June11, 2014)
- 25.Gautam D, Han SJ, Duttaroy A, et al. : Role of the M3 muscarinic acetylcholine receptor in beta-cell function and glucose homeostasis. Diabetes Obes Metab 2007;9Suppl 2:158–169 [DOI] [PubMed] [Google Scholar]
- 26.Laychock SG, Sessanna SM, Lin MH, Mastrandrea LD: Sphingosine 1-phosphate affects cytokine-induced apoptosis in rat pancreatic islet beta-cells. Endocrinology 2006;147:4705–4712 [DOI] [PubMed] [Google Scholar]
- 27.Sridhar GR, Thota H, Allam AR, Suresh Babu C, Siva Prasad A, Divakar C: Alzheimer's disease and type 2 diabetes mellitus: the cholinesterase connection? Lipids Health Dis 2006;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]