Summary
We demonstrate that CXCR4 mRNA contains ARE in its 3′-UTR and regulated by the RNA-binding proteins, TTP and HuR. Normalization of the aberrant TTP-HuR axis resulted in reduced invasion and migration of breast cancer cells toward CXCL12.
Abstract
CXCR4 is a chemokine receptor that is overexpressed in certain cancer types and involved in migration toward distant organs. The molecular mechanisms underlying CXCR4 expression in invasive cancer, particularly posttranscriptional regulation, are poorly understood. Here, we find that CXCR4 harbors AU-rich elements (AREs) in the 3′-untranslated region (3′-UTR) that bind and respond to the RNA-binding proteins, tristetraprolin (TTP/ZFP36) and HuR (ELAVL1). Different experimental approaches, including RNA immunoprecipitation, 3′-UTR reporter, RNA shift and messenger RNA (mRNA) half-life studies confirmed functionality of the CXCR4 ARE. Wild-type TTP, but not the zinc finger mutant, C124R, was able to bind CXCR4 mRNA and ARE. In the invasive breast cancer phenotype, aberrant expression of CXCR4 is linked to both TTP deficiency and HuR overexpression. HuR silencing led to decreased CXCR4 mRNA stability and expression, and significant reduction in migration of the cells toward the CXCR4 ligand, CXCL12. Derepression of TTP using miR-29a inhibitor led to significant reduction in CXCR4 mRNA stability, expression and migration capability of the cells. The study shows that CXCR4 is regulated by ARE-dependent posttranscriptional mechanisms that involve TTP and HuR, and that aberration in this pathway helps cancer cells migrate toward the CXCR4 ligand. Targeting posttranscriptional control of CXCR4 expression may constitute an alternative approach in cancer therapy.
Introduction
Metastasis is one of the hallmarks of cancer and a major cause of mortality in cancer patients. Although the exact mechanisms that dictate the directional movement of tumor cells to distant sites are not well understood, this movement pattern bears close similarity to the chemokine-mediated movement of leukocytes (1). It is well established that chemokines direct the migration of tumor cells that express their respective chemokine receptors to specific sites where the chemoattractants are highly expressed (2). The chemokine ligand CXCL12 and its receptor CXCR4 represent prominent examples of mediators of the interaction between cancer cells and the tumor microenvironment. The CXCR4-CXCL12 axis is a highly dysregulated pathway in invasive cancer (reviewed in ref. 3). CXCR4 is a G-protein-coupled chemokine receptor that promotes the chemotactic movement of breast cancer cells to distant sites of metastasis along a gradient of its ligand, stromal cell-derived factor-1, officially called CXCL12 (4). Both the receptor and ligand are highly expressed in several types of cancer, including breast, prostate, colon and small cell lung cancer (2,5–8). CXCR4 expression has also been correlated with metastatic spread of breast cancers to the lymph nodes (9). Additionally, expression of CXCR4 in breast, colorectal cancer and melanoma is an indicator of poor prognosis (6,10,11). CXCL12 is also highly expressed in sites of tumor metastases, and in breast cancer, these include the bone marrow, lung and liver (2).
Several factors regulate the expression of genes, including posttranscriptional mechanisms that can promote the rapid decay of messenger RNA (mRNA) transcripts. Many gene transcripts are innately unstable due to the presence of AU-rich elements (AREs) in their 3′-untranslated regions (3′-UTR), which are targeted by RNA-binding proteins for decay (12). These include cytokines, chemokines, growth factors, and a repertoire of functionally diverse transcripts that have a transient response nature (13). However, aberrant ARE-mediated pathways can lead to highly stabilized transcripts that promote many pathological conditions such as autoimmune diseases and cancer (14). Dysregulation of these pathways can result from an imbalance in the expression of the RNA-binding proteins HuR and tristetraprolin (TTP) that stabilize or promote decay, respectively, of ARE-harboring mRNA transcripts. In fact, increased HuR expression and TTP deficiency have been associated with a number of tumors, such as breast, colon and prostate cancers (15–20).
Since posttranscriptional regulation of gene expression can be compromised in invasive breast cancer, we examined the posttranscriptional control of CXCR4 in normal and invasive breast cancer cell lines. We found that CXCR4 harbors a functional ARE in its 3′-UTR and, consequently, we found it to be a novel target for the RNA-binding proteins, TTP and HuR. Furthermore, its mRNA stability control is aberrant in invasive breast cancer cells. The study demonstrates a novel role for the TTP/HuR imbalance in chemokine-triggered migration of cancer cells that can be restored by derepressing TTP expression using a miR-29a inhibitor. Targeting pathological CXCR4 ARE-mRNA stabilization may provide an alternative therapeutic approach for treating invasive cancer.
Materials and methods
Cell lines
The breast cancer tumorigenic cell lines MDA-MB-231 and MCF-7, the breast normal-like cell lines MCF12A and MCF10A, HEK293, and Jurkat cells were obtained from ATCC (Rockville, MD). MDA-MB-231, MCF-7 and HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Grand Island, NY) supplemented with 10% fetal bovine serum and antibiotics. Jurkat cells were maintained in RPMI (Life Technologies) supplemented with 10% fetal bovine serum and antibiotics. The normal cell lines MCF12A and MCF10A were cultured in a 1:1 mixture of Ham’s F12 and DMEM supplemented with 10% bovine insulin, 20ng/ml epidermal growth factor and 500ng/ml hydrocortisone (Sigma, St Louis, MO). TTP/zfp36+/+ and TTP/zfp36− /− mouse embryonic fibroblasts were obtained as described previously (21) and were grown in DMEM. HEK293 Tet-On Advanced cells (Clontech, Mountain View, CA) were used in tetracycline-induced expression experiments and were cultured in DMEM supplemented with 10% Tet System Approved FBS (Clontech), 100 μg/ml G418 (Sigma) and 5% Penicillin-Streptomycin (Invitrogen, Carlsbad, CA). All transfections were carried out in reduced serum media using Lipofectamine 2000 (Invitrogen).
mRNA half-life and quantitative reverse transcription–polymerase chain reaction
For half-life experiments, Actinomycin D (ActD, 5 μg/ml; Sigma) was added to the cells for 1, 2, 4 and 6 h prior to extraction of total RNA using Trizol (Sigma). The reverse transcription reaction was performed using 3 μg total RNA, 150ng random primer, 10mM dNTP mixture, 40U/μl RNase OUT and 200U of SuperScript II (Invitrogen). Quantitative PCR was performed as multiplex reactions in a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA) using FAM-labeled TaqMan probes (Applied Biosystems, Foster City, CA) for uPA (PLAU), HuR (ELAV1), enhanced green fluorescent protein (EGFP) or human and mouse CXCR4. VIC-labeled human or mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probes were used as endogenous controls and data from EGFP quantitative PCR experiments were also normalized to a HEX-labeled RFP probe (Metabion). Samples were amplified in triplicate, and quantification of relative expression was performed using the ΔΔCt method. The mRNA half-life determinations were calculated using the one-phase exponential decay method (22) using GraphPad Prism software (GraphPad Software, San Diego, CA). This method has the best fit for mRNA data that tend to decay at a certain rate over a period of time and then reach a plateau; all fits were >R = 0.9.
Western blotting
Western blotting was performed as described previously (18). Primary antibodies used were 1/200 rabbit anti-CXCR4 (Sigma–Aldrich), 1/200 goat anti-HuR (sc-5843; Santa Cruz), 1/200 goat anti-TTP (sc-8458; Santa Cruz), mouse anti β-actin (ab20272) or mouse anti-GAPDH-HRP (ab9482) (1/4000) (Abcam, Cambridge, UK).
miR-29a inhibitor experiments
For miR-29a inhibitor experiments, MDA-MB-231 cells were treated with 50nM control inhibitor or miR-29 inhibitor as a cell-permeable peptide-linked nucleic acid (PNA) with the following sequence RRRQ RRKKR-OOATTTC AGATGGTGCT (Panagene, Daejeon, Korea: OO: two glycol linker) or scrambled control PNA sequence (here, called control PNA). Treatment was for 48 h and was followed by either quantitative PCR, western blotting, mRNA stability or migration experiments.
Immunoprecipitation of RNP complexes
For TTP-IP, MDA-MB-231 cells were seeded in 100 × 20mm culture dishes at a density of 3.6 × 106 cells and incubated overnight. Cells were transfected with 4.5 μg of TTP, or TTP mutant (C124R) plasmid for 24 h. The immunoprecipitation procedure was carried out as described previously (23). RNA was subjected to quantitative reverse transcription–polymerase chain reaction, as described above, using probes for CXCR4 and normalized to GAPDH (Applied Biosystems). For HuR-IP, non-transfected MDA-MB-231 cells were seeded at the same density and lysed upon reaching 90–95% confluence. The lysate was incubated with 25 μg of anti-HuR or normal goat IgG (Santa Cruz). RNA was subjected to quantitative reverse transcription–polymerase chain reaction using probes for CXCR4 or uPA and normalized to GAPDH.
RNA EMSA and supershift
An RNA gel shift assay was carried out as described previously (23). Briefly, a sense RNA oligonucleotide corresponding to CXCR4 3′-UTR was custom synthesized, 5′ end-labeled with biotin, and HPLC purified (Metabion, Germany). The biotin-labeled sense probe was incubated with lysate from either HEK293 cells overexpressing TTP, or the mutant C124R, or control lysate from untransfected cells. The probe was also subjected to a separate assay and incubated with lysate from untransfected MDA-MB-231 cells, and both assays were carried out as described previously (23). The supershift assay was carried out in the same manner, except that 2 μg of anti-HuR antibody (sc-5483) (Santa Cruz) or anti-TTP antibody (sc-8458) (Santa Cruz) was preincubated with MDA-MB-231 or HEK293 lysate, respectively, for 30min before the addition of the labeled probe.
RNA interference
RNA interference studies were performed using three different small interfering RNA (siRNA) duplexes designed for silencing of CXCR4 (NM_003467): siRNA 1 and 2 were as described by Liang et al. (24) and siRNA 3 (sense, 5′-GGUGG UCUAUG UUG GCGUC-3′ and antisense, 5′-GACG CCAACAU AGACCACC-3′). siRNA duplexes designed for silencing of HuR were also used (NM_001419, sense, 5′-GCCU GUUC AGCAGCAUUGG-3′ and antisense, 5′-CCA AUGCUGCUGA ACAGGC-3′). All siRNAs, including non-specific controls, were custom-made by Metabion. The efficiency of siRNA silencing was determined by real-time PCR and western blotting techniques, as described.
Plasmids, 3′-UTR reporter constructs and deletion mutants
Reporter expression vectors for CXCR4 or the control stable bovine growth hormone (BGH) 3′-UTR (control UTR) were obtained by PCR targeting the complete 3′-UTR using the following primers: forward, 5′-GCAG CGGATCCCACAG ATGTAAAGACTT-3′, and reverse, 5′-GCAGCTCT AGAGTTT AACAT GTACTTTTATTAAC-3′. The forward and reverse primers contain the BamHI and XbaI sites, respectively (underlined). The putative ARE and ARE-like sequences correspond to 1498–1561 nt of CXCR4 (NM_001008540) and were synthesized as duplex oligonucleotides with the BamHI and XbaI sites at the termini. These DNA products were cloned into a RPS30-EGFP cellular promoter-driven expression vector (25). Tetracycline-inducible (Tet-On) wild-type and mutant TTP constructs were prepared by PCR of pCR3.1-TTP or pCR3.1-C124R plasmids using the forward primer that includes three copies of the TetO site, 5′-ACCAGGT CCCTATCAGT GATAGAGATC CTCCCTAT CAGTGATAGAGAT CCTCCCTATCAGTGA-3′, and the reverse primer containing the BGH site, 5′–CCATA GAGCCCACC GCAT-3′.
Invasion and migration/chemotaxis assays
For invasion assays, MDA-MB-231 cells transfected with 0.25 μg of luciferase expression cassettes and cotransfected with CXCR4 siRNA or control siRNA for 48 h were reseeded onto the upper chambers of 24-well invasion inserts of 8 μm pore-sized membranes (BD Biosciences) in serum-free DMEM at a density of 3 × 105 cells per well and incubated for 24 h. Six hundred microliters of serum-free DMEM + 0.1% bovine serum albumin or serum-free medium + 10 nM CXCL12/SDF1 (R&D Systems, Minneapolis, MN) was used as the chemoattractant in the lower chambers. The membranes were removed from the inserts and incubated with luciferase lysis buffer (Promega, Madison, WI) for 15min, and then luciferase activity was measured according to the manufacturer’s protocol. For migration experiments, MDA-MB-231 cells were seeded at the same density as for invasion assays onto the upper chamber of 24-well migration plates and 600 μl of serum-free DMEM + 0.1% bovine serum albumin, or serum-free medium + CXCL12 was added to the lower chambers of migration plates and incubated for 24 h. Cell-Titer-Glo reagent (Promega) was then directly added to the lower chamber to measure luminescence associated with the migrating cells. For both invasion and migration assays, a ZENYTH 3100 reader (Anthos Labtec) was used to measure luciferase activity or luminescence, respectively.
Breast cancer patient data and analysis
The Cancer Genome Atlas (TCGA) public resource was mined through the Oncomine web portal, www.oncomine.com. The TCGA program used well-annotated data and ensures TCGA’s rigorous sample requirements that yield high quality data. According to TCGA, all tissues were histologically consistent with breast adenocarcinoma with average of 60% tumor cell nuclei. Further information can be obtained from: http://cancergenome.nih.gov/cancersselected/biospeccriteria. The L2 data (normalized gene expression) for mRNA levels of tissues from ductal breast tumors (N = 394), invasive lobular tumors (N = 36) and normal breast tissues (N = 61) were extracted. The TTP/HuR mRNA ratios and their changes between matched normal breast tissues and the tumor tissues were assessed. The log2 median-centered intensity ratios were used and converted to anti-log for the TTP/HuR mRNA ratios. Pairwise correlations (Spearman’s test) were used to correlate TTP/HuR mRNA ratio with gene-specific log2 median intensity ratios.
Statistical analysis
Data were presented as means ± standard error of the mean. Two-sample Student’s t-tests were used to determine significant differences when comparing two data columns. Furthermore, one-way analysis of variance was used when analyzing three or more data columns. Two-way analysis of variance was used when analyzing two groups of data, each having two data columns.
Results
CXCR4 harbors a functional ARE in its 3′-UTR
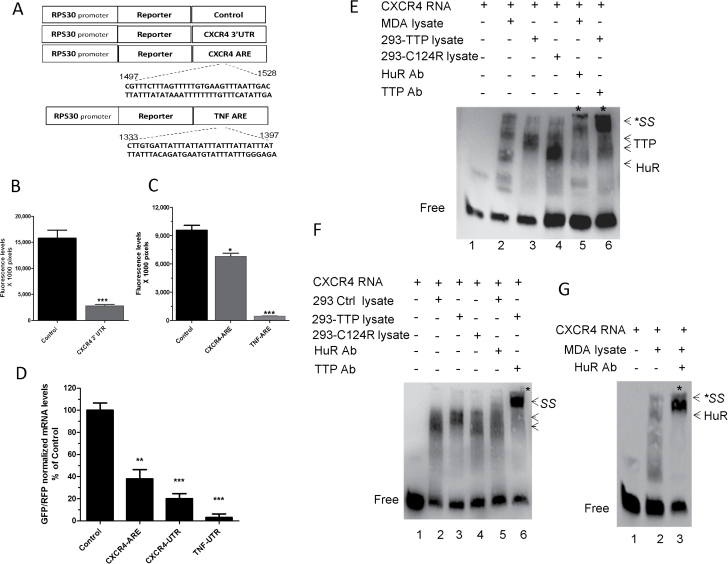
Bioinformatics analysis of CXCR4 3′-UTR revealed the presence of a putative ARE that might be subject to binding and regulation by TTP and HuR. To determine the functionality of the CXCR4 ARE, we transfected an RPS30 promoter-linked EGFP reporter construct containing CXCR4 3′-UTR or its ARE (Figure 1A) into HEK293 cells. This led to an 80% reduction in EGFP reporter expression when the entire CXCR4 3′-UTR was used compared with the control UTR construct (Figure 1B). A 30% reduction in reporter activity was demonstrated when the construct containing the ARE was used (Figure 1C), which may indicate the presence of other or accessory motifs that strengthen the ARE in the complete 3′-UTR construct. TNF ARE was used as a positive control. At the mRNA level, 60 and 80% reductions were observed in mRNA levels expressed from the EGFP-CXCR4 ARE and 3′-UTR reporters, respectively, compared with the control (Figure 1D). These findings indicate that the presence of the ARE, and probably additional motifs, in CXCR4 3′-UTR confers on CXCR4 mRNA an inherent instability attribute.
Fig. 1.
Investigation of ARE functionality in CXCR4 3′-UTR. (A) Schematic representation of the RPS30-EGFP-control 3′-UTR, RPS30-EGFP-CXCR4 3′-UTR and ARE, and RPS30-EGFP-TNF ARE reporter constructs. (B) Regulation of reporter activity by CXCR4 3′-UTR. HEK293 cells were transfected with the RPS30 promoter-linked EGFP reporter plasmid construct containing CXCR4 3′-UTR or a control 3′-UTR (BGH 3′-UTR). GFP fluorescence was measured 24h posttransfection. (C) CXCR4-ARE-mediated regulation of reporter expression. HEK293 cells were transfected with RPS30-EGFP reporter plasmid constructs containing CXCR4 ARE, TNF-ARE or control BGH 3′-UTR. GFP fluorescence was measured 24h posttransfection. (D) mRNA levels of the EGFP reporters; HEK293 cells were transfected with the reporter constructs in B and C for 24h, then RNA was extracted for real-time PCR (quantitative PCR) using a FAM-labeled probe against EGFP and normalized to a VIC-labeled GAPDH then normalized again to a HEX-labeled RFP probe. Data represent mean ± standard error of the mean from three independent experiments *P = 0.01, **P < 0.005 and ***P < 0.001 (Student’s t-test). (E) RNA-electrophoretic mobility shift assay (EMSA) and supershift assay. Biotinylated CXCR4-ARE probe was incubated with HEK293 cell lysate overexpressing TTP or TTP mutant (C124R) or untransfected MDA-MB-231 cell lysate then subjected to non-denaturing polyacrylamide gel and transferred to a nylon membrane. Bands were detected by a chemiluminescent kit. (F) Lysate from HEK293 cells overexpressing TTP, C124R or control plasmid were incubated with CXCR4 biotinylated RNA probe and subjected to RNA-EMSA and supershift assay as described in E. Results are from one experiment representative of at least two independent experiments. (G) Lysate from untransfected MDA-MB-231 cells was incubated with CXCR4 biotinylated RNA probe and subjected to RNA-EMSA and supershift assay for detection of HuR interaction with the probe. Results are representative of two independent experiments. In all RNA-EMSA images, arrows point to the TTP-bound biotinylated RNA complex (middle arrows), HuR-bound complex (lower arrow) or supershifted complexes (upper arrow and asterisks) resulting from preincubation of lysates with anti-TTP or anti-HuR antibodies prior to the addition of CXCR4 RNA probe. Lower bands indicate free probe.
The presence of a functional ARE in CXCR4 3′-UTR prompted us to investigate whether there was an interaction between the RNA-binding proteins TTP and HuR and CXCR4 mRNA at the posttranscriptional level. We performed an electrophoretic mobility shift assay using protein lysate from either HEK293 cells overexpressing TTP, TTP mutant (C124R) or control lysate from untransfected cells or from non-transfected MDA-MB-231 cells, which express high levels of HuR (22). C124R is a non-binding mutant of TTP that exerts a dominant-negative effect (26). The lysate was incubated with biotinylated CXCR4 ARE RNA probe and then subjected to RNA electrophoretic mobility shift assay. Incubation of the CXCR4 mRNA probe with MDA-MB-231 lysate resulted in a prominent complex representing HuR–ARE interaction (Figure 1E, lane 2, upper band). When the probe was incubated with HEK293 cell lysate overexpressing either TTP or C124R (Figure 1E, lanes 3 and 4, respectively), prominent bands were observed representing TTP interaction with the synthetic probe. Addition of anti-HuR to the MDA-MB-231 lysate or anti-TTP to the HEK293 lysate resulted in supershifted bands (lanes 5 and 6, respectively). We repeated the assay for each cell type and presented separately. Incubation of CXCR4 RNA probe with HEK293 lysate produced a complex (Figure 1F, lane 2). When the probe was incubated with the lysate overexpressing TTP, a prominent complex including additional band was seen (Figure 1F, lane 3), both were attenuated when C124R-expressing cell lysate was used (Figure 1F, lane 4) or supershifted upon addition of anti-TTP antibody (Figure 1F, lane 6). Preincubation of anti-HuR with the control lysate resulted in a slight shift (Figure 1F, lane 5), but greater interaction was seen with the MDA-MB-231 cell line, which has a higher basal level of HuR (Figure 1G). These data provide additional evidence for the interaction of the two RNA-binding proteins TTP and HuR with CXCR4 mRNA.
CXCR4 is regulated by TTP
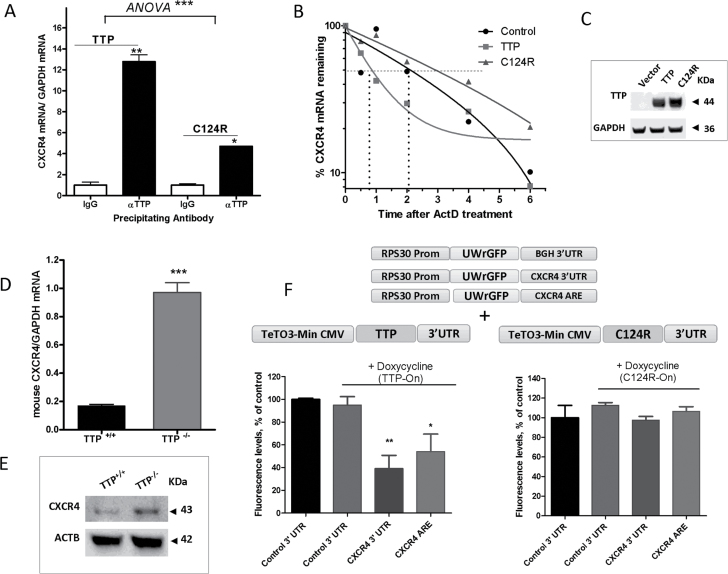
In order to study more closely the association of TTP with CXCR4 mRNA, we performed an RNA immunoprecipitation experiment using MDA-MB-231 cells overexpressing TTP or mutant C124R protein and an anti-TTP antibody. CXCR4 mRNA was highly associated with TTP protein (13-fold, P < 0.01) (Figure 2A), yet weakly with the non-ARE binding TTP mutant, C124R. To determine the functional outcome of TTP association with CXCR4 mRNA, we examined the half-life of CXCR4 mRNA as a result of TTP or C124R overexpression in MDA-MB-231 cells. An almost 2.5-fold decrease in CXCR4 mRNA half-life was observed in TTP overexpressed samples compared with the control (Figure 2B), but no significant effects were detected when the mutant form was overexpressed (Figure 2B). Overexpression of TTP and C124R in MDA-MB-231 cells was verified by western blotting (Figure 2C).
Fig. 2.
TTP regulation of CXCR4 mRNA and protein. (A) Quantitative PCR (qPCR) quantification of CXCR4 mRNA associated with TTP protein. MDA-MB-231 cells were transfected with TTP or C124R expression plasmids for 24h. Cells were lysed, and TTP and C124R proteins were immunoprecipitated using anti-TTP or normal IgG control antibody. Quantification of associated CXCR4 mRNA was performed by qPCR using a FAM-labeled human CXCR4 Taqman expression probe and normalized to a VIC-labeled GAPDH probe. Data are from one experiment representative of two independent experiments *P < 0.05, **P < 0.005 (Student’s t-test), ***P < 0.0001 [analysis of variance (ANOVA)]. (B) TTP regulation of CXCR4 mRNA half-life. MDA-MB-231 cells were transfected with TTP or C124R plasmid then treated the following day with actinomycin D (ActD, 5 μg/ml) for the indicated times then RNA was extracted for qPCR. Results are from one experiment representative of three independent experiments (C) Detection of overexpressed TTP and C124R proteins in MDA-MB-231 cells by western blotting using anti-TTP antibody. (D) qPCR quantification of CXCR4 mRNA in TTP+/+ and TTP− /− mouse embryonic fibroblasts (MEFs) using a FAM-labeled mouse CXCR4 probe and normalized to VIC-labeled mouse GAPDH, ***P < 0.0001 (Student’s t-test). (E) Representative western blot of CXCR4 protein in TTP+/+ and TTP− /− MEFs using anti-CXCR4 antibody. (F and G) TTP regulation of CXCR4 reporter expression. Tetracycline-inducible constructs were obtained by PCR of TTP and C124R plasmids, then 10ng of purified PCR product was transfected into HEK293 Tet-On Advanced cells and cotransfected with 25ng of RPS30 promoter-linked EGFP reporter plasmid constructs containing a control 3′-UTR, CXCR4 3′-UTR or CXCR4 ARE. Doxycycline (0.25 μg) was added 24h after transfection and GFP fluorescence was measured 24h after addition of doxycycline. Data were normalized to their corresponding non-doxycycline-induced samples (taken as 100%). Data are from one experiment representative of at least two independent experiments, * P < 0.05 and **P < 0.001 (Student’s t-test).
In addition, TTP regulation of CXCR4 mRNA was confirmed in TTP+/+ and TTP− /− mouse embryonic fibroblasts; a 5-fold increase in CXCR4 mRNA was demonstrated in the absence of TTP (Figure 2D). CXCR4 protein was also increased in the TTP knockout cell line compared with wild-type mouse embryonic fibroblasts (Figure 2E). Regulation of CXCR4 mRNA was substantiated further by performing Tet-On reporter experiments. Tet-On wild-type and mutant TTP (C124R) constructs were transfected into HEK293 Tet-On cells and cotransfected with RPS30 promoter-linked EGFP reporter constructs containing CXCR4 3′-UTR, CXCR4 ARE or control (stable BGH) 3′-UTR (Figure 2F, upper panel). Induction with the tetracycline analog, doxycycline, resulted in a 60% decrease in CXCR4 3′-UTR reporter activity compared with the non-doxycycline induced reporter and an almost 50% reduction in the presence of CXCR4 ARE alone (Figure 2F). No changes in reporter activity were observed with the mutant construct, C124R (Figure 2F). These findings demonstrate the destabilizing effect of TTP on CXCR4 mRNA and the requirement of the zinc finger domain.
HuR regulation of CXCR4 mRNA
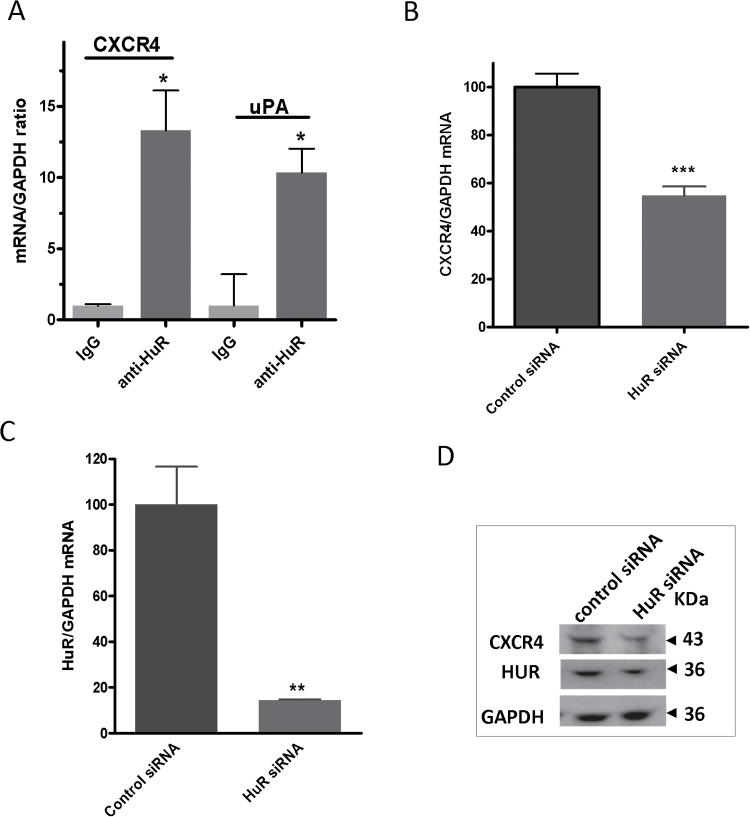
The interaction between HuR and CXCR4 mRNA was also substantiated by RNA immunoprecipitation experiments in MDA-MB-231 cells demonstrating a significant degree of association (13-fold) with HuR protein compared with IgG control (Figure 3A). uPA mRNA was used as a positive control and was highly associated with HuR protein, demonstrating a 10-fold difference (Figure 3A). We next examined the effect of HuR knockdown on CXCR4 expression in the same breast cancer cell line. Cells were transfected with a siRNA specific against HuR, which resulted in >80% reduction in HuR mRNA at 48 h posttransfection (Figure 3C). CXCR4 mRNA was downregulated by almost 50% as a result of HuR silencing (Figure 3B). This reduction was also reflected at the protein level (Figure 3D).
Fig. 3.
HuR regulation of CXCR4 mRNA and protein. (A) qPCR quantification of CXCR4 mRNA associated with HuR protein. MDA-MB-231 cells were lysed and HuR protein was immunoprecipitated using anti-HuR or normal IgG control antibody. Quantification of associated CXCR4 mRNA was performed as described above using a probe specific for human CXCR4 or human uPA as a positive control and both were normalized to human GAPDH. Data are from one experiment representative of two independent experiments, *P < 0.05 (Student’s t-test). (B) Effect of HuR silencing on CXCR4 mRNA. MDA-MB-231 cells were transfected with 25nM HuR siRNA or scrambled siRNA, and CXCR4 mRNA levels were determined by qPCR. Data are from one representative experiment of three independent experiments, ***P < 0.001 (Student’s t-test). (C) HuR silencing in MDA-MB-231 cells. qPCR determination of HuR mRNA levels as a result of HuR silencing described in B. Data are from one representative experiment of three independent experiments, **P < 0.005 (Student’s t-test). (D) Western blotting of CXCR4 and HuR protein levels as a result of HuR silencing in MDA-MB-231 cells.
Aberrant CXCR4 mRNA expression and stability in breast cancer
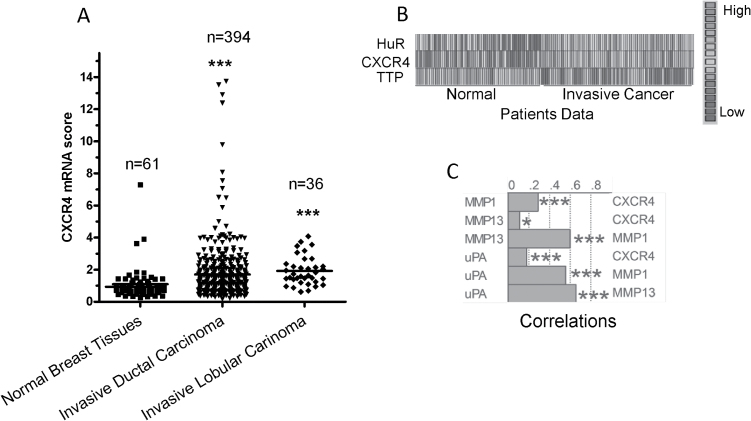
In line with these results, we examined the relationship of CXCR4 with HuR and TTP in breast cancer using clinical data from TCGA through the Oncomine portal. The patient data was mostly of the invasive ductal breast carcinoma as it is the most common type of breast cancer. CXCR4 expression was higher in invasive ductal and invasive lobular carcinoma tissues compared with normal breast tissue (Figure 4A). When the expression of CXCR4 was compared with that of HuR and TTP among normal and invasive cancer patients, both HuR and CXCR4 expression were low in normal and higher in invasive cancer patients, and at the same time both CXCR4 (P < 0.001) and HuR (P < 0.001) were inversely related to TTP (Figure 4B). Furthermore, positive correlations were detected between CXCR4 and other pro-invasion gene products, specifically uPA, MMP1 and MMP13 (Figure 4C).
Fig. 4.
CXCR4 expression in breast cancer patient samples. (A) Comparison of CXCR4 mRNA expression scores of different breast carcinoma samples versus normal matched breast tissue obtained from TCGA data using the Oncomine portal. (B) Heat map for HuR, CXCR4 and TTP in normal and invasive breast cancer patients obtained from TCGA database. (C) Graphical representation of correlation values among the pro-invasion factors and their pairwise correlation with CXCR4 in invasive breast cancer patients. Level 2 gene expression data were obtained from TCGA data using the Oncomine algorithm, ***P < 0.001 and *P < 0.05 (Spearman’s correlation).
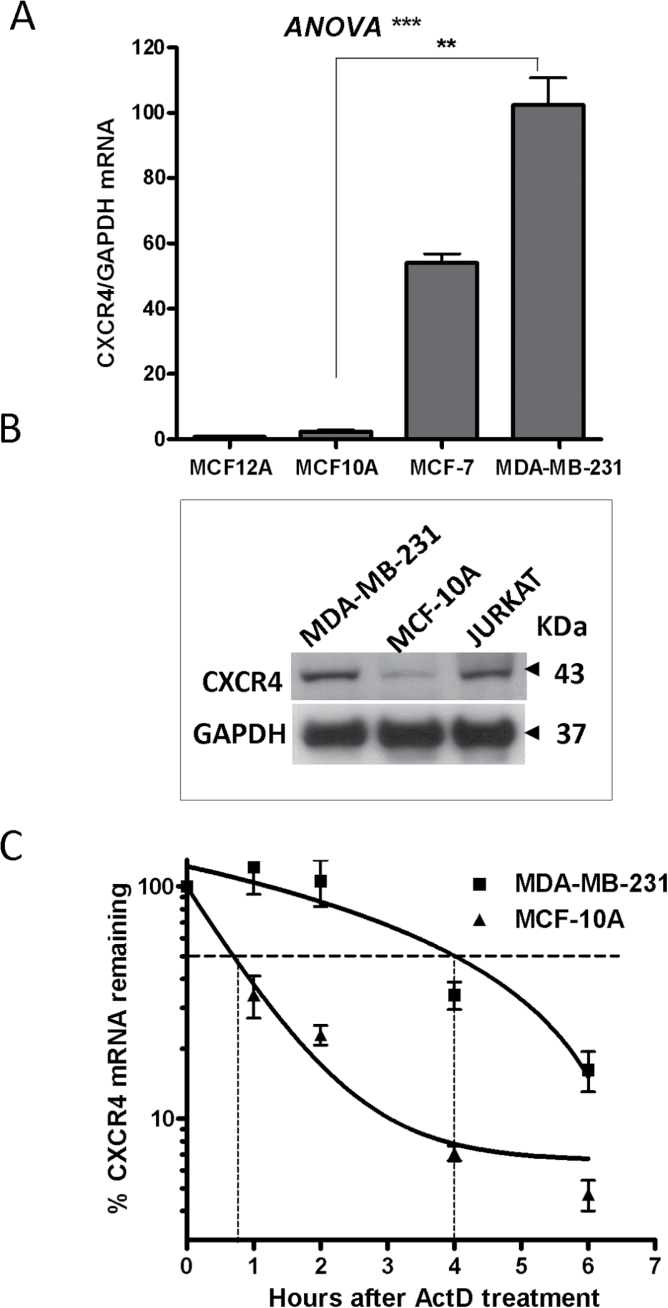
These results were also replicated in several breast cell lines: MCF10A and MCF12A cells representing normal-like breast cells, MCF-7 non-invasive tumor breast cells and the highly invasive MDA-MB-231 cell line (Figure 5). Consistent with their increased migration ability, MDA-MB-231 cells expressed high levels of CXCR4 mRNA, demonstrating a 2-fold difference over the non-invasive MCF-7 tumor cell line and more than a 40-fold and a 130-fold increased expression compared with the normal-like MCF10A and MCF12A, respectively (Figure 5A). CXCR4 protein levels were also higher in MDA-MB-231 cells when compared with their normal counterpart MCF10A (Figure 5B). The half-life of CXCR4 mRNA was also highly stabilized in MDA-MB-231 cells from <1 h in MCF10A cells to 4 h in the invasive cell line (Figure 5C). These results demonstrate an aberrant CXCR4 mRNA stability and overexpression pattern of CXCR4 in breast cancer cells.
Fig. 5.
CXCR4 expression in breast cancer cells. (A) CXCR4 mRNA levels in breast cell lines by quantitative PCR (qPCR) using a human CXCR4 probe normalized to GAPDH. Data are from one representative experiment out of three independent experiments, **P < 0.01 (Student’s t-test), ***P < 0.001 [one-way analysis of variance (ANOVA)]. (B) Western blot of CXCR4 protein in the normal and tumor breast cell lines, MDA-MB-231 versus MCF10A cells; the Jurkat cell line was used as a positive control for CXCR4 expression. (C) CXCR4 mRNA stability determination in MDA-MB-231 versus MCF10A cells. Cells were treated with ActD (5 μg/ml) for the indicated times, and RNA was extracted for qPCR. Data are from one experiment representative of at least three independent experiments.
HuR inhibition and TTP induction reduces CXCR4/CXCL12-mediated invasion and migration
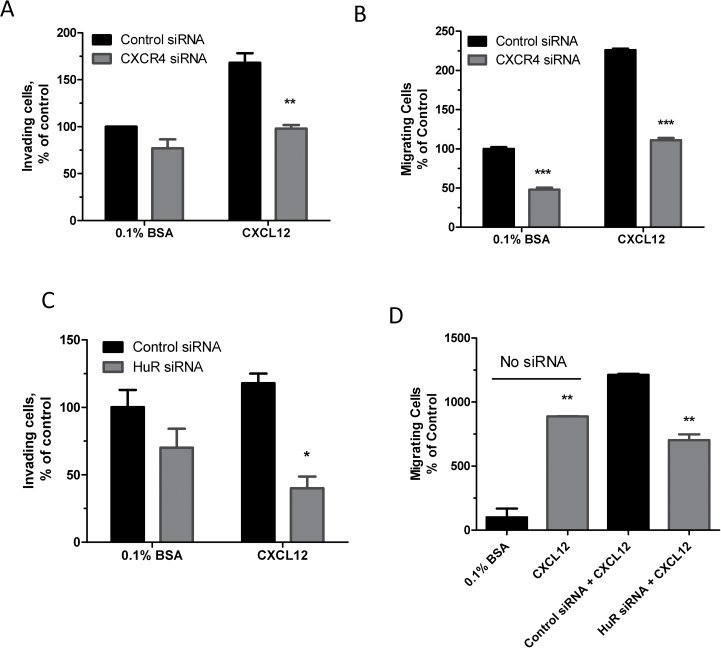
To determine the effect of CXCR4 silencing on the invasiveness and migration potential of MDA-MB-231 cells, we used a specific siRNA directed against CXCR4 mRNA. This resulted in a reduction of ~90% in CXCR4 mRNA (CXCR4 siRNA 3, Supplementary Figure 1, available at Carcinogenesis Online), and it was subsequently used in invasion assays. The siRNA knockdown of CXCR4 led to a 23% reduction in invading cells through the matrigel at basal serum conditions (Figure 6A). The CXCL12, ligand for CXCR4, was used as the chemoattractant to increase the magnitude and specificity of the invasion assay for CXCR4; first, optimal concentration of CXCL12 was first determined using a dose–response chemotaxis assay (Supplementary Figure 2, available at Carcinogenesis Online). A 40% reduction in the invasiveness of MDA-MB-231 cells toward CXCL12 was observed as a result of CXCR4 silencing compared with cells treated with a control siRNA (Figure 6A). Migration of the cells was decreased by >50% when the cells were treated with CXCR4 siRNA at basal serum conditions or CXCL12-treated conditions (Figure 6B). We also sought to determine the effect of HuR silencing on the degree of invasiveness and migratory potential of MDA-MB-231 cells toward CXCL12. Silencing of HuR led to a statistically significant reduction (~60%) in invasiveness of MDA-MB-231 cells in the presence of CXCL12 as the chemoattractant (Figure 6C). Furthermore, a 40% reduction in migration of the breast cancer cells toward CXCL12 was detected when HuR siRNA was used (Figure 6D). The findings indicate that silencing of CXCR4 and HuR both have a significant inhibitory effect on CXCL12-mediated invasion and migration of breast cancer cells.
Fig. 6.
Effect of silencing of CXCR4 and HuR on invasion and migration of MDA-MB-231 cells. (A) Invasiveness of MDA-MB-231 cells as a result of CXCR4 knockdown. MDA-MB-231 cells were transfected with 50nM CXCR4 siRNA or control siRNA and cotransfected with 0.25 μg of luciferase expression constructs for 48h then reseeded in serum-free media in the upper matrigel invasion chambers. CXCL12 or 0.1% bovine serum albumin (BSA) (as a control) was used as the chemoattractant in the lower chambers. Luciferase was measured after 24h. Data represent mean ± standard error of the mean of three independent experiments, **P < 0.005 (Student’s t-test). (B) MDA-MB-231 cells ability to migrate toward CXCL12 as a result of CXCR4 silencing. Cells were transfected with CXCR4 siRNA or control siRNA for 24h and reseeded in serum-free medium in the upper chambers of a migration chamber plate. CXCL12 (10nM) or 0.1% BSA alone was added to the lower chamber and luminescence was measured after 24h. Data are from one experiment representative of two independent experiments, ***P < 0.0001 (Student’s t-test). (C) Effect of HuR knockdown on CXCR4-mediated invasion. MDA-MB-231 cells were transfected with HuR siRNA or control siRNA and reseeded in serum-free media in the upper chambers of a matrigel invasion plate as in A. Data are from one experiment representative of three independent experiments, *P < 0.05 (Student’s t-test). (D) Effect of HuR knockdown on CXCR4-mediated migration. MDA-MB-231 cells were transfected with HuR siRNA or control siRNA and reseeded in serum-free media in the upper chambers of a migration chamber plate. CXCL12 or 0.1% BSA alone was added to the lower chamber, and luminescence was measured after 24h. Data are from one experiment representative of two independent experiments, **P < 0.01 (Student’s t-test).
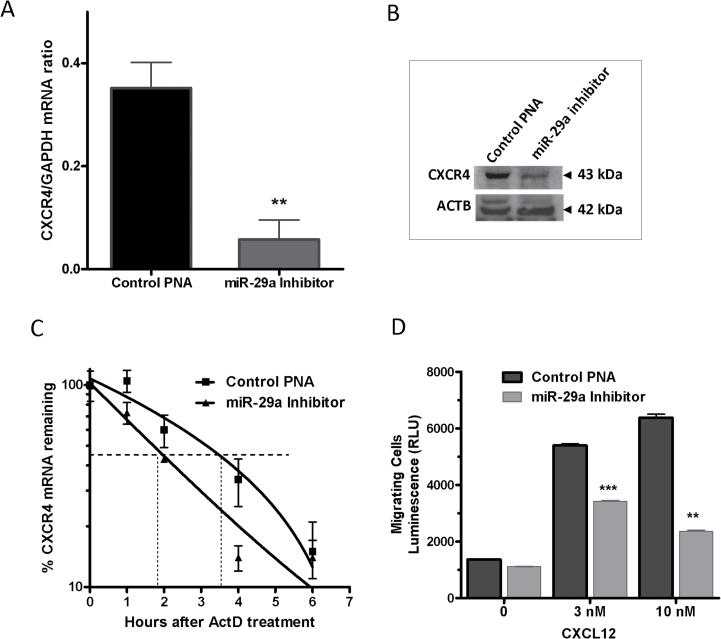
We have previously demonstrated that miR-29a inhibition restores TTP-HuR balance in the MDA-MB-231 cell line (22). Thus, we utilized this inhibitor to induce TTP expression and test the effect on CXCR4 mRNA and protein. Indeed, significant reductions in CXCR4 mRNA (>17-fold, P < 0.001, Figure 7A) and protein (Figure 7B) in MDA-MB231 cells were demonstrated as a result of normalization via miR29a inhibition. The mRNA stability determination for CXCR4 also showed a shorter half-life from 3.5 to 2 h when miR-29a was inhibited in MDA-MB-231 cells (Figure 7C). To determine if miR-29a inhibition in invasive breast cancer cells would impair their CXCR4-mediated ability to migrate, MDA-MB-231 cells were treated with miR-29a inhibitor for 48 h and then subjected to a chemotaxis assay using CXCL12 as the chemoattractant. The ability of the cells to migrate toward CXCL12 was reduced by nearly 35% at a 3nM concentration of CXCL12 and 60% with a 10nM concentration compared with cells treated with a miR inhibitor control (Figure 7D), demonstrating that inhibition of miR-29a reduces the CXCR4-mediated invasive and migratory potential of breast cancer cells.
Fig. 7.
Effect of miR-29a inhibition on CXCR4 mRNA expression and stability. (A) MDA-MB-231 cells were treated with miR-29a inhibitor or control PNA for 48h followed by total RNA extraction for quantitative PCR (qPCR). Data represent mean ± standard error of the mean from two independent experiments, **P < 0.005 (Student’s t-test). (B) Western blot of CXCR4 protein expression as a result of miR-29a inhibition in MDA-MB-231 cells. (C) miR-29a inhibition reduces the half-life of CXCR4 mRNA. MDA-MB-231 cells were treated with miR-29a inhibitor or control for 48h before treatment with ActD (5 μg/ml) for the indicated times followed by total RNA extraction for qPCR. Data are from one experiment representative of two independent experiments. (D) miR-29a inhibitor-mediated attenuation of CXCR4-mediated migration of breast cancer cells. MDA-MB-231 cells were treated with miR-29a inhibitor or control for 48h then reseeded in serum-free media onto the upper chambers of a migration chamber plate. CXCL12 (3 and 10nM) or 0.1% bovine serum albumin alone was added to the lower chamber; luminescence was measured after 24h. Data represent mean ± standard error of the mean from two independent experiments, **P < 0.005, ***P < 0.001 (Student’s t-test).
Discussion
The CXCL12/CXCR4 axis plays an important role in various physiological processes, including lymphoid trafficking, wound healing and in early embryonic development, and expression of both the chemokine and the receptor is found in a wide variety of tissues (27). Furthermore, a deficiency in the chemokine or its receptor can result in malformed vasculature and defects in cardiac and hematopoietic development (28,29). However, the same axis has also been implicated in promoting the directional migration of tumor cells to various sites of metastasis (2,30–32), as both CXCL12 and CXCR4 are overexpressed in different types of cancer and have been shown to correlate with poor prognosis (10,33–36). Here, we found that increased expression of CXCR4 and its functional consequences on breast cancer migration toward its chemokine ligand are caused, at least partly, by aberrant posttranscriptional mechanisms. This CXCR4-mediated disease process can be dampened by inhibiting HuR RNA-stabilizing protein or by induction of TTP mRNA-destabilizing protein.
Although there are reports on the mechanisms regulating CXCR4 signaling and transcription, no known mechanism of posttranscriptional control has been reported. The ARE is one of the most common posttranscriptional control elements that causes default mRNA instability and exists in nearly 4000 transcripts (37). It has been classified in different categories based on its sequence characteristics (38,39). CXCR4 has a Class II ARE (or Group 5) containing one UUAUUUAUA in a U-rich context, but it appears from the reporter and RNA immunoprecipitation data here that other accessory sequences may be needed for stronger effects. For example, the 58 nt U-rich region (64%) is found immediately upstream of the CXCR4 ARE; it has been reported that these types of U-rich region arrangements can enhance the ARE-mediated effects (40). Moreover, the ARE effect is also pronounced at the protein level as compared with the mRNA level, indicating that mRNA decay is significantly operative. There is sufficient evidence regarding the additional role of the ARE in translational repression in addition to its mRNA decay function (41,42). The CXCR4 ARE here has been shown to be recognized, bound by and responded to by TTP and HuR, RNA-binding proteins known to regulate AREs.
The healthy role of AREs can be compromised in cancer, largely because of deficiency of mRNA decay promoting proteins, notably TTP (19,20,43–46), and HuR overexpression (19,47). These irregularities, resulting in an aberrant TTP-HuR axis (22), subsequently lead to mRNA stabilization of key gene products for CXCR4, uPA, uPAR and MMP1/13 mRNAs, and thus promoting tumor cell detachment and migration to distant sites (18,19,48,49). Here, we show that the CXCR4/CXCL12 axis, which can be kept in check by ARE-dependent posttranscriptional mechanisms, can also be perturbed by the aberrant TTP-HuR axis. Our data provide an explanation for the abnormal expression of CXCR4 in invasive breast cancer from a posttranscriptional point of view; that is, low TTP expression combined with upregulation of HuR results in increased CXCR4 mRNA stability and consequently higher levels of protein. The disproportionate expression of TTP and HuR, as well as their target CXCR4, is also observed in invasive tumors of breast cancer patients and probably can promote invasion and metastasis toward organs where CXCL12 is highly expressed such as bone, liver and lung. The role of the CXCR4/CXCL12 axis in breast cancer biology has been explored including elevation of both CXCR4 mRNA and protein levels (2,3,9,11,24,35,50). The purpose of using and analyzing TCGA patient data here is to highlight the correlation between the RNA-binding proteins TTP and HuR and the chemokine receptor CXCR4, whereas the cell line data show direct association and regulation of CXCR4 mRNA by the two proteins. A number of studies have demonstrated appreciable decrease in CXCR4-mediated invasion and metastasis as a result of CXCR4 reduction by monoclonal antibodies, siRNA silencing or small molecular antagonists (2,24,50,51). Our attempt to reduce the CXCR4-mediated migration of MDA-MB-231 cells was by means of a cell-permeable miR-29a PNA; this molecule induces TTP by derepressing TTP (22) and subsequently results in decreased stability of CXCR4 mRNA transcripts. Reduction of TTP by miR-29a inhibitor also leads to a parallel reduction of additional proteolytic factors for degradation of the extracellular matrix, such as uPA and members of the matrix metalloproteinase family (22), a process that is required for invasiveness of cancer cells. Targeting the CXCR4-CXCL12 axis by miR-29a inhibitor may then be an alternative therapeutic approach in invasive cancer.
Supplementary material
Supplementary Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
King Faisal Specialist Hospital and Research Centre (Research Advisory Council-approved Project No. RAC-2110 032).
Supplementary Material
Acknowledgements
We thank Dr P.J.Blackshear and Dr W.S.Lai (NIH) for providing TTP knockout fibroblasts and the C124R mutant TTP. The technical assistance of Ms L.Al-Haj is appreciated.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 3′-UTR
3′-untranslated region
- ARE
AU-rich element
- BGH
bovine growth hormone
- DMEM
Dulbecco’s modified Eagle’s medium
- EGFP
enhanced green fluorescent protein
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- mRNA
messenger RNA
- PNA
peptide-linked nucleic acid
- siRNA
small interfering RNA
- TCGA
The Cancer Genome Atlas
- TTP
tristetraprolin.
References
- 1. Zhu Q., et al. (2012). The role of CXC chemokines and their receptors in the progression and treatment of tumors. J. Mol. Histol., 43, 699–713 [DOI] [PubMed] [Google Scholar]
- 2. Müller A., et al. (2001). Involvement of chemokine receptors in breast cancer metastasis. Nature, 410, 50–56 [DOI] [PubMed] [Google Scholar]
- 3. Domanska U.M., et al. (2013). A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur. J. Cancer, 49, 219–230 [DOI] [PubMed] [Google Scholar]
- 4. Zlotnik A., et al. (2000). Chemokines: a new classification system and their role in immunity. Immunity, 12, 121–127 [DOI] [PubMed] [Google Scholar]
- 5. Taichman R.S., et al. (2002). Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res., 62, 1832–1837 [PubMed] [Google Scholar]
- 6. Zeelenberg I.S., et al. (2003). The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res., 63, 3833–3839 [PubMed] [Google Scholar]
- 7. Burger M., et al. (2003). Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene, 22, 8093–8101 [DOI] [PubMed] [Google Scholar]
- 8. Allinen M., et al. (2004). Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell, 6, 17–32 [DOI] [PubMed] [Google Scholar]
- 9. Kato M., et al. (2003). Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res., 5, R144–R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scala S., et al. (2005). Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin. Cancer Res., 11, 1835–1841 [DOI] [PubMed] [Google Scholar]
- 11. Xu T.P., et al. (2013). The impact of chemokine receptor CXCR4 on breast cancer prognosis: a meta-analysis. Cancer Epidemiol., 37, 725–731 [DOI] [PubMed] [Google Scholar]
- 12. Hitti E., et al. (2012). Sequence variations affecting AU-rich element function and disease. Front. Biosci. (Landmark Ed)., 17, 1846–1860 [DOI] [PubMed] [Google Scholar]
- 13. Bakheet T., et al. (2006). ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res., 34(Database issue), D111–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khabar K.S. (2010). Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell. Mol. Life Sci., 67, 2937–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Denkert C., et al. (2004). Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin. Cancer Res., 10, 5580–5586 [DOI] [PubMed] [Google Scholar]
- 16. Dixon D.A., et al. (2001). Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest., 108, 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niesporek S., et al. (2008). Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression. Int. J. Oncol., 32, 341–347 [PubMed] [Google Scholar]
- 18. Al-Souhibani N., et al. (2010). The RNA-binding zinc-finger protein tristetraprolin regulates AU-rich mRNAs involved in breast cancer-related processes. Oncogene, 29, 4205–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young L.E., et al. (2009). The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology, 136, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brennan C.M., et al. (2009). The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res, 69, 5168–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lai W.S., et al. (2006). Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol., 26, 9196–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Al-Ahmadi W., et al. (2013). miR-29a inhibition normalizes HuR over-expression and aberrant AU-rich mRNA stability in invasive cancer. J. Pathol., 230, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Ahmadi W., et al. (2009). Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation. Nucleic Acids Res., 37, 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang Z., et al. (2005). Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res., 65, 967–971 [PMC free article] [PubMed] [Google Scholar]
- 25. Hitti E., et al. (2010). A versatile ribosomal protein promoter-based reporter system for selective assessment of RNA stability and post-transcriptional control. RNA, 16, 1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lai W.S., et al. (2002). Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem., 277, 9606–9613 [DOI] [PubMed] [Google Scholar]
- 27. Rossi D., et al. (2000). The biology of chemokines and their receptors. Annu. Rev. Immunol., 18, 217–242 [DOI] [PubMed] [Google Scholar]
- 28. Tachibana K., et al. (1998). The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature, 393, 591–594 [DOI] [PubMed] [Google Scholar]
- 29. Zou Y.R., et al. (1998). Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature, 393, 595–599 [DOI] [PubMed] [Google Scholar]
- 30. Kucia M., et al. (2005). Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells, 23, 879–894 [DOI] [PubMed] [Google Scholar]
- 31. Yagi H., et al. (2011). A synthetic biology approach reveals a CXCR4-G13-Rho signaling axis driving transendothelial migration of metastatic breast cancer cells. Sci. Signal., 4, ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burger J.A., et al. (2006). CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood, 107, 1761–1767 [DOI] [PubMed] [Google Scholar]
- 33. Muller A., et al. (2006). Chemokine receptors in head and neck cancer: association with metastatic spread and regulation during chemotherapy. Int. J. Cancer, 118, 2147–2157 [DOI] [PubMed] [Google Scholar]
- 34. Cabioglu N., et al. (2005). Chemokine receptor CXCR4 expression in breast cancer as a potential predictive marker of isolated tumor cells in bone marrow. Clin. Exp. Metastasis, 22, 39–46 [DOI] [PubMed] [Google Scholar]
- 35. Salvucci O., et al. (2006). The role of CXCR4 receptor expression in breast cancer: a large tissue microarray study. Breast Cancer Res. Treat., 97, 275–283 [DOI] [PubMed] [Google Scholar]
- 36. Koshiba T., et al. (2000). Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: a possible role for tumor progression. Clin. Cancer Res., 6, 3530–3535 [PubMed] [Google Scholar]
- 37. Halees A.S., et al. (2008). ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res., 36(Database issue), D137–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng S.S., et al. (1996). Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol., 16, 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bakheet T., et al. (2001). ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res., 29, 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu N., et al. (1997). Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol., 17, 4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tiedje C., et al. (2012). The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet., 8, e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qi M.Y., et al. (2012). AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol. Cell. Biol., 32, 913–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sohn B.H., et al. (2010). Functional switching of TGF-beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology, 138, 1898–1908 [DOI] [PubMed] [Google Scholar]
- 44. Griseri P., et al. (2011). A synonymous polymorphism of the Tristetraprolin (TTP) gene, an AU-rich mRNA-binding protein, affects translation efficiency and response to Herceptin treatment in breast cancer patients. Hum. Mol. Genet., 20, 4556–4568 [DOI] [PubMed] [Google Scholar]
- 45. Rounbehler R.J., et al. (2012). Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell, 150, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Upadhyay R., et al. (2013). Genetic polymorphisms in RNA binding proteins contribute to breast cancer survival. Int. J. Cancer, 132, E128–E138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Denkert C., et al. (2006). Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod. Pathol., 19, 1261–1269 [DOI] [PubMed] [Google Scholar]
- 48. Tran H., et al. (2003). Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell. Biol., 23, 7177–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yuan Z., et al. (2011). Knockdown of human antigen R reduces the growth and invasion of breast cancer cells in vitro and affects expression of cyclin D1 and MMP-9. Oncol. Rep., 26, 237–245 [DOI] [PubMed] [Google Scholar]
- 50. Hassan S., et al. (2011). CXCR4 peptide antagonist inhibits primary breast tumor growth, metastasis and enhances the efficacy of anti-VEGF treatment or docetaxel in a transgenic mouse model. Int. J. Cancer, 129, 225–232 [DOI] [PubMed] [Google Scholar]
- 51. Ramsey D.M., et al. (2013). Halting metastasis through CXCR4 inhibition. Bioorg. Med. Chem. Lett., 23, 20–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.