Summary
We show that only a third of CRC risk alleles identified in European-ancestry genome-wide association studies are also associated with risk in AAs. Some genetic risk factors in AAs are independent of those in whites.
Abstract
Genome-wide association studies (GWAS) in colorectal cancer (CRC) identified five regions near transforming growth factor β-related genes BMP4, GREM1, CDH1, SMAD7 and RPHN2. The true risk alleles remain to be identified in these regions, and their role in CRC risk in non-European populations has been understudied. Our previous work noted significant genetic heterogeneity between African Americans (AAs) and European Americans (EAs) for single nucleotide polymorphisms (SNPs) identified in GWAS. We hypothesized that associations may not have been replicated in AAs due to differential or independent genetic structures. In order to test this hypothesis, we genotyped 195 tagging SNPs across these five gene regions in 1194 CRC cases (795 AAs and 399 EAs) and 1352 controls (985 AAs and 367 EAs). Imputation was performed, and association testing of genotyped and imputed SNPs included ancestry, age and sex as covariates. In two of the five genes originally associated with CRC, we found evidence for association in AAs including rs1862748 in CDH1 (ORAdd = 0.82, P = 0.02) and in GREM1 the SNPs rs10318 (ORRec = 60.1, P = 0.01), rs11632715 (ORRec = 2.36; P = 0.004) and rs12902616 (ORRec = 1.28, P = 0.005), the latter which is in linkage disequilibrium with the previously identified SNP rs4779584. Testing more broadly for associations in these gene regions in AAs, we noted three statistically significant association peaks in GREM1 and RHPN2 that were not identified in EAs. We conclude that some CRC risk alleles are shared between EAs and AAs and others are population specific.
Introduction
Genome-wide association studies (GWAS) in colorectal cancer (CRC) have identified associations in over 40 genetic regions through analysis of common single nucleotide polymorphisms (SNPs) (1–10). Of particular interest are SNPs in five regions that contain genes involved in transforming growth factor β (TGFβ) signaling, including BMP4, GREM1, CDH1, SMAD7 and RHPN2 (11). The TGFβ pathway has been implicated previously in CRC pathogenesis because SMAD4, TGFβR2 and TGFβR1 are commonly mutated somatically during carcinogenesis (12), and germline mutations in SMAD4 and BMPR1A cause juvenile polyposis syndromes that predispose to CRC (13).
Although the genes of the TGFβ signaling pathway are good candidates for germline CRC susceptibility, GWAS only provide proxies for the true functional risk variants, which are yet to be identified, and they may be less than ideal for fine mapping. Several attempts have been made to localize the functional risk alleles more precisely by genotyping more SNPs in large European-ancestry sample sets for some of the regions that exhibited genetic associations. In the analysis of TGFβ-related genes, these efforts pinpointed 12 SNPs that exhibited maximal effects, constituting 8 independent risk associations. For example, the SMAD7 SNPs rs4939827 and rs12953717 are in linkage disequilibrium (LD) with rs58920878, which has allele-specific effects on transcription of SMAD7 (14). CDH1 SNPs rs4939827 and rs12953717 are in LD with rs7199991, which is genetically associated with four expression quantitative trait loci that correlate with expression of the upstream gene ZFP90. RHPN2 SNPs rs9929218 and rs1862748 are in LD with rs28626308, which results in a non-synonymous change in the RhoGTP-binding domain of the RHPN2 protein (15). In BMP4 (rs4444325 and rs1957636) and GREM1 (rs10318, rs16969681 and rs11632715), multiple, independent risk alleles have been reported (16). The evidence that any of these SNPs are true risk variants is inconclusive, leaving fundamental questions unanswered, namely, what are the true CRC risk variants, their effect sizes and their mechanisms of action?
Replication of risk-associated SNPs in non-European populations could provide better localization of risk alleles if the risk alleles are shared between different continental populations. In our initial work, we tested whether SNPs found to be associated in European-ancestry GWAS were good markers for CRC susceptibility in African American (AA) CRC (17,18). We replicated associations at three loci (SNPs in CDH1, GREM1 and in the 8q24 region), but there was significant heterogeneity between the European American (EA) and AA odds ratios (ORs), and overall the European-derived SNPs made poor markers for CRC susceptibility in AAs. We propose two possible explanations for this lack of replication. The first is that the SNPs found to be associated with CRC in EAs are shared between AAs and EAs; but because LD decreases more rapidly in AAs than in EAs, SNPs associated in EAs are not good markers for the true CRC risk variants in AAs. We call this explanation the differential genetic structure hypothesis. Alternatively, risk alleles are not shared between different continental populations, rather they are population specific. We call this explanation the independent genetic structure hypothesis. In the present report, we explore these hypotheses to better characterize the genetic structure of CRC risk.
Materials and methods
Human subjects and samples
Cases and controls were obtained from two institutions, University of Chicago Medicine (UCM), consisting of both AAs and EAs, and the University of North Carolina (UNC), consisting of AAs only. Characteristics of these subjects are shown in Supplementary Table 1, available at Carcinogenesis Online. In total, we included DNA from 1194 CRC cases (803 UCM and 391 UNC) and 1352 controls (935 UCM and 417 UNC).
For UCM cases and controls, two series of DNA samples were included: (i) a retrospective series obtained from formalin-fixed paraffin-embedded tissue and (ii) a prospective series obtained from blood samples as described previously (17,18). For both series, controls were cancer free at the time of inclusion. For the retrospective series, cases consisted of individuals with CRC who underwent surgical resection between 1994 and 2008 ascertained retrospectively from the Cancer Center and Pathology Department databases. Individuals known to have hereditary syndromes (familial adenomatous polyposis and Lynch syndrome) or inflammatory bowel disease were excluded. Hospital-based control samples were ascertained through our Pathology Department database and included cancer-free individuals who had thyroidectomies and amputations. Controls were matched to cases by age at diagnosis, 10-year birth cohort, gender and race as recorded in the database. Germline DNA for cases and controls was prepared from archived formalin-fixed surgical specimens from the paraffin block tissue repository. For each case, a block of normal colorectal, thyroid or soft tissue from the surgical margins was pulled. Five sections 10-µ thick were cut or plugs were punched from each block for DNA extraction.
For the prospective series, blood samples from additional UCM cases and controls were obtained. Cases were recruited in oncology clinics since 2006. The control subjects included individuals found to have a normal screening colonoscopy or cancer-free individuals obtained from the UC Translational Research Initiative in the Department of Medicine (TRIDOM). TRIDOM is an ongoing, large-scale, clinic-based sample collection and study to investigate the relationship of biomarkers with health status, disease status and disease progression. Subjects over the age of 18 were recruited and consented from various UCM outpatient clinics beginning in 2005. Consented individuals had 10 cc of peripheral blood drawn, and deidentified samples were banked. The age at time of sample collection was used as the age for each control.
Germline DNAs were prepared using the Gentra Puregene kit (Qiagen) according to the manufacturer’s instructions. For formalin-fixed, paraffin-embedded tissues, the paraffin was first removed with octane–methanol, and the proteinase K extraction step was extended to 3 days, adding fresh enzyme on each day, followed by heating the sample at 95°C for 15 min prior to protein precipitation.
Samples from UNC cases and controls were obtained through a large-scale, population-based case–control study of colon and rectal cancer, conducted in a 33-county area in central and eastern North Carolina. Cases were drawn at random from all CRC cases reported to the North Carolina Central Cancer Registry. Controls were randomly selected from North Carolina Division of Motor Vehicle records, based on sampling probabilities within blocks defined by 5-year age group, sex and race, using the technique of randomized recruitment (19). The details of this study have been published previously (20). Additional covariates including tobacco and alcohol use, non-steroidal anti-inflammatory use, red meat and fiber intake, total calories and body mass index were included in the analyses only for the UNC series as these data are not available for the UCM series.
The UCM and UNC studies were approved by their respective institutional review boards, and where appropriate, subjects provided written informed consent.
SNP selection and genotyping
For this study, we selected the five gene regions identified in the original European-ancestry CRC GWAS. The regions included the TGFβ-related genes BMP4, CDH1, GREM1, RHPN2 and SMAD7 (Table I). For each gene, the region from which SNPs were selected was defined from 5kb upstream of the start site of transcription to 5kb downstream of the termination site, and it also included all SNPs within the LD block that harbored the susceptibility allele as determined using Haploview (21) and the CEPH HapMap data. LD blocks are defined according to the default Haploview method that defines a block if 95% of the informative SNP pairs are in strong LD (22). Tagging SNPs for each region were selected using the method of Carlson et al. (23) as implemented in Haploview using an r 2 cut-off of 0.80 and minor allele frequency cut-off of 0.05. We repeated the tagging SNP analysis in the Yoruba population using genotype data from HapMap Phase 3 (http://hapmap.ncbi.nlm.nih.gov/). We selected a total of 214 tagging SNPs across the five genes.
Table I.
List of single nucleotide polymorphisms in TGFβ-related genes associated with CRC in European-ancestry populations
| Gene | Chr | SNP | BP position | OR | Risk allele | Risk allele frequency |
|---|---|---|---|---|---|---|
| BMP4 | 14q22.2 | rs4444235 | 54410919 | 1.12 | C | 0.46 |
| rs1957636 | 54560018 | 1.08 | A | 0.40 | ||
| GREM1 | 15q13.3 | rs10318 | 33025979 | 1.18 | T | 0.19 |
| rs4779584 | 32994756 | 1.19 | T | 0.18 | ||
| rs16969681 | 32993111 | 1.18 | T | 0.09 | ||
| rs11632715 | 33004247 | 1.12 | A | 0.47 | ||
| CDH1 | 16q22.1 | rs9929218 | 68820946 | 0.88 | A | 0.29 |
| rs1862748 | 68832943 | 0.88 | T | 0.31 | ||
| SMAD7 | 18q21.1 | rs4939827 | 46453463 | 0.85 | T | 0.49 |
| rs12953717 | 46453929 | 1.19 | T | 0.31 | ||
| RHPN2 | 19q13.1 | rs10411210 | 33532300 | 0.79 | T | 0.10 |
| rs7259371 | 33534641 | 0.86 | A | 0.18 |
ORs and allele frequencies are from Houlston et al. (1), Tomlinson et al. (7) and Tomlinson et al. (16). BP, base pair from genome build 104.0; Chr, chromosome position.
SNP genotyping assays were developed using Genotyper 2.0, and 195 assays were devised based on nine multiplex PCRs. We genotyped these 195 SNPs using the Sequenom MassARRAY platform as described previously (17,18). Quality control procedures to ensure high genotype quality were performed in several steps. First, we evaluated individual and SNP missingness for each plex. If an individual’s missing rate was greater than 20%, which would suggest poor genotype quality for this subject in this particular plex, we set all of his or her genotypes to missing in this plex. Individuals with total missingness greater than 30%, which would occur for example if missingness was greater than 20% in two or more plexes, were excluded from further analyses. We used this relatively permissive genotype missingness rate to accommodate the high level of missingness found in two plexes in which genotype quality was less optimal. SNPs with minor allele frequency ≤ 0.025, missing ≥ 15% of their genotypes, or which had Hardy–Weinberg equilibrium P values ≤ 0.003 were excluded from additional analyses. After performing QC measures, there were a total of 361 EA cases and 347 EA controls genotyped on 125 markers, with an average missingness per individual of 4.9%. For AAs, 622 cases and 819 controls remained with genotype data on 153 markers with an average missingness per individual of 4.3%.
Genetic ancestry estimation
In order to control for confounding based on ancestry differences, West African ancestry was estimated in all cases and controls using 100 ancestry informative markers, which have been previously genotyped on the Sequenom platform (17). Global individual ancestry (% West African and % European) was calculated from the genotype data using the Bayesian Markov Chain Monte Carlo method implemented in the program STRUCTURE 2.1 (24). STRUCTURE 2.1 assumes an admixture model using prior population information and independent allele frequencies. The Markov Chain Monte Carlo model was run using K = 3 populations (58 Europeans, 67 Native Americans and 62 West Africans) and a burn-in length of 30 000 iterations followed by 70 000 replications. West African ancestry estimates were used as covariates in the regression models for analyses with the AA sample set.
Genotype imputation
In order to improve our ability to resolve the causative risk allele and to more accurately evaluate association for ungenotyped SNPs, we performed imputation on each gene for each ancestral group separately. Imputation was performed using IMPUTE2 version 2.3.0 using the integrated phase 1 version 3 of the 1000 Genomes reference dataset (25). We imputed genotypes over a 5-megabase region with the gene of interest being centered in the region. Association analyses were restricted to SNPs within a region containing the gene plus 20kb upstream of the transcription start site and 20kb downstream of the transcription stop site (cf. Supplementary Table 2, available at Carcinogenesis Online). In addition, we included only those SNPs with IMPUTE2 info scores ≥ 0.50 and minor allele frequencies ≥ 0.01.
Statistical analysis
Gene-based association testing.
We tested for association between SNPs (both genotyped and imputed) and CRC in the combined UCM and UNC AA series and separately in the UCM EAs. We used SNPTEST v2.4.1 (26) to perform logistic regression on the imputed and genotyped data using additive, dominant and recessive models and fit age and sex as covariates for both EA and AA analyses. Proportion of West African ancestry was used as a covariate for the AA analysis only. Independence of SNP effects was assessed by examining the amount of LD between SNPs (r 2) and by testing for association of each SNP conditional on other significant SNPs within the same gene using SNPTEST’s–condition_on argument.
Replication analysis.
One of the main aims of this study was to determine whether SNPs consistently identified as CRC-associated variants in EA populations also play a role in CRC in AAs. Because it is plausible that the SNP identified in EAs is serving as a surrogate for the true underlying risk variant, we tested both the reference SNPs associated with CRC in EAs and those SNPs with r 2 values greater than 0.80 in the EAs. Many of the SNPs we are attempting to replicate may represent the same signal within the gene they reside. For example, in European-ancestry populations, rs4779584 and rs10318 in GREM1 have an r 2 = 0.49, rs9929218 and rs186278 in CDH1 have an r 2 = 0.83, rs4939827 and rs12953717 in SMAD7 have an r 2 = 0.59 and rs10411210 and rs7259371 in RHPN2 have an r 2 = 0.45. To account for testing not only the originally reported SNP but all SNPs in r 2 ≥ 0.80 with it, we calculated an empirical critical value to determine whether any of the tested SNPs supported evidence for association, and thus replication. This was done by permuting case–control status and repeating the same set of tests 2000 times. The distribution of the minimum P value was then determined, and the 5th percentile of this distribution was set as the critical value.
Gene-wide multiple testing.
We assessed statistical significance for SNP tests within genes after accounting for multiple testing in two ways. For each gene-ancestry combination, we calculated a gene-based P value for each SNP using pACT (27) that accounted for the correlation between the SNPs within each gene and the correlation between the three genetic models tested. Second, we calculated q values using the method of Benjamini and Hochberg (28) for the tests performed in each gene-ancestry group, and again for all genes combined within an ancestry group.
Testing tumor site and lifestyle factors.
We tested for SNP effect size heterogeneity in colon cancer versus rectal cancer by treating colon cancer samples as cases and rectal cancer samples as controls and performing logistic regression as described in Gene-based association testing section. We investigated whether lifestyle factors influenced the risk of CRC in AAs, and if so, whether the CRC–SNP genotype associations could be explained by these differences or whether stronger evidence for CRC–SNP associations could be found by conditioning on important lifestyle factors. The variables we explored included food energy, dietary fiber, alcohol consumption, body mass index, non-steroidal anti-inflammatory drug use, smoking, red meat consumption and amount of cigarettes smoked per day. We plotted the distribution of these variables in cases and controls and calculated the Wilcoxon Rank Sum test P value. The Wilcoxon Rank Sum test was used because the distribution of these traits was not normal. Smoking status differences in case and controls were tested using a chi-squared test of homogeneity. We regressed each trait on case–control status and on the other traits. To test the effects of the lifestyle factor on SNP associations with CRC, we compared the analytic results from logistic regression models including sex, age and proportion African ancestry as covariates with and without the lifestyle factor as a covariate.
Results
Testing the differential genetic structure hypothesis
If the differential genetic structure hypothesis was correct, then we should be able to identify SNPs that are associated with CRC in both EAs and AAs. We would expect these SNPs to be in LD with the originally identified SNP in EAs but not in LD with the original SNP in AAs. Accordingly, we looked for SNPs with these characteristics in five TGFβ-related genes BMP4, GREM1, CDH1, SMAD7 and RHPN2 that have shown consistent association with CRC in studies of European-ancestry populations (see Table I). To do this, we first identified SNPs in high LD (r 2 ≥ 0.8) in EAs with the original variants reported from GWAS. In the five genes, we identified between 1 and 58 SNPs (107 total) that were in LD with the original GWAS variant, and these SNPs were tested as candidates for the true risk-associated allele. We calculated empirical critical values to maintain a type I error rate of 0.05 for each set of SNPs that tag the European-ancestry CRC-associated SNPs (see Materials and Methods).
Three of the SNPs originally identified in European-ancestry GWAS showed evidence for association in the AA sample using a critical value that only corrected for the three genetic models tested (Table II). Similar to the results we reported earlier (17), rs1862748 in CDH1 (ORAdd = 0.82, P = 0.02) and rs10318 in GREM1 (ORRec = 60.1, P = 0.01; ORAdd = 1.47, P = 0.02) were significantly associated with CRC in AAs. The SNP rs11632715 in GREM1 (ORRec = 2.36, P = 0.004), which was reported later as an association independent of other GREM1 SNPs (16), was also significantly associated with AA CRC. The SNP rs76211684, which is in LD with rs11632715, was found to be slightly more significantly associated (ORRec = 2.63; P = 0.002), but it has a similar allele frequency and OR to rs11632715. The GREM1 SNP rs12902616 (ORRec = 1.28, P = 0.005), which is in LD with the SNP rs4779584 previously identified in European-ancestry GWAS, was significant after correcting for testing three models performed on it and the other 17 SNPs in GREM1 that had r 2s ≥ 0.80 with rs4779584 and with rs10318. No other SNPs in LD with the original SNPs showed statistically significant evidence of association (all the results from this statistical analysis are shown in Supplementary Table 3, available at Carcinogenesis Online). Thus, only 2 of the 5 genes tested that are consistently found to be associated with CRC in European-ancestry populations are also associated with risk in AAs.
Table II.
P values and odds ratios of SNPs associated with CRC in European-ancestry populations or of SNPs in LD with those variantsa
| Replicationb | GWASc | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | EA | Freq | OR | |||||||||||||
| Gene | Reference SNP (rs)d | SNP (rs) in LD | Position | All | Case Freq | Cont Freq | Best OR | Min P | Best Mod | Case Freq | Cont Freq | Best OR | Min P | Best Mod | ||
| BMP4 | 4444235 | 54410919 | C | 0.31 | 0.34 | 0.87 | 0.892 | Dom | 0.43 | 0.43 | 1.10 | 0.287 | Dom | 0.46 | 1.11 | |
| 1957636 | 54560018 | A | 0.74 | 0.75 | 1.00 | 0.478 | Rec | — | — | — | — | — | 0.40 | 1.08 | ||
| 1957606 | 54526105 | A | 0.75 | 0.75 | 1.00 | 0.422 | Rec | — | — | — | — | — | ||||
| GREM1 | 10318 | 33025979 | A | 0.05 | 0.03 | 60.05 | 0.011* | Rec | 0.13 | 0.14 | 1.34 | 0.291 | Rec | 0.18 | 1.18 | |
| 4779584 | 32994756 | C | 0.69 | 0.68 | 1.12 | 0.040 | Rec | 0.48 | 0.51 | 0.86 | 0.855 | Rec | 0.18 | 1.19 | ||
| 12902616 | 32997175 | G | 0.82 | 0.78 | 1.28 | 0.005* | Rec | 0.59 | 0.61 | 0.82 | 0.791 | Rec | ||||
| 11632715 | 33004247 | A | 0.30 | 0.29 | 2.36 | 0.004* | Rec | 0.32 | 0.33 | 0.99 | 0.506 | Rec | 0.47 | 1.12 | ||
| 76211684 | 33002938 | T | 0.31 | 0.29 | 2.63 | 0.002* | Rec | 0.32 | 0.33 | 1.00 | 0.505 | Rec | ||||
| CDH1 | 9929218 | 68820946 | A | 0.32 | 0.30 | 0.97 | 0.429 | Rec | 0.29 | 0.33 | 0.68 | 0.065 | Rec | 0.29 | 0.88 | |
| 8044058 | 68807088 | A | 0.23 | 0.23 | 1.34 | 0.138 | Rec | 0.24 | 0.29 | 0.54 | 0.015 | Rec | ||||
| 1862748 | 68832943 | T | 0.17 | 0.20 | 0.82 | 0.023* | Add | 0.82 | 0.29 | 0.81 | 0.036 | Add | 0.31 | 0.88 | ||
| 4783685 | 68834107 | T | 0.08 | 0.10 | 0.72 | 0.018* | Dom | 0.26 | 0.32 | 0.74 | 0.005 | Add | ||||
| SMAD7 | 4939827 | 46453463 | C | 0.36 | 0.35 | 1.09 | 0.561 | Dom | 0.50 | 0.52 | 0.80 | 0.090 | Add | 0.49 | 0.85 | |
| 12953717 | 46453929 | T | 0.33 | 0.31 | 1.26 | 0.103 | Rec | 0.42 | 0.44 | 0.97 | 0.562 | Rec | 0.42 | 1.19 | ||
| RHPN2 | 10411210 | 33532300 | T | 0.42 | 0.42 | 0.86 | 0.153 | Dom | 0.09 | 0.13 | 0.74 | 0.036 | Add | 0.10 | 0.79 | |
| 12459751 | 33538783 | G | 0.79 | 0.78 | 0.86 | 0.102 | Dom | 0.93 | 0.89 | 0.66 | 0.021 | Add | ||||
| 7259371 | 33534641 | A | 0.69 | 0.68 | 1.03 | 0.596 | Rec | 0.21 | 0.24 | 0.88 | 0.208 | Rec | 0.18 | 0.86 | ||
| 2042190 | 33532716 | C | 0.73 | 0.74 | 1.00 | 0.512 | Add | 0.19 | 0.23 | 0.55 | 0.054 | Rec | 0.18 | 0.86 | ||
aSNP analytic data were included in this table if (1) the SNP was a reference SNP identified in European-ancestry GWAS for colorectal cancer or (2) the SNP was in linkage disequilibrium (r 2 > 0.8) with a reference SNP and obtained a smaller P value in the analysis. Add, additive genetic model; All, allele; Cont, control; Dom, dominant genetic model; freq, frequency; Rec, recessive genetic model.
bReference SNPs are listed in Table I. The GREM1 SNP rs16969681 was not included in Table II because the imputation quality score was < 0.5.Three genetic models were tested. The best model (mod) and best OR are the ones associated with the smallest P value (min p).
cThese data are from the reported GWAS studies in individuals of European descent (see Table I).
dReference SNPs are listed in Table I. The GREM1 SNP rs16969681 was not included in the Table because the imputation quality score was < 0.5.
*Statistically significant P value (in bold) at the 0.05 level after controlling for multiple testing.
Gene-based analysis
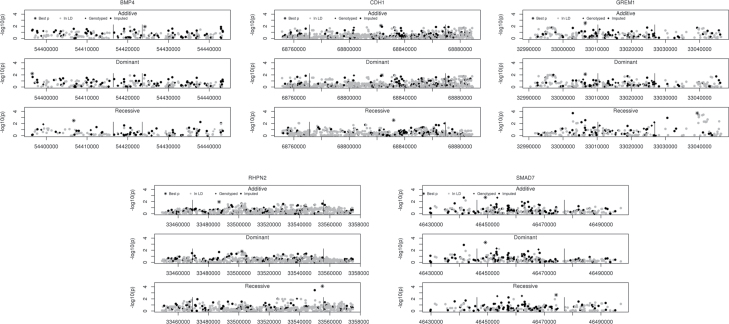
Given the prior evidence for the five genes investigated here being involved in CRC in EAs and given their role in TGFβ signaling, it is reasonable to hypothesize that some previously unidentified variant(s) within these genes may be involved in CRC in AAs. We pursued this question by testing both directly genotyped and imputed SNPs in the five candidate genes discussed above. In total, we tested 2105 SNPs. Figure 1 shows association results for the five gene regions in AAs.
Fig. 1.
Mini Manhattan plots for each of the genes analyzed in African Americans with colorectal cancer. For each gene, three genetic models were tested. Along the x-axis is the base pair position in the genome and along the y-axis is the −log10 of the P value. The darkly shaded symbols represent SNPs genotyped in the study and the lightly shaded symbols represent SNPs whose genotypes were imputed. The asterisks represent the SNP with the smallest P value.
In two genes, GREM1 and RHPN2, we noted several association peaks (P < 10− 3) (Table III). In GREM1, there were two association peaks of imputed SNPs noted under a recessive model. One peak was located 5′ to GREM1 centered on rs148375239 [ORRec (SE) = 17.33 (24.14), P = 1.9 × 10− 4, P corrected = 0.024; ORAdd (SE) = 1.40 (0.20), P = 0.02]; rs148375239 was gene-wide significant after correcting for the number of SNPs, their LD and the three models tested. The second peak was located 3′ to GREM1 and included four SNPs that were gene-wide significant (rs17816285, rs8031380, rs7496578 and rs4337272). SNP rs17816285 showed the strongest association [ORRec (SE) = 3.13 (1.03), P = 1.9 × 10− 4, P corrected = 0.024]. The other three SNPs in the peak had r 2 ≥ 0.94 with rs17816285, whereas the SNP in the other peak, rs148375239, was not in LD with this SNP (r 2 = 0.01). Conditional tests revealed that the signals from these two SNPs are independent of one another, as well as being independent of the previously identified GREM1 risk-associated variants (Supplementary Table 4, available at Carcinogenesis Online).
Table III.
Novel SNP associations identified in AA colorectal cases and controls
| Gene | SNP | Position | MAF cases | MAF controls | P value Add | P value Dom | P value Rec | P value corrected | OR Add | OR Dom | OR Rec |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GREM1 | rs17816285 | 33039298 | 0.29 | 0.26 | 0.018 | 0.185 | 0.0002 | 0.024 | 1.30 | 1.20 | 3.13 |
| rs148375239 | 33002864 | 0.17 | 0.14 | 0.019 | 0.084 | 0.0002 | 0.024 | 1.40 | 1.33 | 17.33 | |
| rs8031380 | 33041130 | 0.28 | 0.25 | 0.021 | 0.172 | 0.0003 | 0.042 | 1.29 | 1.21 | 3.18 | |
| rs7496578 | 33039620 | 0.28 | 0.25 | 0.027 | 0.212 | 0.0004 | 0.044 | 1.28 | 1.19 | 3.12 | |
| rs4337272 | 33039821 | 0.376 | 0.348 | 0.024 | 0.187 | 0.000381 | 0.045 | 1.29 | 1.20 | 3.19 | |
| RHPN2 | rs113984415 | 33555034 | 0.16 | 0.19 | 0.022 | 0.163 | 0.00008 | 0.045 | 0.76 | 0.83 | 0.13 |
Add, additive genetic model; Dom, dominant genetic model; MAF, minor allele frequency; Rec, recessive genetic model.
In RHPN2, one imputed SNP, rs113984415, localized to the 5′ region, was gene-wide significant in AAs under a recessive model [ORRec (SE) = 0.13 (0.09), P corrected = 8 × 10− 5] and is independent of the previously reported rs10411210 and rs12459751 SNPs (r 2 < 0.20, Supplementary Table 4, available at Carcinogenesis Online). The RHPN2 SNP rs113984415 had a substantially smaller minor allele frequency in EAs than in AAs, 1.7% versus 17.4%, respectively; consequently, the effect of this SNP is population restricted. The GREM1 SNPs were all imputed successfully in the EA samples; however, none conferred risk for CRC in the EAs. For example, the allele frequencies of rs17816285, the most significantly associated GREM1 SNP in AAs, were 0.29 in AA cases and 0.25 in AA controls, but they were 0.25 in EA cases and 0.24 in EA controls. The allele frequencies of rs148375239—the other independently associated GREM1 SNP in AAs—were 0.167 in AA cases and 0.143 in AA controls, but they were 0.169 in both EA cases and controls. If these findings are confirmed, it would suggest that at least a subset of CRC risk alleles is specific to the AA population.
Analysis of tumor site and diet/lifestyle factors
We investigated whether any of the previously associated or newly identified variants showed significant differences in OR based on tumor site. Only a single SNP demonstrated nominal significance: rs113984415 was significantly associated in colon cancer (OR = 0.04, P = 1 × 10− 6) but showed no evidence of being associated with rectal cancer (OR = 0.64, P = 0.49) (Supplementary Table 5, available at Carcinogenesis Online).
The means of three correlated lifestyle factors—energy intake, fiber intake and meat consumption—were significantly different between cases and controls when using test-wise type I error rate of 0.05 (P energy = 0.03, P fiber = 0.01, P meat = 2 × 10− 5) (Supplementary Figure 1, available at Carcinogenesis Online). When all three were regressed on case–control status, only meat consumption remained significant. Thus, we compared SNP–CRC association test results with and without red meat consumption as a covariate (Supplementary Table 6, available at Carcinogenesis Online). The ORs and P values for the GREM1 SNP rs10318 were much smaller when meat consumption was included in the model. The full results of this analysis can be found in the Supplementary Material, available at Carcinogenesis Online.
Discussion
GWAS and subsequent fine-mapping studies in European-ancestry populations have identified genetic associations in candidate genes involved in TGFβ signaling, including BMP4, GREM1, CDH1, SMAD7 and RHPN2 (1,3,5,15,16). In our earlier work, we directly genotyped a sample of AA CRC cases and controls and tested the previously identified SNPs in these five gene regions but were only able to replicate the SNPs rs10318 in GREM1 and rs1862748 in CDH1 (18). In order to better characterize the genetic structure of CRC risk, we undertook the current study to test whether our failure to replicate the other variants was due to differential LD structure causing EA CRC-associated SNPs to no longer serve as good proxies for the true risk variant. In addition, we tested whether independent risk variants existed in these regions in AAs.
In our test of the differential genetic structure hypothesis, we were able to confirm our previous replication of the GREM1 SNP rs10318 and the CDH1 SNP rs1862748. The association signal at rs1862748 was stronger in the present analysis because we had additional genotype information through imputation of missing genotypes and because we considered recessive and dominant genetic models in addition to the log additive model, which was the only model we tested in our first replication attempt. We were also able to replicate the second GREM1 SNP, rs4779584, by identifying another SNP in LD with it, rs12902616, (r 2 EA = 0.82, r 2 AA = 0.72), that showed significant association using a dominant model (P = 0.005). Also, we were able to replicate the GREM1 SNP rs11632715, which in Europeans is reported to be an association independent of rs10318 and rs4779584. We did not replicate any SNPs in BMP4, SMAD7 or RHPN2. These results agree with a recently reported study in AA CRC (29), with the exception of an AA-associated SNP in SMAD7, which we did not identify as associated in the present sample set.
In addition to determining whether or not CRC-associated risk variants identified in European-ancestry populations are equally relevant for AA populations, studying AAs could also allow us to better localize the causative risk variant. We used ENCODE annotation served by HaploReg (30) to investigate if the functional evidence for the variant identified in the AAs is more compelling than the original EA reported variant. Both the original European-ancestry GREM1 variant rs4779584 and its most significant proxy in AAs, rs12902616, are located within intriguing transcription factor (TF) binding sites (IRF and PBX1, respectively); however, they do not appear to alter the binding affinity. The CDH1 SNP, rs1862748, is located in enhancer sequences associated with histone marks, but the histone marks are not found in colon cancer cells. Interestingly, the most significant AA variant in LD with rs1862748, rs4783685, is in a binding site for the YingYang1 (YY1) TF, which has been reported to be overexpressed in colon cancer (31).
In our test of the independent genetic structure hypothesis, we identified three previously unreported SNPs in GREM1 and RHPN2 that appear to be specific to AA CRC risk. Neither of these GREM1 SNPs is in LD with any of the previously identified European-ancestry CRC SNPs in GREM1 (maximum r 2 = 0.16 in AAs), nor was the RHPN2 SNP in LD with the previously identified European-ancestry risk variant in RHPN2 (r 2 = 0). Although none of the GREM1 SNPs were associated with histone marks, they did alter the binding motifs of several TFs. For example, rs8031380 alters ERG-1 (Early Growth Response-1) binding motif. The ERG-1 gene is a tumor suppressor that can be induced by tolfenamic acid, an anticancer non-steroidal anti-inflammatory drug that promotes apoptosis in colon cancer cells (32). The GREM1 SNP, rs7496578, alters NRF-2 and TCF11 binding motifs, both of which play a role in oxidative stress response. Kelch-like ECH-associated protein 1 (Keap1) in the NRF-2 pathway is differentially expressed in response to oxidative stress in normal and colon cancer cells (33). The GREM1 SNP rs17816285 is in LD (r 2 AA = 0.93) with rs10519740, which localizes to a histone-marked enhancer sequence in human skeletal muscle myoblasts and is associated with the differential expression of six genes (CCDC43, LOC400713, CA5B, CA5BL, C8orf70 and TPRKB) in HapMap Yoruba expression quantitative trait locus studies but not in HapMap European expression quantitative trait locus studies (34). The RHPN2 AA risk variant rs113984415 is contained in sequences that define TF binding motifs and promoter-associated histone modifications in 9 ENCODE cell types. Additionally, the novel spliceosomal factor, ZNF263, which is able to induce alternative splicing (35), binds at this TF binding motif. rs113984415 is nearly West African specific (minor allele frequency = 0.34 in HapMap Yoruba and 0.017 in our Chicago EA controls). These exciting results underline the mounting evidence that AAs possess a set of AA-specific risk alleles in human disease, e.g. in Alzheimer’s disease (36), obesity (37) and warfarin dosing (38). In some instances, the AA-specific alleles reside in previously identified genes.
Although we were successful in replicating CRC risk-associated SNPs in two of five genes identified in European-ancestry populations, an important question is why the remaining risk-associated SNPs failed to replicate. Possible explanations include (i) the unreplicated variants do not confer risk in AAs because increased risk depends on interaction with ethnicity-restricted environmental or genetic factors; (ii) all the variants tested do indeed confer risk in AAs, but we were insufficiently powered to identify association with all of them; (iii) the associations in European-ancestry populations represent synthetic associations, in which one SNP is associated with multiple, much less common risk alleles, as hypothesized by Goldstein and colleagues (39) and finally, (iv) the SNPs in the three unreplicated genes represent false positives in the European-ancestry populations. It is unlikely that the SNPs in the three unreplicated genes are false positives given the sample sizes (>25 000 subjects) on which the associations are based (1,2,9,16). It has been argued that synthetic associations are more the exception than the rule (40,41) although the extent to which this hypothesis has been tested in European-ancestry CRC associations is unclear. With regard to power, using the OR values for the 12 SNPs in Table I, we calculated that the likelihood that we would replicate four or fewer of the 12 EA variants is 0.48; however, the unreplicated SNPs had higher power than average and the probability that none of these variants were among the four replicated SNPs is <0.01. Finally, although the possibility that gene-by-gene or gene-by-environment interaction could explain the lack of replication, testing this hypothesis will require a larger and more comprehensive dataset with respect to number of samples, the proportion of the genome interrogated and potentially relevant non-genetic factors. This last hypothesis is extremely intriguing and has many public health and clinical implications.
In summary, we have found three novel CRC risk-associated variants in two known susceptibility genes that appear to be specific, or nearly so, to AAs, and we have found that four variants in two genes previously identified in European-ancestry populations serve as markers for CRC risk in AAs as well. Additional studies will need to be conducted to confirm our novel results, but if successful they will join an increasing number of population-specific variants, providing further credence to the need for personalized and population-based medicine.
Supplementary Material
Supplementary Tables 1–6 and Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute of the National Institutes of Health under award numbers (R01CA140804 to N.A.E., U01CA153060 to N.A.E., K08CA142892 to S.S.K., CA066635 to R.S.S., and P30DK034987 to R.S.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
Acknowledgements
The authors are grateful to W.Buikema and staff of the University of Chicago DNA Sequencing Facility for assistance with the genotyping.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AA
African American
- Add
additive
- CRC
colorectal cancer
- CEPH
Centre d’etude du polymorphism humain
- Dom
, dominant
- EA
European American
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- OR
odds ratio
- Rec
recessive
- SNP
single nucleotide polymorphism
- TF
transcription factor
- UCM
University of Chicago Medicine
- UNC
University of North Carolina.
References
- 1. Houlston R.S., et al. ; Colorectal Cancer Association Study Consortium; CoRGI Consortium; International Colorectal Cancer Genetic Association Consortium. (2008). Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet., 40, 1426–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Houlston R.S., et al. ; COGENT Consortium; CORGI Consortium; COIN Collaborative Group; COINB Collaborative Group. (2010). Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet., 42, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broderick P., et al. ; CORGI Consortium. (2007). A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet., 39, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 4. Gruber S.B., et al. (2007). Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol. Ther., 6, 1143–1147 [DOI] [PubMed] [Google Scholar]
- 5. Tenesa A., et al. (2008). Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet., 40, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomlinson I., et al. CORGI Consortium. (2007). A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet., 39, 984–988 [DOI] [PubMed] [Google Scholar]
- 7. Tomlinson I.P., et al. ; CORGI Consortium; EPICOLON Consortium. (2008). A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet., 40, 623–630 [DOI] [PubMed] [Google Scholar]
- 8. Zanke B.W., et al. (2007). Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet., 39, 989–994 [DOI] [PubMed] [Google Scholar]
- 9. Peters U., et al. ; Colon Cancer Family Registry and the Genetics and Epidemiology of Colorectal Cancer Consortium. (2013). Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology, 144, 799–807.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunlop M.G., et al. Colorectal Tumour Gene Identification (CORGI) Consortium; Swedish Low-Risk Colorectal Cancer Study Group; COIN Collaborative Group. (2012). Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet., 44, 770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tenesa A., et al. (2009). New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat. Rev. Genet., 10, 353–358 [DOI] [PubMed] [Google Scholar]
- 12. Lampropoulos P., et al. (2012). TGF-beta signalling in colon carcinogenesis. Cancer Lett., 314, 1–7 [DOI] [PubMed] [Google Scholar]
- 13. Howe J.R., et al. (1998). Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science, 280, 1086–1088 [DOI] [PubMed] [Google Scholar]
- 14. Pittman A.M., et al. (2009). The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res., 19, 987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carvajal-Carmona L.G., et al. (2011). Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum. Mol. Genet., 20, 2879–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tomlinson I.P., et al. COGENT Consortium; CORGI Collaborators; EPICOLON Consortium. (2011). Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet., 7, e1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kupfer S.S., et al. (2009). Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis, 30, 1353–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kupfer S.S., et al. (2010). Genetic heterogeneity in colorectal cancer associations between African and European americans. Gastroenterology, 139, 1677–1685, 1685.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weinberg C.R., et al. (1991). Randomized recruitment in case-control studies. Am. J. Epidemiol., 134, 421–432 [DOI] [PubMed] [Google Scholar]
- 20. Sansbury L.B., et al. (2006). COX-2 polymorphism, use of nonsteroidal anti-inflammatory drugs, and risk of colon cancer in African Americans (United States). Cancer Causes Control, 17, 257–266 [DOI] [PubMed] [Google Scholar]
- 21. Barrett J.C., et al. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265 [DOI] [PubMed] [Google Scholar]
- 22. Gabriel S.B., et al. (2002). The structure of haplotype blocks in the human genome. Science, 296, 2225–2229 [DOI] [PubMed] [Google Scholar]
- 23. Carlson C.S., et al. (2004). Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet., 74, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Falush D., et al. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abecasis G.R., et al. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marchini J., et al. (2007). A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913 [DOI] [PubMed] [Google Scholar]
- 27. Conneely K.N., et al. (2007). So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am. J. Hum. Genet., 81, 1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benjamini Y., et al. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B, 57, 289–300 [Google Scholar]
- 29. Wang H., et al. (2013). Fine-mapping of genome-wide association study-identified risk loci for colorectal cancer in African Americans. Hum. Mol. Genet., 22, 5048–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ward L.D., et al. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40(Database issue), D930–D934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chinnappan D., et al. (2009). Transcription factor YY1 expression in human gastrointestinal cancer cells. Int. J. Oncol., 34, 1417–1423 [PubMed] [Google Scholar]
- 32. Lee S.H., et al. (2008). ESE-1/EGR-1 pathway plays a role in tolfenamic acid-induced apoptosis in colorectal cancer cells. Mol. Cancer Ther., 7, 3739–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang L.C., et al. (2013). Immunohistochemical study of the Nrf2 pathway in colorectal cancer: Nrf2 expression is closely correlated to Keap1 in the tumor and Bach1 in the normal tissue. Appl. Immunohistochem. Mol. Morphol., 21, 511–517 [DOI] [PubMed] [Google Scholar]
- 34. Gamazon E.R., et al. (2010). SCAN: SNP and copy number annotation. Bioinformatics, 26, 259–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams D.J., et al. (2001). ZNF265–a novel spliceosomal protein able to induce alternative splicing. J. Cell Biol., 154, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reitz C., et al. Alzheimer Disease Genetics Consortium. (2013). Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ϵ4,and the risk of late-onset Alzheimer disease in African Americans. JAMA, 309, 1483–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen G., et al. (2012). Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans. Hum. Mol. Genet., 21, 4530–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perera M.A., et al. (2013). Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet, 382, 790–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dickson S.P., et al. (2010). Rare variants create synthetic genome-wide associations. PLoS Biol., 8, e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anderson C.A., et al. (2011). Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol., 9, e1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wray N.R., et al. (2011). Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biol., 9, e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.