Summary
We have identified relevant genes behind the GWAS SNPs for colorectal cancer at three loci using eQTLs analysis. A detailed analysis of the regions revealed new candidate functional SNPs for two loci, based on larger correlation with gene expression.
Abstract
In this study, we aim to identify the genes responsible for colorectal cancer risk behind the loci identified in genome-wide association studies (GWAS). These genes may be candidate targets for developing new strategies for prevention or therapy. We analyzed the association of genotypes for 26 GWAS single nucleotide polymorphisms (SNPs) with the expression of genes within a 2 Mb region (cis-eQTLs). Affymetrix Human Genome U219 expression arrays were used to assess gene expression in two series of samples, one of healthy colonic mucosa (n = 47) and other of normal mucosa adjacent to colon cancer (n = 97, total 144). Paired tumor tissues (n = 97) were also analyzed but did not provide additional findings. Partial Pearson correlation (r), adjusted for sample type, was used for the analysis. We have found Bonferroni-significant cis-eQTLs in three loci: rs3802842 in 11q23.1 associated to C11orf53, COLCA1 (C11orf92) and COLCA2 (C11orf93; r = 0.60); rs7136702 in 12q13.12 associated to DIP2B (r = 0.63) and rs5934683 in Xp22.3 associated to SHROOM2 and GPR143 (r = 0.47). For loci in chromosomes 11 and 12, we have found other SNPs in linkage disequilibrium that are more strongly associated with the expression of the identified genes and are better functional candidates: rs7130173 for 11q23.1 (r = 0.66) and rs61927768 for 12q13.12 (r = 0.86). These SNPs are located in DNA regions that may harbor enhancers or transcription factor binding sites. The analysis of trans-eQTLs has identified additional genes in these loci that may have common regulatory mechanisms as shown by the analysis of protein–protein interaction networks.
Introduction
Genome-wide association studies (GWAS) have been successful in identifying susceptibility loci for cancer and other diseases, but no progress has been made regarding the functional mechanisms underlying the associations. In colorectal cancer (CRC), 26 susceptibility single nucleotide polymorphisms (SNPs) in 23 different loci have been identified in GWAS to date (Supplementary Table 1, available at Carcinogenesis Online) (1–12). Most of them are located in intergenic positions and the genes responsible for the risk modification are unknown. The identification of these genes is important because they may be considered targets for developing new strategies for prevention or therapy (13).
The combination between high throughput genotyping and gene expression profiling technologies allows studying genome-wide associations between genetic polymorphisms and gene expression levels, known as expression quantitative trait loci (eQTL). The identification of eQTL has been proposed as a method to find genes underlying the associations with disease risk (14). The eQTL analysis also has been proposed as a tool to improve the power of GWAS (15) or to engineer genetic-gene expression networks and discover new mechanisms or pathways related to disease (16).
Most analyses of eQTL have used lymphoblastoid cell lines (14), which may not be optimal when the interest is in explaining risk in specific target tissues. Global eQTL analyses of diverse tissues have been done in liver (17), kidney (18) and brain (19), among others. The Genotype-Tissue Expression (GTEx) project (20) aims to create a comprehensive public atlas of gene expression and regulation across multiple human tissues (http://www.broadinstitute.org/gtex). Regarding colon cancer, the interest of analyzing eQTL for GWAS SNPs has been recognized (21) and some of the articles reporting GWAS SNPs have analyzed expression levels in reduced sets of tumors or lymphoblastoid cell lines to document a potential functional role of the SNPs (1,3,5,9). More recently, Loo et al. have found interesting associations using expression data assessed in colonic mucosa, either from tumor or normal mucosa adjacent to tumor, though the limited sample size provided low power to identify small associations (22).
In this study, we analyze cis- and trans-eQTL for GWAS SNPs to identify candidate genes responsible for CRC susceptibility. We combine two series of samples, one of healthy colonic mucosa and other of normal mucosa adjacent to colon cancer. In parallel, we have also analyzed the effect in tumor mucosa, but these data are more difficult to interpret because the expression in tumors is more heterogeneous and is highly altered by diverse mechanisms.
Materials and methods
Subjects and samples
Colon tumor and paired adjacent normal mucosa tissue samples used in this study were selected from a series of cases with a new diagnosis of colon adenocarcinoma attending the University Hospital of Bellvitge in Barcelona between January 1996 and December 2000. Patients included were diagnosed of stage II, microsatellite stable colon cancer, were surgically treated and had not received adjuvant chemotherapy. Adjacent mucosa was obtained from the proximal surgical margins and was at least 10 cm distant from the tumor lesion. Healthy colon mucosa samples were obtained during colonoscopy between February and May 2010. These samples come from a series of unselected patients who underwent a colonoscopy indicated for screening or suspicion of colonic pathology but no colonic lesions were observed. Biopsies were obtained from left and right colon. For this study, we selected randomly approximately half from each site (Supplementary Table 2, available at Carcinogenesis Online).
All subjects provided written informed consent to participate in the study and the ethics committee of the hospital cleared the protocol. Additional information about the study can be found at http://www.colonomics.org.
The eQTL analysis was focused on expression data assessed in normal mucosa. Though we initially selected 100 patients and 50 healthy controls, the final sample size after quality control of the data was 144: 47 from healthy donors and 97 adjacent normal mucosa from patients. Gene expression in tumors (n = 97) was also analyzed, and the results compared with those of normal mucosa. Also, for completeness and because we have previously demonstrated in these same samples that the expression in some genes is different between adjacent normal and healthy mucosa (23), we have performed the analyses separated for each tissue (Supplementary File 1, available at Carcinogenesis Online).
DNA and RNA extraction
DNA was extracted from colon mucosa specimens using the phenol–chloroform protocol. Total RNA was isolated from tissue samples using a miRCURY™ RNA Isolation Kit (Exiqon) according to manufacturer’s protocol, quantified by NanoDrop® ND-1000 Spectrophotometer (Nanodrop technologies, Wilmington, DE) and stored at −80°C. The quality of these RNA samples was assessed with the RNA 6000 Nano Assay (Agilent Technologies, Santa Clara, CA). RNA integrity numbers showed good quality (mean = 8.1 for tumors, 7.5 for adjacent normal and 8.2 for healthy normal). RNA purity was measured with the ratio of absorbance at 260nm and 280nm (mean = 1.96, SD = 0.04), with no differences among tissue types.
Genotype and selection of SNPs
Twenty-six risk SNPs identified in GWAS studies up to January 2013 (Supplementary Table 1, available at Carcinogenesis Online, US National Human Genome Research Institute (NHGRI) Catalog of Published GWAS; http://www.genome.gov/GWASstudies) were considered for analysis. Genotypes were obtained hybridizing genomic DNA extracted from colonic mucosa in Affymetrix Genome-Wide Human SNP 6.0 array (Affymetrix, Santa Clara, CA), which includes nearly 1 M SNP markers. Genotype calling was performed with Corrected Robust Linear Model with Maximum Likelihood Classification algorithm as implemented in R/Bioconductor package crlmm.
Genotypes for 17 SNPs had not been directly analyzed in the array and were imputed using IMPUTE2 (24) after haplotyping with SHAPEIT (25). The 1000 genomes panel (March 2012 version) was used as reference (http://www.1000genomes.org). Genotypes were attributed to the largest posterior probability if that was >0.6 and were defined missing otherwise. All SNPs, with the exception of rs4444235 could be imputed with certainty >0.98 (Supplementary Table 1, available at Carcinogenesis Online).
Expression data
Expression data were generated with Affymetrix Human Genome U219 Array Plate platform (Affymetrix, Santa Clara, CA). Three 96-array plates were used for this purpose, and a blocked experimental design was implemented to avoid biases due to potential plate effects (i.e. all plates contained the same proportion of healthy mucosa, normal and tumor samples). After evaluating the quality of all 250 CEL files using Affymetrix standard quality parameters (e.g. level of background noise, labeling and hybridization efficiency and RNA degradation), four arrays (two normal–tumor pairs) were excluded from the data set. Therefore, a final data set of 246 arrays was used for subsequent analyses. Raw data were normalized using the Robust Multiarray Average algorithm implemented in the affy package of R/Bioconductor. The gene expression data set is available at Gene Expression Omnibus with GEO series accession number GSE44076. Expression levels of a set of genes of this microarray has been validated with quantitative PCR and shown excellent correlation coefficients (data not shown).
Prior to the analysis of eQTL, expression probe sets were mapped to genes. For genes with more than one probe set in the array, a principal component analysis was used to capture the largest common variability extracting the first component. For each GWAS SNP, a region of 2Mb upstream and downstream was used to identify candidate cis-genes. Supplementary File 1, available at Carcinogenesis Online, shows the list of genes explored for each SNP.
Statistical analysis
To reduce the number of tests performed, while maintaining high power to identify eQTL, only the additive genetic model was considered. Genotypes were coded as the number of variant alleles (0, 1, 2) and this variable considered as quantitative. The additive model is known to capture most of the dominant and recessive effects (26). Partial Pearson correlation coefficients (adjusted for group: healthy/affected) were used for the analysis of normal tissue. The hypothesis test for this analysis is equivalent to that of a linear model adjusted for group. Previously, a rough exploration was performed of gene expression data to exclude large asymmetries in log-transformed gene expression distribution values, which could bias the analyses. A test for interaction with tissue group was used to confirm that the results were homogeneous irrespective of tissue type.
To avoid a large number of false positive results, a minimum significance level of 0.01 was used to consider an association relevant for reporting. In addition, to account for multiple comparisons, a Bonferroni correction was applied, taking into account the number of genes analyzed in the 4Mb region times 26, the number of independent loci SNPs analyzed. An additional analysis was performed for trans-eQTL, searching for genes associated to the GWAS SNPs outside the ±2Mb region in the whole genome. For these analyses, the Bonferroni correction used accounted for all annotated genes and SNPs tested (P < 1e-7 were considered significant).
Once a gene was identified in a locus as eQTL of the GWAS SNP, a search for functional SNPs was performed, analyzing those in linkage disequilibrium (LD) with the GWAS SNP that explained a larger fraction of variance of the gene expression. LD was calculated with SNAP web tool (27).
Bioinformatics methods
Genes identified by eQTL analysis were further analyzed using bioinformatics tools to assess their potential role in cancer development. Transcription factor regulatory networks reconstructed with the ARACNe algorithm were explored (28). The motif enrichment analysis in gene promoters was performed through the positional weight matrices collected in Jaspar (29) and Transfac (30) databases. To study the binding sites enrichment near the transcription start site, the Matching algorithm was used (31). Putative functional relationships within eQTL at protein level were assessed through the construction and analysis of protein–protein interaction networks (PPINs). BIANA software was used to retrieve these PPINs (32). BIANA builds networks by selecting interacting partners for an initial set of seed proteins (i.e. the relevant proteins), combining data from public databases. In this analysis, only human and experimentally determined interactions were considered.
Candidate functional SNPs were explored in the UCSC Genome Browser for marks of functional relevance in the ENCODE data tracks, like DNAase I hypersensitivity, open chromatin by formaldehyde-assisted isolation of regulatory elements and histone modifications in some cell lines.
Results
We identified 20 SNP—gene expression associations in nine loci with nominal P value <0.01 for genes within 2 Mb of the GWAS SNPs (Table I). In three loci, these associations were significant after Bonferroni correction (11q23.1 with genes C11orf53, COLCA1, COLCA2, 12q13.12 with gene DIP2B and Xp22.2 with genes SHROOM2 and GPR143). A detailed analysis for each locus analyzed can be found in Supplementary File 1, available at Carcinogenesis Online.
Table I.
cis-eQTL analysis for significant associations between GWAS SNPs and expression of genes within ±2 Mb
| Locus | SNP | Gene symbol | Gene name | r* | P value | Bonferroni$ |
|---|---|---|---|---|---|---|
| 1q41 | rs6691170 | TLR5 | Toll-like receptor 5 | −0.17 | 0.040 | N |
| 1q41 | rs6691170 | MARC1 | Mitochondrial amidoxime reducing component 1 | 0.18 | 0.031 | N |
| 1q41 | rs6691170 | SLC30A10 | Solute carrir family 30. Member 10 | 0.24 | 0.0035 | N |
| 11q13.4 | rs3824999 | FCHSD2 | FCH and double SH3 domains 2 | 0.24 | 0.0034 | N |
| 11q23.1 | rs3802842 | COLCA1 | Chromosome 11 open reading frame 92 | −0.57 | 1.3E-16 | Y |
| 11q23.1 | rs3802842 | C11orf53 | Chromosome 11 open reading frame 53 | −0.59 | 8.2E-18 | Y |
| 11q23.1 | rs3802842 | COLCA2 | Chromosome 11 open reading frame 93 | −0.62 | 4.4E-21 | Y |
| 11q123.1 | rs3802842 | TEX12 | Testis expressed 12 | 0.24 | 0.0036 | N |
| 12q13.12 | rs11169552 | KRT5 | Keratin 5 | 0.25 | 0.0021 | N |
| 12q13.12 | rs11169552 | KRT6C | Keratin 6C | 0.21 | 0.0091 | N |
| 12q13.12 | rs11169552 | DIP2B | DIP2 disco-interacting protein 2 homolog B (Drosophila) | 0.41 | 1.1E-7 | Y |
| 16q22.1 | rs9929218 | GFOD2 | Glucose-fructose oxidoreductase domain containing 2 | 0.24 | 0.0039 | N |
| 16q22.1 | rs9929218 | ATP6V0D1 | ATPase. H+ transporting. Lysosomal 38kDa. V0 subunit d1 | 0.22 | 0.0076 | N |
| 16q22.1 | rs9929218 | ZFP90 | Zinc finger protein 90 homolog | −0.27 | 0.0009 | N |
| 18q21.1 | rs4939827 | LIPG | Lipase. Endothelial | 0.22 | 0.0064 | N |
| 20p12.3 | rs961253 | PROKR2 | Prokineticin receptor 2 | −0.22 | 0.0084 | N |
| 20q13.33 | rs4925386 | C20orf201 | Chromosome 20 open reading frame 201 | −0.22 | 0.0094 | N |
| 20q13.33 | rs4925386 | FLJ16779 | Uncharacterized LOC100192386 | −0.22 | 0.0081 | N |
| Xp22.3 | rs5934683 | GPR143 | G protein-coupled receptor 1434 | −0.40 | 1.5E-7 | Y |
| Xp22.3 | rs5934683 | SHROOM2 | Shroom family member 2 | −0.47 | 1.5E-10 | Y |
*Pearson partial correlation coefficient.
$Significant after Bonferroni correction (P value < 0.05/genes in the ±2Mb region/26).
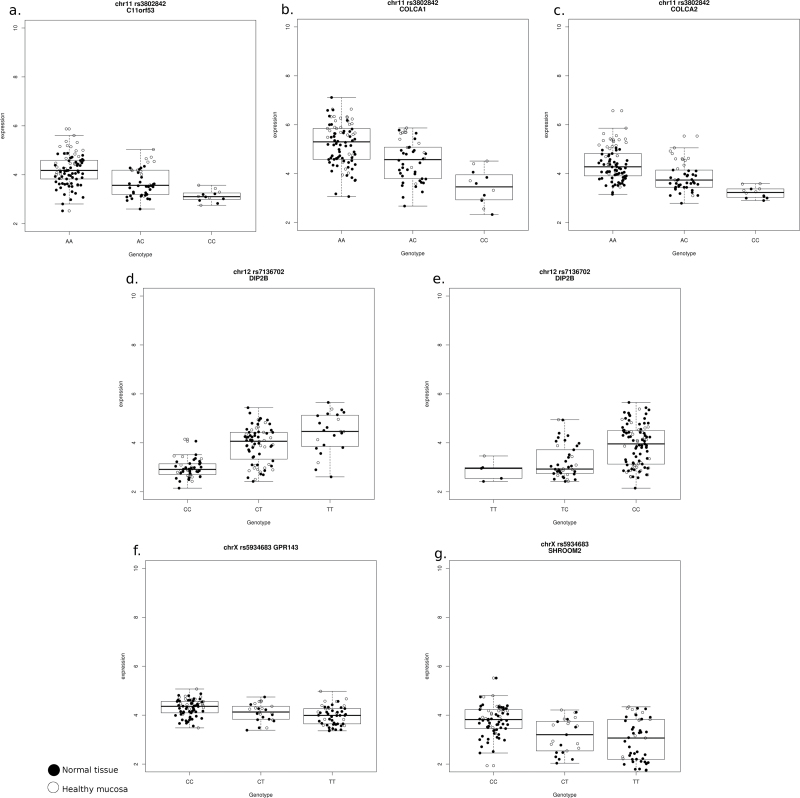
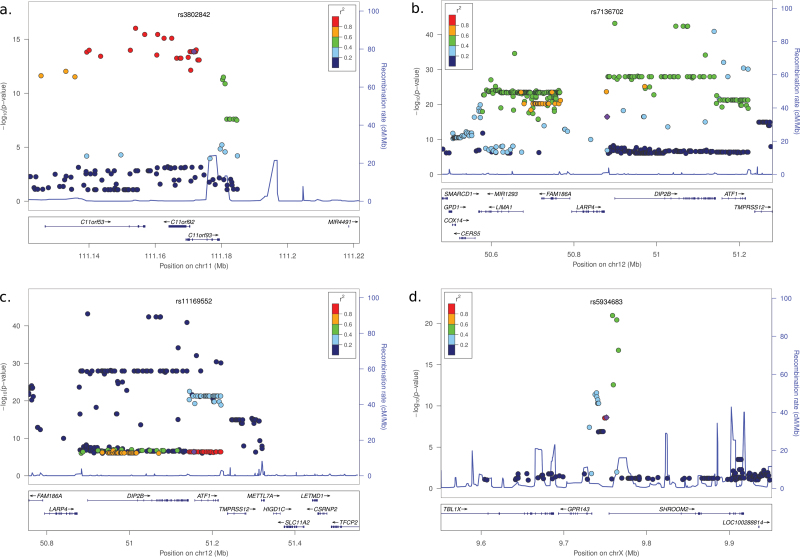
The strongest association was in locus 11q23.1 tagged by rs3802842. This SNPs was associated with the expression of three genes: C11orf53, COLCA1 and COLCA2. The association was very strong and similar for the three genes (r = 0.59, 0.62 and 0.57, respectively, Figure 1a–c). In fact, the expression of the three genes was highly correlated (Pearson’s r between 0.70 and 0.76, Supplementary Figure 1, available at Carcinogenesis Online) and it was impossible to identify one best candidate among them. The association was significant for both normal tissue types but not for the tumor (Supplementary File 1, available at Carcinogenesis Online). This locus corresponds to a small region with many SNPs in high LD. We analyzed the SNPs in the region (Figure 2a) in relation to the expression of a summary meta-gene derived from the first principal component of the three genes. Interestingly, the SNP that explained the highest proportion of variance of the meta-gene was rs7130173 (r = 0.67, Supplementary Figure 2 and File 2, available at Carcinogenesis Online).
Fig. 1.
Box-plot of cis-eQTL for significant associations analyzed in normal colon. White dots indicate healthy colonic mucosa and black dots indicate adjacent normal mucosa from patients. The y-axis shows the normalized (log2) mRNA expression level for the gene calculated from the U219 Affymetrix array.
Fig. 2.
Detailed distribution of the SNPs for loci 11q23.1, 12q13.12 and Xp22.3. Each dot represents a SNP. The purple dot indicates the GWAS SNP. For other SNPs the colors indicate the LD r 2 respect the GWAS SNP. The left y-axis indicates the −log(P value) from the association of the SNPs with the expression of the identified gene. The right y-axis indicates the recombination rate, represented with a blue line in the graph. The figure was generated with LocusZoom (46).
The second locus with an eQTL identified was rs11169552 at 12q13.12, which was associated to DIP2B expression (r = 0.40). In this locus, the meta-analysis by Houlston et al. (4) also had identified rs7136702 as a second candidate, though this SNP was not included in NHGRI database. In fact, the correlation between rs7136702 and DIP2B expression was stronger (r = 0.63) than that for rs11169552 (Figure 1d and e). SNP rs7136702 is 18.6 kb upstream of DIP2B promoter and rs11169552 in the 3′ region of DIP2B (Figure 2b and c). These SNPs are in weak LD (r 2 = 0.043). To explore if the effect on the expression of DIP2B was modified by the two SNPs independently, we performed a combined analyses followed by a conditional analysis. Supplementary Figure 4, available at Carcinogenesis Online, shows that the expression of DIP2B was increased when the T allele was present for SNPs rs7136702, independently of rs11169552 genotype. The conditional analysis confirmed that rs7136702 was significant even when adjusted for rs11169552 (P < 2e-16), but the latter was not significant when adjusted for rs7136702 (P = 0.01). The correlation of genotypes with DIP2B expression was similar for all tissue types, including tumor, but was not significant after Bonferroni correction when analyzed in healthy mucosa only (Supplementary File 1, available at Carcinogenesis Online). A detailed analysis of other imputed SNPs in the region that were in LD with these two showed that rs11169552 was in complete LD with a large set of SNPs located within gene ATF1, but the expression of ATF1 was not related to the SNP. SNP rs7136702, however, was quite unique regarding LD in the region of DIP2B. The best proxy had r 2 < 0.8 and was 150 kb upstream of DIP2B in a region with other genes. The analysis of DIP2B in relation to all the SNPs in the region within 500 kb identified five new SNPs for which the correlation with DIP2B expression was over 0.85 (Supplementary File 2 and Figure 5, available at Carcinogenesis Online). The most interesting of these SNPs was rs61927768, which is located in the promoter region of DIP2B, at 40 bp to the transcription start site (r = 0.86). SNP rs61927768 was in LD with rs7136702 (r 2 = 0.58) but LD of rs11169552 was weaker (r 2 =0 .10). A conditional analysis of the effect of rs61927768 with the GWAS SNP rs7136702 showed a stronger association for rs61927768 with DIP2B expression than any other SNP in the region (Supplementary Figure 6, available at Carcinogenesis Online). Since this SNP was in the promoter region of DIP2B, we analyzed its potential impact on the transcription factor binding. Indeed, the SNP was located inside of the enrichment peak of transcription factors binding motifs summarized in Jaspar and Transfac databases. The transcription factor SP1 was found with a high similarity score between its binding motif and the genomic sequence near rs61927768 in DIP2B promoter (Supplementary Figure 7, available at Carcinogenesis Online).
The third locus with eQTL was Xp22.3 that related rs5934683 to SHROOM2 and GPR143. The expression of these genes increased in carriers of the variant allele (C) compared with the ancestral allele (T). Similar to the locus on chromosome 11, the expression of these two genes was highly correlated (r = 0.73, Supplementary Figure 8, available at Carcinogenesis Online), though the variability for SHROOM2 was larger than that for GPR143 (Figure 1f and g). The correlation was similar for all tissue types when analyzed separately, including tumor, but was only significant after Bonferroni correction in adjacent normal mucosa (Supplementary File 1, available at Carcinogenesis Online). We analyzed the expression of SHROOM2 and GPR143 in relation to other SNPs in the region to identify candidates to causal SNPs for this locus. The best candidates were rs957490 and a deletion in position chrX:9758012 (r = 0.74, Supplementary Figure 9 and File 2, available at Carcinogenesis Online). Both candidates were in complete LD and located at about 7 kb downstream of the GWAS SNP, within the first intron of SHROOM2 (Figure 2d). The LD of these candidates with the GWAS SNP was r 2 = 0.3.
Other loci for which the eQTL analysis may have identified relevant genes are shown in Table I. However, because they were not significant after Bonferroni correction, these might be just false positive results. It is also worth noting that several other genes were associated to gene expression only when tumor tissues were analyzed (Table II). This was not related to a lack of expression of the genes in the normal tissue (data not shown). We have not considered these findings relevant because none was significant after Bonferroni correction, and the lack of effect in the normal tissue suggested that they were false positive results (Supplementary File 1 and Figure 10).
Table II.
Tumor tissue eQTL analysis for significant association between GWAS SNPs and expression of genes within ±2 Mb
| Locus | SNP | Gene symbol | Gene name | Tumor | Normal | ||
|---|---|---|---|---|---|---|---|
| r* | P value | r $ | P value | ||||
| 1q41 | rs6687758 | DUSP10 | Dual specificity phosphatase 10 | 0.27 | 0.0083 | 0.13 | 0.12 |
| 6p21.2 | rs1321311 | SCUBE3 | Signal peptide, CUB domain. EGF-like 3 | −0.27 | 0.0071 | 0.07 | 0.43 |
| 11q13.4 | rs3824999 | SLCO2B1 | Solute carrier organic anion transporter family, member 2B1 | −0.27 | 0.0073 | −0.01 | 0.94 |
| 11q13.4 | rs3824999 | SERPINH1 | Serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | −0.29 | 0.0042 | −0.05 | 0.53 |
| 11q23.1 | rs3802842 | TEX12 | Testis expressed 12 | −0.30 | 0.0031 | 0.24 | 0.0036 |
| 12q13.12 | rs7136702 | TUBA1C | Tubulin. Alpha 1c | 0.29 | 0.0046 | 0.06 | 0.51 |
| 12q13.12 | rs11169552 | SMAGP | Small cell adhesion glycoprotein | −0.26 | 0.0088 | −0.14 | 0.087 |
| 16q22.1 | rs9929218 | TSNAXIP1 | Translin-associated factor X interacting protein 1 | 0.28 | 0.0055 | −0.05 | 0.59 |
| 16q22.1 | rs9929218 | HSF4 | Heat shock transcription factor 4 | 0.28 | 0.0067 | 0.05 | 0.54 |
| 16q22.1 | rs9929218 | NUTF2 | Nuclear transport factor 2 | 0.32 | 0.0017 | 0.09 | 0.27 |
| 20p12.3 | rs961253 | GPCPD1 | Glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) | −0.30 | 0.0032 | 0.03 | 0.73 |
| 20p12.3 | rs961253 | CRLS1 | Cardiolipin synthase 1 | −0.28 | 0.0062 | 0.08 | 0.34 |
| 20p12.3 | rs961253 | LINC00654 | Long intergenic non-protein coding RNA 654 | −0.29 | 0.0037 | −0.03 | 0.75 |
| 20p12.3 | rs961253 | FERMT1 | Fermitin family member 1 | −0.26 | 0.0090 | 0.15 | 0.08 |
| 20p13.33 | rs2423279 | MCM8 | Minichromosome maintenance complex component 8 | −0.29 | 0.0038 | −0.03 | 0.69 |
| 20p13.33 | rs2423279 | TRMT6 | tRNA methyltransferase 6 homolog (S. cerevisiae) | −0.29 | 0.0043 | −0.02 | 0.80 |
*Pearson correlation coefficient.
$Pearson partial correlation coefficient.
trans-eQTL analysis
Finally, an analysis of trans-eQTL was performed in which correlations were explored with expression in all genes available in the array. Interestingly, the same loci in which we had identified significant cis-eQTL showed additional associations with other genes (Table III). Because these results could be related to common regulation of the cis-eQTL genes and the other genes, we explored transcriptional networks and PPINs. Our analysis of transcriptional networks estimated with the ARACNe algorithm did not show any results.
Table III.
trans-eQTL analysis for significant associations between the GWAS SNPs and all the genes
| Locus | SNP | Gene symbol | Gene name | r* | P value | Bonferroni$ |
|---|---|---|---|---|---|---|
| 11q23.1 | rs3802842 | GNG13 | Guanine nucleotide binding protein (G protein). Gamma 13 | −0.44 | 5.4E-9 | Y |
| 11q23.1 | rs3802842 | BMX | BMX non-receptor tyrosine kinase | −0.39 | 6.0E-7 | N |
| 11q23.1 | rs3802842 | HTR3E | 5-hydroxytryptamine (serotonin) receptor 3E. Ionotropic | −0.37 | 1.9E-6 | N |
| 11q23.1 | rs3802842 | SH2D6 | SH2 domain containing 6 | −0.37 | 2.1E-6 | N |
| 11q23.1 | rs3802842 | PSTPIP2 | Proline-serine-threonine phosphatase interacting protein 2 | −0.36 | 3.4E-6 | N |
| 11q23.1 | rs3802842 | LRMP | Lymphoid-restricted membrane protein | −0.35 | 1.2E-5 | N |
| 11q23.1 | rs3802842 | AZGP1 | Alpha-2-glycoprotein 1. Zinc-binding | −0.33 | 3.2E-5 | N |
| 12q13.12 | rs11169552 | HAL | Histidine ammonia-lyase | −0.41 | 9.2E-8 | Y |
| 12q13.12 | rs11169552 | TERF2 | Telomeric repeat binding factor 2 | 0.37 | 3.0E-6 | N |
| 12q13.12 | rs11169552 | HSP90AB1 | Heat shock protein 90kDa alpha (cytosolic). Class B member 1 | 0.36 | 4.3E-6 | N |
| 12q13.12 | rs11169552 | SIAH2 | Siah E3 ubiquitin protein ligase 2 | 0.36 | 5.0E-6 | N |
| 12q13.12 | rs11169552 | BCL7A | B-cell CLL/lymphoma 7A | 0.36 | 5.7E-6 | N |
| 12q13.12 | rs11169552 | MED28 | Mediator complex subunit 28 | 0.35 | 1.0E-5 | N |
| 12q13.12 | rs11169552 | AHSA1 | AHA1. Activator of heat shock 90kDa protein ATPase homolog 1 (yeast) | 0.34 | 2.3E-5 | N |
| 12q13.12 | rs11169552 | CCDC71 | Coiled-coil domain containing 71 | 0.33 | 3.1E-5 | N |
| 12q13.12 | rs11169552 | POM121 | POM121 transmembrane nucleoporin | 0.33 | 3.7E-5 | N |
*Pearson partial correlation coefficient.
$Significant after Bonferroni correction (P value < 1e-7).
PPINs retrieved with BIANA software were more successful. For SNP rs3802842 at locus 11q23.1, the proteins of five out of seven trans-eQTL significant genes (AZGP1, LRMP, BMX, PSTPIP2, GNG13) and the cis-eQTL c11orf53 interacted with each other through linker proteins (Supplementary Figure 11, available at Carcinogenesis Online). The analysis of locus 12q13.12 also revealed that the proteins of all trans-eQTL genes (AHSA1, BCL7A, POM121, TERF2, HAL, MED28, SIAH2, CCDC71, HSP90AB1) and the cis-eQTL DIP2B interacted with each other within the retrieved network (Supplementary Figure 12, available at Carcinogenesis Online).
Discussion
We have examined 26 SNPs identified in GWAS of CRC for association with the expression of neighboring genes. Three of these SNPs were cis-eQTLs of one or more genes. In a second step, we have identified nearby SNPs in LD with the GWAS hits that explain a larger proportion of the expression variation and are proposed as putative causal SNPs at these loci.
Three genes were involved in locus 11q23.1—C11orf53, COLCA1 and COLCA2—associated to rs3802842. This SNP was one of the early CRC GWAS SNPs identified by Tenesa et al. (9) in the Scottish GWAS with an odds ratio of 1.11. It has later been extensively replicated in several large studies and meta-analyses (33–35). The protein products of these genes have been poorly characterized and have unknown function. The suggestion that rs3802842 is an eQTL for these genes has been recently reported (36,37). The risk allele was associated with a decreased gene expression, suggesting a tumor suppressor effect. Our data showed that the expression levels of the three genes were highly correlated, probably because they share common regulatory mechanisms.
Our search in the region has identified rs7130173 as the SNP that explained the highest proportion of variance of the common gene expression of the three genes extracted with principal components analysis (Supplementary Figure 2 and File 1, available at Carcinogenesis Online). This SNP is in LD with rs3802842 (r 2 = 0.95) and is located at 17 kb upstream, in an intron of C11orf53. Several ENCODE signals (DNAase I hypersensitivity, open chromatin by formaldehyde-assisted isolation of regulatory elements, and histone modifications in some cell lines) suggest that an enhancer might be at that position (Supplementary Figure 13, available at Carcinogenesis Online). Interestingly, Biancolella et al. (36) have performed functional studies and also concluded that rs7130173 is the most likely causal variant in the region. These authors also report rs10891246 as an alternative causal candidate. In our analysis, this SNP is highly correlated with gene expression (r = 0.66) but slightly less than rs7130173 (r = 0.67).
DIP2B has been identified as the relevant gene related to GWAS SNPs at locus 12q13.12. The initial report of this locus by Houlston et al. (4) provided two risk SNPs, rs11169552 located at 5′ and rs7136702 located at 3′, with low LD between them. The first one showed a higher association in the GWAS, but a subsequent fine-mapping study has suggested the possibility of two independent loci at 12q13.12 (38). Other study in adenoma has also observed a more significant association for rs7136702 than for rs11169552 (39). Our eQTL analysis has shown that the correlation of DIP2B expression is stronger with rs7136702. Furthermore, we have identified alternative candidate SNPs in the region in LD with rs7136702 that show even higher correlation with DIP2B expression. One of them, rs61927768, located in the promoter region of DIP2B, just at 40 bp upstream of the transcription start site, might be responsible for the functional effect in the locus because our bioinformatics analysis identified that it might affect the binding of multiple transcription factors with compatible motifs (Supplementary Figure 6, available at Carcinogenesis Online). Although this in silico analysis needs validation, the information is valuable. ENCODE data at this position reveal high DNA activity, with DNAase I hypersensitivity and open chromatin shown by formaldehyde-assisted isolation of regulatory elements analysis (Supplementary Figure 14, available at Carcinogenesis Online).
Mutations in the DIP2B gene (a CGG-repeat expansion) have been associated to mental retardation in relation to FRA12A, a folate-sensitive fragile site, though this may be unrelated to CRC susceptibility. DIP2B protein has a DMAP1 binding domain, and it has been suggested that it may be related to DNA methylation, a more plausible mechanism of involvement in CRC susceptibility (40).
The third significant locus, Xp22.3, has related rs5934683 to SHROOM2 and GPR143. Similar to the 11q23.1 locus, the expression levels of these two genes are highly correlated though the expression in the colon of SHROOM2 is much higher than that of GPR143 (Supplementary Figure 7, available at Carcinogenesis Online). Because rs5934683 is located in the promoter region of SHROOM2, most of the data point to this gene as the relevant one. Both SHROOM2 and GPR143 have been related to retinal pigmentation and it is known that congenital hypertrophy of retinal pigment epithelium lesions are typical of the familial adenomatous polyposis syndrome (41).
SHROOM2 expression already had been related to rs5934683 genotypes in normal colon and CRC tissue (3). Here we have identified other polymorphisms that show higher correlations with SHROOM2 and GPR143 expressions (Supplementary File 2, available at Carcinogenesis Online). They are located in the first intron of SHROOM2, but we have not found strong evidence to suggest one of them as better candidate than rs5934683, and this region requires in-depth functional studies.
Our study is not the first to attempt identifying eQTL for CRC GWAS SNPs but is the first that uses healthy colon mucosa for this purpose with a large enough sample size. Previous study by Loo et al. (22) used 40 samples of tumor and adjacent normal mucosa and identified four eQTL in three loci. Two eQTL were restricted to tumor tissue: ATP5C1 in locus 10p14 and DLGAP5 in locus 14q22.2; and two restricted to adjacent mucosa: NOL3 and DDX28 in locus 16q22.1. We have not been able to replicate their findings in this study. The most likely reason is that their findings might be false positive results. Though they calculated a false discovery rate as a multiple comparisons correction, the associations found were not strongly significant and none would have passed Bonferroni correction. Their sample size was relatively small and random variation related to sample heterogeneity may have hindered them finding our associations. Some of the associations reported by Loo et al. (22) were restricted to tumor tissue. Altough these effects could be related to tumor progression, probably tumor heterogeneity adds to the variability increasing the likelihood of a false positive result when the sample size is small. Our significant results generally had good correlation both in tumor and normal tissue, but not always. For example, the associations for genes in locus 11q23.1 were not observed in tumors. We have identified some associations restricted to tumor tissue (Table II), but consider them likely false positive results because the Bonferroni adjusted P values are not significant.
The analysis of trans-eQTL is more prone to false positive results because the number of comparisons increases substantially. We have done these analyses as an exploratory exercise because the statistical power we have is limited. It is interesting that the only loci that have shown some near-significant results were the same that we had identified with strong cis-eQTL: 11q23.1 and 12q13.12. We have hypothesized that these long-distance associations might be related to physical interactions between the cis-eQTL and the trans-eQTL genes. Indeed, we could find that both sets of genes were linked in PPIN either directly or with just one intermediate protein. PPINs provides a framework to study a hypothetical signaling pathway from cis-eQTL to trans-eQTL that could participate in early stages of carcinogenesis. At locus 11q23.1, the cis-eQTL C11orf53 interacted with the trans-eQTL BMX through a fragment of the Alzheimer Amyloid Precursor protein. Previous reports have described the implication of Alzheimer Amyloid Precursor protein in colon cancer. Interestingly, a work by Venkataramani et al. (42) postulate an inhibition of proliferation via downregulation of Alzheimer Amyloid Precursor protein when cancer cells were treated with valproic acid, an histone deacetylase inhibitor. This downregulation was not directed but mediated by the chaperone GRP78 that was also present in our retrieved network. Regarding the trans-eQTL associations for locus 12q13.12, the retrieved PPIN showed that DIP2B could interact through linker proteins with the nine trans-eQTLs identified. Within this PPIN, it is noteworthy that a common linker protein—histone H3—binds the cis-eQTL DIP2B with trans-eQTLs POM121, HSP90AB1 and CCDC71, suggesting that the first steps in CRC carcinogenesis mediated by this susceptibility gene may be activated by epigenetic mechanisms. In the same network, the direct interaction between AHSA1 and HSP90AB1 was also of interest because it could imply a co-operative role of these proteins in the cell.
This study has some limitations. The expression levels have been measured with a microarray platform. The simultaneous measurement of the transcriptome may be suboptimal for some genes and this may be related to the low number of loci with eQTL found. We have performed some technical validation experiments with quantitative PCR and the correlation was excellent. Though absolute expression levels cannot be measured with microarrays, relative levels and comparison of groups should be correct, other than the effect of random measurement error.
We have combined in the analysis samples of healthy mucosa and adjacent normal tissue from patients with cancer. Our aim was to increase statistical power. However, this joint analysis might be a source of false positive results because we have previously analyzed that some genes are overexpressed in the adjacent normal mucosa as a reaction to the presence of the tumor (23). To decrease this risk, we have used partial correlation analysis, adjusting for tissue type, and have verified the homogeneity of the effect for all significant results. The correlations of DIP2B and the genes in locus Xp22.3 were not significant after Bonferroni correction when the analyses were restricted to healthy mucosa, but this was probably related to the small sample size of the group (n = 47).
Also, the genotypes have been analyzed with a microarray platform and we have used imputed SNP data for many of our analysis. SNP imputation is currently a well-accepted technique to fine-map and explore non-genotyped SNPs. The Affymetrix Human SNP array 6.0 used in our study has nearly 1 million SNPs and all the quality scores were adequate for the imputed SNPs analyzed (Supplementary Table 1, available at Carcinogenesis Online). This, however, does not rule out some misclassification and reduced statistical power to detect additional eQTL because no eQTL has been identified for most of the GWAS loci. Our sample size can detect correlations >0.26 with 90% power, but smaller correlations may be of interest. The power to detect trans-eQTL associations was smaller because the multiple comparisons problem is severe then.
Finally, our study only included colon specimens from stage II microsatellite stable patients. This selection could raise concern about generalizability of the results. However, we have previously analyzed that the expression levels are very similar in colon and rectal tumors (43) and this has been confirmed in the The Cancer Genome Atlas (TCGA) study (44).
Implications
The outcomes of GWAS studies so far have had little impact in public health. The value for risk prediction or stratification of current SNPs is limited, but new intervention opportunities could appear if susceptibility genes were identified. This has been demonstrated in prostate cancer, where the gene beta-microseminoprotein was identified and now the detection of its protein in blood has been proposed for prostate cancer screening complementing prostate-specific antigen (45). In this study, we have identified candidate cancer genes in three GWAS loci, and for two of them we propose new functional SNPs responsible for the increased risk of CRC. However, further experimental validations are needed to establish the role of these SNPs and the function of the genes identified.
Supplementary material
Supplementary Tables 1 and 2, Figures 1–14 and Files 1 and 2 can be found at http://carcin.oxfordjurnals.org/ and at http://www.colonomics.org/eqtl
Funding
Catalan Institute of Oncology; Instituto de Salud Carlos III (PI08-1635, PI08-1359, PS09-1037, PI11-01439, CB06/02/2005, and Acción Transversal del Cancer); Departament d'Universitats, Recerca i Societat de la Informació (DURSI) de la Generalitat de Catalunya (2009SGR1489); Fundación Científica de la Asociación Española Contra el Cáncer (AECC); European Commission (FP7-COOP-Health-2007-B: HiPerDART); National Institutes of Health (U19 CA148107–CORECT).
Supplementary Material
Acknowledgements
Pilar Medina, Carmen Atencia and Isabel Padrol helped with the clinical annotation of the samples used in this study. Ferran Martinez provided technical support for the bioinformatics work. A.C. was supported by a PFIS fellowship of the ISCIII. SNP genotyping services were provided by the Spanish “Centro Nacional de Genotipado” (CEGEN-ISCIII, www.cegen.org). Sample collection was supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncología de Catalunya.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CRC
colorectal cancer
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- PPIN
protein–protein interaction network
- SNP
single nucleotide polymorphism.
References
- 1. Broderick P., et al. ; CORGI Consortium. (2007). A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet., 39, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 2. Cui R., et al. (2011). Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut, 60, 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunlop M.G., et al. ; Colorectal Tumour Gene Identification (CORGI) Consortium; Swedish Low-Risk Colorectal Cancer Study Group; COIN Collaborative Group. (2012). Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet., 44, 770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Houlston R.S., et al. ; COGENT Consortium; CORGI Consortium; COIN Collaborative Group; COINB Collaborative Group. (2010). Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet., 42, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Houlston R.S., et al. ; Colorectal Cancer Association Study Consortium; CoRGI Consortium; International Colorectal Cancer Genetic Association Consortium. (2008). Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet., 40, 1426–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia W.H., et al. ; Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Colon Cancer Family Registry (CCFR). (2013). Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet., 45, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peters U., et al. (2012). Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet., 131, 217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peters U., et al. ; Colon Cancer Family Registry and the Genetics and Epidemiology of Colorectal Cancer Consortium. (2013). Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology, 144, 799–807.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tenesa A., et al. (2008). Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet., 40, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tomlinson I., et al. ; CORGI Consortium. (2007). A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet., 39, 984–988 [DOI] [PubMed] [Google Scholar]
- 11. Tomlinson I.P., et al. ; CORGI Consortium; EPICOLON Consortium. (2008). A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet., 40, 623–630 [DOI] [PubMed] [Google Scholar]
- 12. Zanke B.W., et al. (2007). Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet., 39, 989–994 [DOI] [PubMed] [Google Scholar]
- 13. Tenesa A., et al. (2009). New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat. Rev. Genet., 10, 353–358 [DOI] [PubMed] [Google Scholar]
- 14. Cheung V.G., et al. (2010). Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol., 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheung V.G., et al. (2009). Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet., 10, 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang H.P., et al. (2012). Coanalysis of GWAS with eQTLs reveals disease-tissue associations. AMIA Summits Transl. Sci. Proc., 2012, 35–41 [PMC free article] [PubMed] [Google Scholar]
- 17. Chambers J.C., et al. ; Alcohol Genome-wide Association (AlcGen) Consortium; Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study; Genetic Investigation of Anthropometric Traits (GIANT) Consortium; Global Lipids Genetics Consortium; Genetics of Liver Disease (GOLD) Consortium; International Consortium for Blood Pressure (ICBP-GWAS); Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC). (2011). Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet., 43, 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wheeler H.E., et al. (2009). Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet., 5, e1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zou F., et al. ; Alzheimer’s Disease Genetics Consortium. (2012). Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet., 8, e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. GTEx Consortium. (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pardini B., et al. (2012). Gene expression variations: potentialities of master regulator polymorphisms in colorectal cancer risk. Mutagenesis, 27, 161–167 [DOI] [PubMed] [Google Scholar]
- 22. Loo L.W., et al. (2012). cis-Expression QTL analysis of established colorectal cancer risk variants in colon tumors and adjacent normal tissue. PLoS One, 7, e30477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanz-Pamplona R., et al. (2014). Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol. Cancer, 13, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marchini J., et al. (2007). A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913 [DOI] [PubMed] [Google Scholar]
- 25. Delaneau O., et al. (2013). Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods, 10, 5–6 [DOI] [PubMed] [Google Scholar]
- 26. Freidlin B., et al. (2002). Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum. Hered., 53, 146–152 [DOI] [PubMed] [Google Scholar]
- 27. Johnson A.D., et al. (2008). SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics, 24, 2938–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Margolin A.A., et al. (2006). ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics, 7(suppl. 1), S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sandelin A., et al. (2004). JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res., 32, D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matys V., et al. (2006). TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res., 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sekhon J.S. (2011). Multivariate and propensity score matching software with automated balance optimization: the matching package for R. J. Stat. Softw., 42, 1–52 [Google Scholar]
- 32. Garcia-Garcia J., et al. (2010). Biana: a software framework for compiling biological interactions and analyzing networks. BMC Bioinformatics, 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pittman A.M., et al. ; CORGI Consortium; EPICOLON Consortium. (2008). Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum. Mol. Genet., 17, 3720–3727 [DOI] [PubMed] [Google Scholar]
- 34. von Holst S., et al. (2010). Association studies on 11 published colorectal cancer risk loci. Br. J. Cancer, 103, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zou L., et al. (2012). Replication study in Chinese population and meta-analysis supports association of the 11q23 locus with colorectal cancer. PLoS One, 7, e45461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biancolella M., et al. (2014). Identification and characterization of functional risk variants for colorectal cancer mapping to chromosome 11q23.1. Hum. Mol. Genet., 23, 2198–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peltekova V.D., et al. (2014). Identification of genes expressed by immune cells of the colon that are regulated by colorectal cancer-associated variants. Int. J. Cancer, 134, 2330–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spain S.L., et al. (2012). Refinement of the associations between risk of colorectal cancer and polymorphisms on chromosomes 1q41 and 12q13.13. Hum. Mol. Genet., 21, 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carvajal-Carmona L.G., et al. ; APC Trial Collaborators; APPROVe Trial Collaborators; CORGI Study Collaborators; Colon Cancer Family Registry Collaborators; CGEMS Collaborators. (2013). Much of the genetic risk of colorectal cancer is likely to be mediated through susceptibility to adenomas. Gastroenterology, 144, 53–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winnepenninckx B., et al. (2007). CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am. J. Hum. Genet., 80, 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Díaz-Llopis M., et al. (1988). Congenital hypertrophy of the retinal pigment epithelium in familial adenomatous polyposis. Arch. Ophthalmol., 106, 412–413 [DOI] [PubMed] [Google Scholar]
- 42. Venkataramani V., et al. (2010). Histone deacetylase inhibitor valproic acid inhibits cancer cell proliferation via down-regulation of the alzheimer amyloid precursor protein. J. Biol. Chem., 285, 10678–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanz-Pamplona R., et al. (2011). Gene expression differences between colon and rectum tumors. Clin. Cancer Res., 17, 7303–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Network. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haiman C.A., et al. (2013). Levels of beta-microseminoprotein in blood and risk of prostate cancer in multiple populations. J. Natl. Cancer Inst., 105, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pruim R.J., et al. (2010). LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.