Summary
Increased abundance of HERV-K gag mRNA in PBMC was found to be associated with a prostate cancer diagnosis irrespective of race/ethnicity and a patient’s age, indicating its potential as a diagnostic biomarker in addition to PSA.
Abstract
Aberrant expression of subgroup k human endogenous retroviruses (HERV-K) has been observed in prostate cancer. This subgroup is unique because it encodes sequences in the human genome containing open reading frames for near intact retroviruses. We hypothesized that HERV-K reactivation could serve as a non-invasive early disease detection marker for prostate cancer. We evaluated HERV-K gag messenger RNA (mRNA) expression in blood samples of African-American and European-American men using a case–control design via quantitative real-time PCR. Additionally, we examined HERV-K envelope protein expression in prostate tumors by immunohistochemistry. HERV-K envelope protein was commonly upregulated in prostate tumors, but more so in tumors of African-American than European-American patients (61% versus 40%, P < 0.01). Examining HERV-K gag expression in peripheral blood mononuclear cells (PBMC) from 294 cases and 135 healthy men, we found that the abundance of HERV-K gag message was significantly higher in cases than controls and was associated with increased plasma interferon-γ. Men with gag expression in the highest quartile had >12-fold increased odds {odds ratio = 12.87 [95% confidence interval 6.3–26.25]} of being diagnosed with prostate cancer than those in the lowest quartile. Moreover, our results showed that HERV-K expression may perform better as a disease biomarker in older than younger men (whereas the sensitivity of prostate-specific antigen (PSA) testing decreases with age) and in men with a smoking history compared with never smokers. Combining non-invasive HERV-K testing with PSA testing may improve the efficacy of prostate cancer detection specifically among older men and smokers who tend to develop a more aggressive disease.
Introduction
It is estimated that prostate cancer will account for 28% of all new cancer diagnoses in USA men in 2013 with ~30 000 deaths expected, ranking it as the second leading cause of cancer death in men in the country (1). Epidemiological studies identified aging, disease family history, race/ethnicity and obesity and diet as being significant risk factors for the development of the disease (2). Other studies suggest that inflammation and infections contribute to disease development (3,4). More recently, the reactivation of endogenous retroviruses in the HERV-K family has been associated with a prostate cancer diagnosis (5–7).
Human endogenous retroviruses (HERVs) represent the remnants of ancient germline infections by exogenous retroviruses (reviewed in ref. 8) and today compose ~8% of the human genome (9). For the most part, they have become defective over time via the accumulation of inactivating mutations and through silencing by epigenetic mechanisms such as DNA methylation (10). The HERV-K (HML-2) subgroup (hereafter referred to as HERV-K) is unique among HERVs, in that a proportion of its constituent proviruses retain complete open reading frames for all retroviral genes (8). Furthermore, a selection of these proviruses are human specific and polymorphic (11).
The genetic structure of an intact HERV-K provirus consists of open reading frames for the retroviral genes: gag, pro, pol and env, flanked by two long terminal repeats that regulate their expression (12). HERV-K consists of two major types defined by the presence or absence of a 292 base pair deletion at the junction of the pol and env genes, which fuses their reading frames (8). Type 1 HERV-K proviruses harbor the deletion, whereas type 2 remains intact (8). Four HERV-K transcripts have been described to date: full length (gag) messenger RNA (mRNA), singly spliced env mRNA, doubly spliced rec/np9 mRNA and the ‘hel’ transcript that lacks any known function (13). High levels of HERV-K mRNA and protein have been observed in a variety of cancers, including germ cell, breast, ovarian, lymphoma and melanoma, but a causal link to any of these diseases remains to be identified (14,15). HERV-K transcripts have also been detected in prostate cancer cell lines (16) and tissues (6) and a humoral response to the HERV-K gag protein has been observed in sera from prostate cancer patients (5). Furthermore, this immune response correlated with disease progression (5), indicating that an inflammatory immune response to HERV-K, which does not eradicate the HERV-K expressing tumor, may promote disease progression.
We hypothesized that HERV-K reactivation could serve as a non-invasive early disease detection marker for prostate cancer and therefore evaluated HERV-K expression in tumor and blood samples. Because peripheral blood mononuclear cells (PBMC) can provide a suitable surrogate to an individual’s health status (17) and distinct PBMC gene expression profiles have been observed in a number of non-hematological cancers (18), we analyzed HERV-K mRNA expression in PBMC from prostate cancer patients and healthy volunteers using a case–control design. We also evaluated whether HERV-K reactivation may occur differently in patients of American and European ancestry. Using this approach, we observed differences in HERV-K reactivation between African-American and European-American men and found that blood-based HERV-K expression is a candidate early detection biomarker for prostate cancer.
Materials and methods
Subject recruitment and collection of PBMC
Selection criteria for cases (n = 270) and population-based controls (n = 91) that participated in the National Cancer Institute (NCI) study: prostate cancer patients were eligible for the study when a diagnosis of prostate cancer has been made within 2 years prior to recruitment; resided in Maryland and adjacent counties in Pennsylvania, Delaware, Virginia, or District of Columbia, if they were born in the USA; were either African-American or European-American by self-report; had a working home phone number; were physically and mentally able of performing the interview; were not severely ill; spoke English fluently and were able to give informed consent and did not reside in an institution such as a prison, nursing home or shelter. Male population controls were frequency matched on age and race to cases and had the same eligibility criteria with the exception that they could not have a personal history of cancer, radiation therapy or chemotherapy. Controls resided in the greater Baltimore area and adjacent counties in Maryland. Selection criteria for cases (n = 23) and hospital-based controls (n = 44) that participated in the Georgetown study: Prostate cancer patients were eligible for the study when a diagnosis of prostate cancer has been made by the attending physician, and these patients resided in the District of Columbia or its adjacent states of Maryland or Virginia. Patients were self-reported as African-American and were physically and mentally capable of providing informed consent. Male hospital controls were frequency matched on age to cases and had the same eligibility criteria as cases with the exception that they could not have a personal history of cancer. No significant difference was found between the age at diagnosis/recruitment in Georgetown Cohort versus the NIH cohort (P = 0.29) overall or when comparing African-Americans alone (P = 0.87).
Blood was drawn at time of recruitment. PBMC were collected from both prostate cancer patients (n = 294) and men without a diagnosis of the disease (n = 135). The cells were isolated from whole blood by standard ficoll–hypaque density gradient centrifugation and stored at −80°C. Men with prostate cancer were recruited between 2004 and 2008 under two Institutional Review Board (IRB)-approved protocols (NCI IRB #05-C-N021 and Georgetown University IRB #2003–013) and had a prostate cancer diagnosis within the last 2 years prior to recruitment (median time between diagnosis and recruitment = 206 days, range 0–714 days). These patients had prostate cancer at time of recruitment and came to the hospital for consultation or to seek treatment including prostatectomy, radiation therapy, or androgen ablation therapy. The subjects were recruited at four hospitals: the Veterans Affairs Medical Center and the University of Maryland Medical Center in Baltimore City, the Department of Urology at the Georgetown University Hospital and the Washington DC Veterans Affairs Medical Center. All completed an informed consent. Controls were either population-based controls (n = 91) recruited under the NCI IRB approved protocol #05-C-N021 (NCT00342771) (19), or they were men without a previous cancer diagnosis (by self-report) visiting the Georgetown University Hospital (n = 44), accompanying other people or coming for a routine checkup. The latter were recruited under the Georgetown University IRB-approved protocol #2003–013. All controls completed an informed consent. Prostate-specific antigen (PSA) test results were available for 287 of the 294 prostate cancer patients; they were not available for the controls. Both cases and controls completed interviewer-administered questionnaires but only the NCI-based study collected information on smoking history from study participants. Information on smoking was available for 359 subjects (270 cases, 89 controls) and was categorized into current, former, and current smokers, or into pack-years. A never smoker was defined as a subject who did not currently smoke and also smoked <100 cigarettes in his lifetime. A past smoker did not smoke cigarettes in the 6 months prior to enrolment. Race/ethnicity was self-reported.
RNA Isolation from PBMC and detection of HERV-K gag mRNA
Total RNA was isolated using the TRIZOL reagent according to the manufacturer’s instructions. Five hundred nanograms of RNA was reverse transcribed and the complementary DNA was added to the quantitative real-time PCR assays. Previously published primers were used to amplify HERV-K gag transcripts (F, 5′-AGC AGG TCA GGT GCC TGT AAC ATT-3′; R, 5′-TGG TGC CGT AGG ATT AAG TCT CCT-3′) (20). In addition, primers specific for 18s were used as an internal standard reference. Data were collected using the ABI PRISM® 7500 Sequence Detection System. Normalized expression was calculated using the comparative CT method and fold changes were derived from the 2−ΔΔCt values (21).
Detection of HERV-K Env type I and Env type II transcripts in a subset of cases and controls
RNA was treated with TURBO DNAse (Ambion, Biosciences, Ireland) for 30 min at 37°C to eliminate genomic DNA. DNase was inactivated using 50mM ethylene diamine tetra-acetic acid at 75°C for 10 min. First strand complementary DNA synthesis was performed on 10ng of RNA using a Tetro cDNA Synthesis Kit (Bioline, MyBio, Ireland) primed with random hexamers. Quantitative PCR took place in a StepOne Plus real-time PCR system (Applied Biosystems, Biosciences, Ireland) together with Sensifast SYBR Hi-ROX (Bioline, MyBio, Ireland) and the following primers, 5′-CTAT TTCTTCGGACCTGTTCTTG-3′; env1 forward, 5′-GGAG ATGGTAA CACC AGTCACAT-3′; env1 reverse, 5′-GGATAACGATACCCAATGGAAAT-3′; env2 forward, 5′-CAAAATGGTGACGTCAGAAGAA-3′; env2 reverse, 5′-CAGG CATAG GGAGACTTACCAC-3′. Thermal cycling consisted of enzyme activation (95°C for 2 min), followed by 40 cycles of both denaturation and annealing/extension (95°C for 5 s and 60°C for 15 s, respectively). Subsequent melt curve analysis was carried out using the following conditions: 95°C for 15 min, 60°C for 1 min and 95°C for 15 min. Gene expression levels in all samples were normalized to an 18S rRNA reference gene (RefSeq accession number NR_003286) using the delta Ct method. Values were displayed as the mean of duplicate samples.
Measurement of cytokines in human plasma samples
Heparinized plasma was collected from prostate cancer patients and population-based controls in the NCI study and stored at −80°C. Plasma interferon-γ (INFγ), IP10, tumor necrosis factor-α, and interleukin-1β (IL-1β) concentrations were determined at a Leidos Biomedical Research Inc/NCI core facility using the human electrochemiluminescence immunoassays from Mesoscale Discovery (Gaithersburg, MD) under standardized conditions. Ultrasensitive multiplex electrochemiluminescence immunoassay plates were custom designed and were analyzed on the MesoScale Discovery 6000 instrument, following manufacturer’s assay and analysis protocols.
Immunohistochemistry for HERV-K envelope expression
Immunohistochemistry (IHC) for HERV-K envelope protein expression was performed on formalin-fixed, paraffin-embedded tissue sections using standard protocols. We performed IHC on whole section tumors to examine protein localization. These tumors were obtained from patients recruited into the NCI study (IRB #05-C-N021). For IHC scoring, a tissue microarray (race/ethnicity) was obtained from the NCI Cooperative Prostate Cancer Tissue Resource. Two cores were scored per case on the tissue microarray and the average score between the two was calculated. Most tumors (304 out of 310) on the TMAs were of acinar adenocarcinoma histology, whereas the others had ductal carcinoma histology. Slides were deparaffinized, blocked with normal serum according to the VECTASTAIN® ABC protocol, and incubated with 1:200 diluted mouse monoclonal 6H5 antibody raised against the HERV-K envelope protein. This antibody was purified from a hybridoma cell supernatant and the specificity for the HERV-K envelope protein was demonstrated as described previously (22). After washing steps and incubation with a biotinylated secondary antibody, sections were incubated with VECTASTAIN® ABC reagent containing an antibiotin antibody labeled with peroxidase and stained with peroxidase substrate solution for desired stain intensity.
Cell lines
RWPE1, DU145, PC-3, 22Rv1 and CWR22 were sourced from the American Type Culture Collection (Manassas, VA) and cultured according to recommendations. In brief, CWR22 and 22Rv1 were cultured in RPMI 1640 medium with L-glutamine (Sigma #R8758) and supplemented with 10% fetal bovine serum Sigma #F7524). DU145 was cultured in minimum essential medium (1×) with Earle’s (Gibco #22561-021) supplemented with 10% fetal bovine serum. PC-3 was cultured in F12 nutrient mixture (HAM) medium, with L-glutamine (Gibco #21765-029) supplemented with 10% fetal bovine serum. RWPE1 was cultured in keratinocyte medium, supplemented with epidermal growth factor and Bovine Pituitary Extract (Gibco #17005–042). All five cell lines were authenticated by LGC standards (United Kingdom) in May 2013 via short tandem repeat profiling and were found to be the correct cell lines.
Western blot analysis
Cells were seeded in 10cm3 dishes at a cell density of 1 × 106 per dish and grown for 3 days. Cells were rinsed twice with cold phosphate-buffered saline and lysed directly on the dish with cold radioimunoprecipitation assay buffer (Thermo-Scientific Pierce, Ireland #89900) supplemented with protease inhibitors (Thermo-Scientific Pierce, Ireland, #78410), scraped and spun at 14 000 g for 15min at 4°C. Supernatant was collected and stored at −20°C for western blot analysis of protein expression. Extracted protein was quantified using a bicinchoninic acid assay kit. HERV-K env and HERV-K gag levels were detected through use of a primary anti-HERV-K env mouse monoclonal antibody (clone 6H5, Dr Feng Wang-Johanning) (22,23) and an anti-HERV-K gag mouse monoclonal antibody (LifeSpan Biosciences, Seattle, Washington, DC, #LS-C65287), respectively. Both antibodies were diluted 1:1000 in 5% skimmed milk reconstituted in 1× Tris-buffered saline (pH = 8) 0.1% Tween. These dilutions were added to the transfer membrane and shaken overnight at 4°C, following a 1h room temperature blocking in 5% skimmed milk in Tris-buffered saline. Mouse monoclonal anti-β-actin antibody (Thermo-Scientific Pierce, Ireland #10624754) was used to confirm even protein loading. Secondary antibodies used were Goat Anti-Mouse horseradish peroxidase (Thermo-Scientific Pierce, Ireland #31430) and detection was imaged on the Alpha Imager imaging system.
Statistical analysis
Data analysis was performed using the Stata/SE 11 (Stata Corp, College Station, TX) and GraphPad Prism 5 (GraphPad Software, San Diego, CA) statistical software packages. All statistical tests were two sided. P < 0.05 was considered statistically significant. The χ2 and Fisher’s exact tests and univariate and multivariable logistic regression were used to analyze dichotomized data and to calculate odds ratios (ORs). The multivariable models were adjusted for age at diagnosis and race/ethnicity. An interaction test was performed in the logistic regression model to assess statistical interactions between variables. The Mann–Whitney test was used to compare the differences of both plasma cytokine levels and HERV-K Ct-based expression values between groups.
Results
HERV-K gag mRNA is elevated in the PBMC of prostate cancer patients
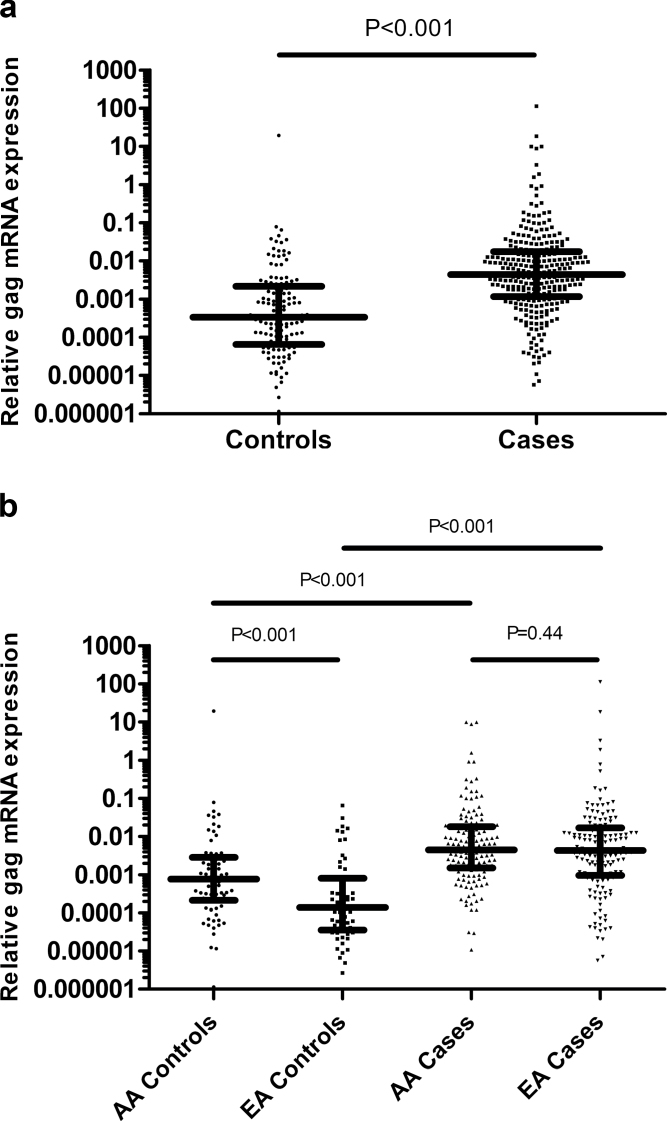
We compared HERV-K gag expression in total RNA isolated from PBMC collected from men without a cancer diagnosis (n = 135) and prostate cancer patients (n = 294) using quantitative real-time PCR. The characteristics of the controls and cases are shown in Supplementary Table 1, available at Carcinogenesis Online. Figure 1A shows that although HERV-K gag mRNA is detectable in the PBMC of both controls and prostate cancer patients, HERV-K gag levels were significantly elevated in patients compared with controls. Univariate and multivariable logistic regression analyses further showed that HERV-K gag mRNA expression is associated with a diagnosis of prostate cancer irrespective of whether the analysis was conducted using gag expression as a continuous variable or comparing high versus low values of HERV-K gag (when using either median or quartile values of HERV-K gag as cutoffs; Table I). These data highlight the robustness of the association between HERV-K marker expression and prostate cancer. For example, an above median gag expression in PBMC was associated with a 6-fold increased odds of having a prostate cancer diagnosis in the multivariable analysis [OR = 6.02 (95% confidence interval 3.73–9.72)], compared with below median gag expression. In addition, our findings revealed a significant dose relationship between expression levels of HERV-K in PBMC and the likelihood of being diagnosed with prostate cancer (Table I). Notable, those men with an HERV-K gag expression in the highest quartile had >12-fold increased odds [OR =12.87 (95% confidence interval 6.3–26.25)] of being diagnosed with prostate cancer compared with men in the lowest quartile of PBMC HERV-K expression. After adjustment for age at diagnosis and race/ethnicity, this relationship remained, and men with HERV-K gag expression levels in the highest quartile had a 17.3-fold increased odds of being diagnosed with prostate cancer in the multivariable logistic regression analysis [OR =17.3 (95% confidence interval 8.1–37.0)]. Next, we examined the effects of treatment on HERV-K expression in the PBMC of prostate cancer patients but found no significant difference in expression between men who had received prior treatment and those without it (Supplementary Figure 1, available at Carcinogenesis Online). Additional analyses showed that HERV-K gag expression in the PBMC of prostate cancer patients did not correlate with PSA levels at diagnosis (Spearman’s rho = −0.01, P = 0.92). Thus, they appear to be independent markers. We also examined if the time from diagnosis to blood draw had an effect on HERV-K gag expression in patients but did not find a significant relationship between these two variables (Supplementary Figure 2, available at Carcinogenesis Online). Likewise, HERV-K gag expression did not differ significantly depending on Gleason score at diagnosis (Supplementary Figure 3, available at Carcinogenesis Online). In contrast, an analysis by stage showed that HERV-K gag expression was significantly higher in stage II patients than in stage I (P = 0.034; Supplementary Figure 4, available at Carcinogenesis Online), whereas no difference was found between patients with low-stage disease (stages I or II) and the few patients with high-stage disease (stages III or IV).
Fig. 1.
Comparison of HERV-K gag mRNA expression in PBMC isolated from the blood of healthy controls (n = 135) and prostate cancer patients (n = 294) using previously published gag mRNA primer set. (a) HERV-K gag mRNA was significantly elevated in PBMC isolated from prostate cancer patients compared with the control population (Mann–Whitney test, P < 0.001). (b) HERV-K gag mRNA is significantly elevated in PBMC from African-American (AA) controls compared with European-American (EA) controls (Mann–Whitney test, P < 0.001), whereas there was no significant difference in the levels of HERV-K gag between African-American and European-American patients (Mann–Whitney test, P = 0.44). HERV-K gag was significantly elevated in patients compared with population controls in both African-Americans (Mann–Whitney test, P < 0.001) and European-Americans (Mann–Whitney test, P < 0.001).
Table I.
Logistic regression analysis of the association between HERV-K gag mRNA levels in PBMC and a prostate cancer diagnosis
| Univariate analysisa | Multivariable analysis Ib | Multivariable analysis IIc | Multivariable analysis IIId | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% confidence interval) | P value | n | OR (95% confidence interval) | P value | n | OR (95% confidence interval) | P value | n | OR (95% confidence interval) | P value | w | |
| Logistic regression with HERV-K gag coded as a continuous variable (per Ct value) | ||||||||||||
| Gag mRNA | 1.33 (1.24–1.43) | <0.0001 | 429 | 1.36 (1.26–1.47) | <0.0001 | 427 | 1.49 (1.35–1.65) | <0.0001 | 358 | 1.67 (1.42–1.96) | <0.0001 | 148 |
| Logistic regression comparing high versus low HERV-K gag expressione | ||||||||||||
| Low gag | 1 | 1 | 1 | 1 | ||||||||
| High gag | 5.85 (3.67–9.36) | <0.0001 | 429 | 6.02 (3.73–9.72) | <0.0001 | 427 | 10.35 (5.43–19.8) | <0.0001 | 358 | 24.52 (8.61–69.8) | <0.0001 | 148 |
| Logistic regression to evaluate a dose response effect after stratification of HERV-K (HML-2) gag expression into quartilesf | ||||||||||||
| 1st quartile | 1 | 48 | 1 | 48 | 1 | 42 | 1 | 30 | ||||
| 2nd quartile | 1.38 (0.61–3.11) | 0.433 | 57 | 1.58 (0.69–3.62) | 0.280 | 57 | 1.53 (0.61–3.81) | 0.363 | 43 | 2.25 (0.50–10.1) | 0.288 | 20 |
| 3rd quartile | 4.14 (1.98–8.67) | <0.0001 | 98 | 5.90 (2.69–13.0) | <0.0001 | 96 | 7.15 (3.01–16.9) | <0.0001 | 80 | 7.80 (2.05–29.7) | 0.003 | 32 |
| 4th quartile | 12.87 (6.30–26.3) | <0.0001 | 226 | 17.3 (8.07–37.0) | <0.0001 | 226 | 36.4 (14.7–90.2) | <0.0001 | 193 | 94.9 (22.0–408) | <0.0001 | 66 |
| P trend | <0.0001 | P trend | <0.0001 | P trend | <0.0001 | P trend | <0.0001 | |||||
a294 cases and 135 controls.
bAdjusted for age at diagnosis and race/ethnicity.
cAdjusted for age at diagnosis, race/ethnicity and smoking status.
dAdjusted for age at diagnosis, race/ethnicity, smoking status and plasma IFNγ and IP10.
eHERV-K gag levels were dichotomized using the median expression in the control population.
fHERV-K gag levels were divided into quartiles based on HERV-K gag quartile distribution in the control population.
HERV-K gag mRNA is elevated in African-American controls compared with European-American controls
African-American men are at an increased risk of developing prostate cancer and are also at an increased risk of diagnosis with advanced disease, compared with European-American men. We therefore examined whether race/ethnicity influences the association of HERV-K gag mRNA with prostate cancer. The analysis showed that gag expression is associated with a prostate cancer diagnosis in both population groups (Supplementary Table 2, available at Carcinogenesis Online), but as shown in Figure 1B, African-American population controls expressed significantly more HERV-K gag mRNA in their PBMC than European-American population controls. Levels of HERV-K gag message were not different by race/ethnicity in the case population, suggesting that aberrant HERV-K gag expression in PBMC of prostate cancer patients is similar irrespective of race/ethnicity.
Association of HERV-K gag mRNA with a prostate cancer diagnosis increases with age and is most robust in older men
The sensitivity of PSA testing decreases with age, partially due to an increase in the prevalence of benign prostate hyperplasia, which also elevates PSA. To determine whether HERV-K gag expression is predictive of prostate cancer across all age group, we performed a stratified analysis comparing the association of HERV-K gag according to age groups. We stratified men into three similar-sized groups, ages 41–59 (n = 128), ages 60–69 (n = 179) and ages ±70 (n = 120). The findings from the univariate logistic regression analysis in Table II show that although HERV-K gag is predictive of prostate cancer across all ages, the strength of the association increased with age. HERV-K gag was most predictive in men aged ±70, indicating that combining HERV-K testing with PSA testing may improve the efficacy of prostate cancer detection in these older men. This remained true in the multivariable analysis after adjusting for race/ethnicity. To determine if there is a modifying effect of age at diagnosis on the association between HERV-K gag expression and prostate cancer, we performed a statistical interaction test, first on the three individual age categories with HERV-K gag quartile expression but found that the P interaction was not significant (P values ranged between 0.40 and 0.96). We then dichotomized age into high/low with the median as cutoff and assessed the modifying effect of the age variable on the association between HERV-K gag expression and prostate cancer within each HERV-K expression quartile (quartiles as described in Table II). The test showed that age at diagnosis has a statistically significant modifying effect on the association of HERV-K gag with prostate cancer within each HERV-K gag quartile expression (Q2 P interaction = 0.049; Q3 P interaction = 0.019; Q4 P interaction = 0.017). These results are suggestive, but not definitive, of a stronger association of HERV-K with prostate cancer with increasing age.
Table II.
Age-stratified logistic regression analysis of the association between HERV-K gag mRNA levels and prostate cancer
| Univariate analysis | Multivariable analysisa | ||||||
|---|---|---|---|---|---|---|---|
| OR (95% confidence interval) | P value | N | OR | 95% confidence interval | P value | n | |
| Men aged 41–59 | |||||||
| 1st quartileb | 1 | 12 | 1 | 12 | |||
| 2nd quartile | 1.12 (0.25–4.91) | 0.880 | 18 | 1.22 (0.27–5.60) | 0.795 | 18 | |
| 3rd quartile | 3.15 (0.76–13.00) | 0.113 | 26 | 4.67 (1.02–21.38) | 0.047 | 26 | |
| 4th quartile | 9.8 (2.56–37.55) | 0.001 | 72 | 11.96 (2.92–48.92) | 0.001 | 72 | |
| P trend | <0.0001 | P trend | <0.0001 | ||||
| Men aged 60–69 | |||||||
| 1st quartile | 1 | 23 | 1 | 23 | |||
| 2nd quartile | 1.07 (0.32–3.63) | 0.912 | 22 | 1.54 (0.42–5.54) | 0.513 | 22 | |
| 3rd quartile | 4.15 (1.43–12.04) | 0.009 | 45 | 5.53 (1.79–17.09) | 0.003 | 45 | |
| 4th quartile | 13.29 (4.58–38.57) | <0.0001 | 89 | 19.52 (6.13–62.12) | 0.001 | 89 | |
| P trend | <0.0001 | P trend | <0.0001 | ||||
| Men aged ±70 | |||||||
| 1st quartile | 1 | 13 | 1 | 13 | |||
| 2nd quartile | 3.0 (0.40–18.24) | 0.233 | 17 | 2.96 (0.48–18.06) | 0.239 | 17 | |
| 3rd quartile | 8.25 (1.49–45.42) | 0.015 | 25 | 9.07 (1.59–51.71) | 0.013 | 25 | |
| 4th quartile | 22.00 (4.33–111.7) | <0.0001 | 65 | 23.94 (4.57–125.4) | <0.0001 | 65 | |
| P trend | <0.0001 | P trend | <0.0001 | ||||
aAdjusted for race/ethnicity.
bHERV-K gag levels were divided into quartiles based on HERV-K gag quartile distributions in the control population.
Association of HERV-K gag mRNA expression with the risk of prostate cancer is modified by smoking status
The smoking status was available for 270 cases and 89 population-based controls in this cohort (total n = 359), categorized as current smoker, former smoker or never smoker. Moreover, pack-years smoked information was available for 358 of them. Although smoking status or pack-years did not directly correlate with the level of HERV-K gag expression in the blood samples, smoking status was found to modify the association of HERV-K gag expression with a diagnosis of prostate cancer. As shown in Table III, the strength of the association between HERV-K gag and disease was found to be highest in current smokers, whereas lowest in never smokers. When stratified by pack-years of smoking, there was an increase in the strength of the association between HERV-K gag expression and disease with an increase in pack-years of tobacco exposure (Supplementary Table 3, available at Carcinogenesis Online). Because of these modifying effects of smoking on the association between HERV-K gag expression and disease, we performed an interaction analysis. A test for interaction between smoking status and HERV-K gag expression in prostate cancer development with HERV_K gag expression categorized as high/low (dichotomized at the median) indicated no statistically significant interaction between HERV-K gag and former smoker status (P interaction = 0.40) or HERV-K and current smoker status (P interaction = 0.26). We also performed an interaction test between pack-years exposure (dichotomized at the median) and gag expression levels (dichotomized at the median). The detected interaction (or modifying effect of tobacco exposure) did not reach statistical significance (P interaction = 0.096). Therefore, although HERV-K gag in PBMC is more closely associated with prostate cancer among heavy smokers than light or never smokers, there may not be a direct interaction between HERV-K expression and smoking exposure on prostate cancer risk. A larger and more appropriately powered study would be required to test this and show that such an interaction exists.
Table III.
Logistic regression analysis of the association between HERV-K gag mRNA levels and prostate cancer by smoking status
| Univariate analysis | Multivariable analysisa | |||||
|---|---|---|---|---|---|---|
| OR (95% confidence interval) | P value | N | OR (95% confidence interval) | P value | n | |
| Never smokers | ||||||
| Gag mRNA (continuous) | 1.41 (1.21–1.63) | <0.0001 | 121 | 1.43 (1.22–1.67) | <0.0001 | 121 |
| Gag mRNAb (median) | 8.07 (3.03–21.5) | <0.0001 | 121 | 9.38 (3.33–26.4) | <0.0001 | 121 |
| Former smokers | ||||||
| Gag mRNA (continuous) | 1.44 (1.27–1.63) | <0.0001 | 173 | 1.48 (1.29–1.69) | <0.0001 | 173 |
| Gag mRNA (median) | 13.8 (6.27–30.4) | <0.0001 | 173 | 17.4 (7.29–41.8) | <0.0001 | 173 |
| Current smokers | ||||||
| Gag mRNA (continuous) | 1.93 (1.31–2.87) | 0.001 | 64 | 2.35 (1.33–4.16) | 0.003 | 64 |
| Gag mRNA (median) | 25.5 (4.56–142) | 0.001 | 64 | 30.2 (4.40–208) | 0.001 | 64 |
aAdjusted for age at diagnosis and race/ethnicity.
bHERV-K gag levels were dichotomized using the median expression in the control population.
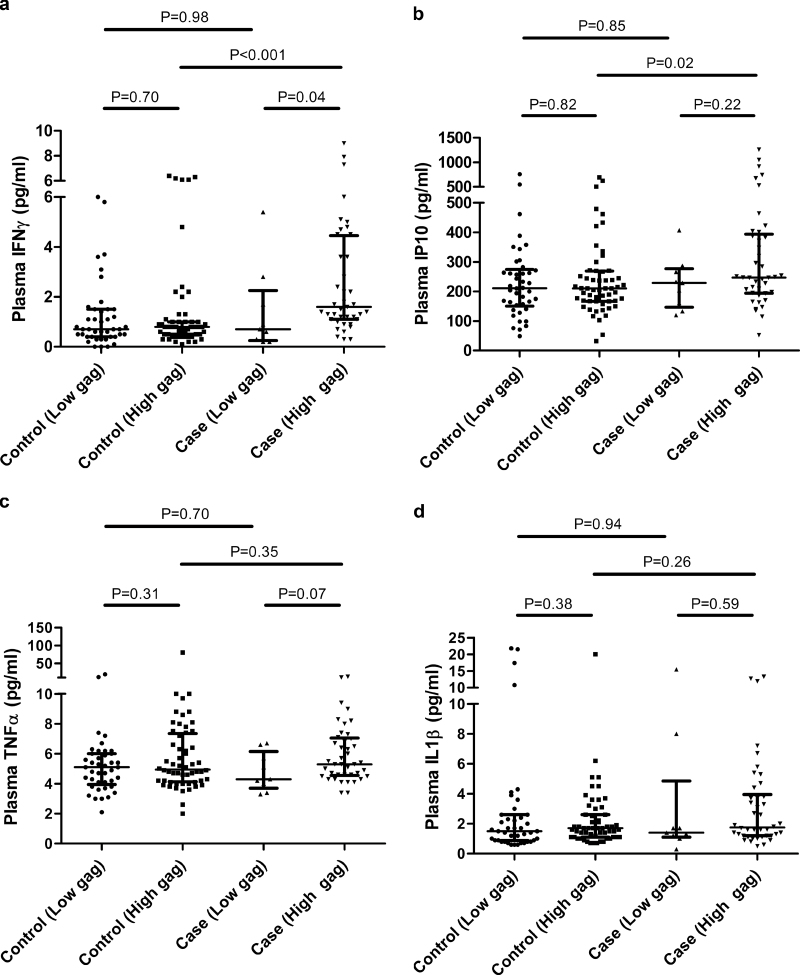
Prostate cancer patients with elevated HERV-K gag show increased serum expression of viral response IFNγ and IP10 expressions
To determine whether elevation of HERV-K gag mRNA in PBMC from prostate cancer results in the secretion of viral response INFs and inflammatory cytokines, we measured the expression of IFNγ, IP10, tumor necrosis factor-α and IL-1β in plasma of a subset of prostate cancer cases and controls. We found that the plasma levels of the viral response cytokines, IFNγ (Figure 2A) and its downstream effector IP10 (Figure 2B) were significantly elevated in the case population with high levels of HERV-K gag mRNA expression. We did not find the same elevated levels of these cytokines in cases with low gag expression, or among the control population. Neither tumor necrosis factor-α (Figure 2C) or IL-1β (Figure 2D) was elevated in patients with high levels of HERV-K gag mRNA. Multivariable logistic regression (Model 3) in Table I demonstrates that after adjusting for IFNγ and IP10 levels, HERV-K remains significantly associated with prostate cancer. Additionally we examined the impact of IFNγ or IP10 on the association of HERV-K gag mRNA with prostate cancer (Supplementary Table 4, available at Carcinogenesis Online). The univariate logistic regression showed that high levels of HERV-K gag mRNA are more strongly associated with prostate cancer in patients with high IFNγ or IP10, compared with those with low IFNγ or IP10. Notable, IFNγ or IP10 level above the median in the absence of high levels of HERV-K gag was not found to be associated with prostate cancer. These associations upheld in the multivariable analysis after adjusting for age at diagnosis, race/ethnicity and smoking status.
Fig. 2.
Association between high HERV-K gag mRNA expression and elevated plasma IFNγ and IP10 levels. Plasma IFNγ (a) and its downstream mediator IP10 (b) were significantly increased in patients with high gag expression compared with controls with high gag expression (Mann–Whitney test IFNγ, P < 0.001; Mann–Whitney test IP10, P = 0.02). Additionally, cases with high gag expression had significantly higher levels of plasma IFNγ compared with cases with low gag expression. No significant association of gag expression with tumor necrosis factor-α (c) or IL-1β (d) was observed.
Detection of HERV-K env type I and type II mRNA transcripts in a subset of the PBMC
We designed primers specific to the env of HERV-K type I and HERV-K type II viruses. Type I and type II are distinguished by a 292 bp deletion in the env gene (type I). Supplementary Figure 5, available at Carcinogenesis Online, shows that both type I and type II env mRNA were elevated in the PBMC of the cancer patients, indicating that the HERV-K reactivation may arise from multiple HERV-K loci in prostate cancer patients.
HERV-K env protein is expressed in prostate tumors
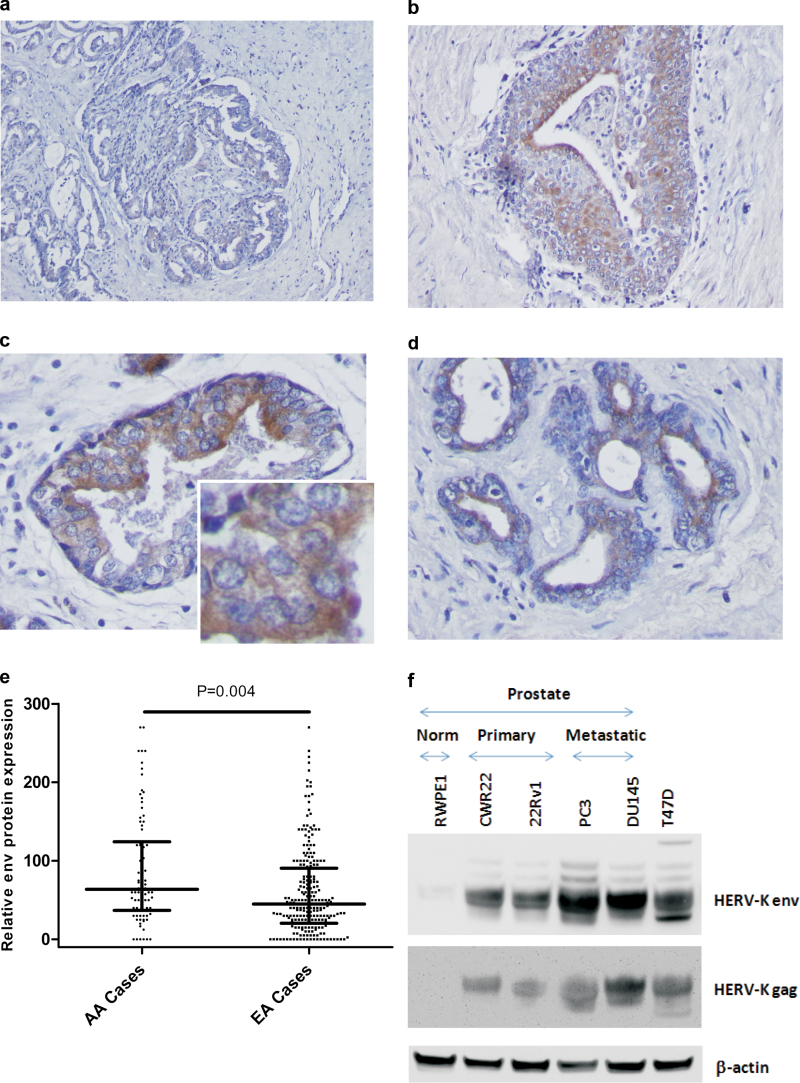
We used IHC to assess whether prostate tumors express the HERV-K env protein using whole tissue sections to examine protein localization and a tissue microarray for scoring. Consistent with our findings analyzing HERV-K expression levels in PBMC, expression of the HERV-K env protein in tumors varied considerably among patients. Figure 3A–D shows that HERV-K env protein expression is cytoplasmic and membrane located, and also localizing toward the lumen of the gland. Env protein expression levels were significantly higher in African-American than European-American patients (Figure 3E), and 61% of the African-American patients presented with tumors that had high env expression (above median), whereas only 40% of the tumors from European-American patients fell into the same category (P < 0.001). In contrast, we did not detect aberrant HERV-K env expression in prostate tissues from patients with benign prostate hyperplasia (data not shown). Western blot analysis of cell extracts further corroborated the presence of both HERV-K gag and HERV-K envelope protein in human prostate cancer cells, whereas expression was low to absent in the non-tumorigenic RWPE1 cells (Figure 3F).
Fig. 3.
HERV-K expression in human prostate adenocarcinomas. Shown is IHC analysis of four invasive adenocarcinomas for expression of HERV-K envelope (env) protein using the monoclonal anti-envelope antibody, 6H5. (a) Scattered positivity for HERV-K env expression in the tumor epithelium, showing a low to moderate antigen expression as indicated by the brown chromogen deposits. (b–d) Locally intensive staining for env expression in the tumor epithelium. Staining shows a cytosolic to membrane distribution with a more intensive staining of cancer cells toward the luminal side of the cancerous gland (c and d). a and b: Magnification: ×100. c and d: Magnification: ×400. Inset: higher resolution image for the env protein-positive tumor epithelium. Counterstain: Hematoxylin. (e) Immunostaining for env protein in human prostate tumors using a tissue microarray that included tumors from both African-American (n = 105) and European-American patients (n = 272). On average, tumors from African-American patients showed a higher expression of the HERV-K env protein than tumors from European-Americans (Student’s t-test, P < 0.001). (f) HERV-K gag and env protein expression were detected in human prostate cancer cell lines (CWR22, 22Rv1, PC-3 and DU145) but were not detected in the non-tumorigenic human prostate cell line, RWPE1. An extract from the HERV-K-positive T47D human breast cancer cell line were included as a positive control.
Discussion
Our study makes the novel observation that HERV-K mRNA expression is aberrantly increased in PBMC from prostate cancer patients compared with healthy male controls. Thus, the evaluation of blood-based HERV-K expression may serve an early disease detection biomarker in prostate cancer. Moreover, the expression as of HERV-K as an early disease biomarker may perform better in older than younger men, whereas the sensitivity of PSA testing decreases with age. Thus, combining HERV-K testing with PSA testing may improve the efficacy of prostate cancer detection in this age group. Other studies have observed the presence of autoantibodies to HERV-K gag in sera from prostate cancer patients and described the aberrant expression of HERV-K mRNA and proteins in prostate cancer tissues (5,6,24). Hence, our observation of aberrantly increased blood-based expression of HERV-K in prostate cancer patients is consistent with these previous findings.
PSA testing is the gold standard for prostate cancer screening. The traditional cutoff for an abnormal PSA level is 4.0ng/ml. One of the major problems is that PSA has poor discriminating ability in men with symptomatic benign prostate hyperplasia versus those with prostate cancer (25). Additionally, PSA levels rise as men grow older, which can lead to increased false positive PSA tests and unnecessary biopsies (26). Consistent with these data, PSA levels displayed a slight albeit non-significant increase with age at diagnosis in our patient cohort (Spearman’s rho = 0.09, P = 0.13), whereas HERV-K gag expression did not increase with either the age at diagnosis for the cases (Spearman’s rho = −0.01, P = 0.91) or the age of recruitment for the controls (Spearman’s rho = −0.04, P = 0.61). Currently, the American Urological Association only recommends the use of PSA screening in asymptomatic men aged between 55 and 69 (27). Our data suggest that blood-based HERV-K is a candidate biomarker for the detection of prostate cancer, potentially with a focus on older men. Future research must determine its predictive value in conjunction with PSA testing.
Incidence and mortality rates of prostate cancer are significantly higher in men of African ancestry compared with men from other population groups in the USA, the Caribbean and the United Kingdom (28). This cancer health disparity may relate to unknown causative factors that influence disease pathology in men of African ancestry and induce a more aggressive disease among them. Therefore, we examined HERV-K expression in tumors and blood samples from both African-American and European-American patients. Indeed, we found that HERV-K expression in prostate tumors is significantly higher in the African-American patients, compared with European-American patients, based on the immunohistochemical analysis of HERV-K envelope protein expression in these tissues. However, we did not find that HERV-K message levels are significantly different in PBMC from these two patient groups. Accordingly, HERV-K as biomarker should perform similarly among these patients. However, we found that baseline levels of HERV-K mRNA in PBMC of healthy controls were greater in African-Americans than European-Americans. Currently, we do not know why African-American men may express higher baseline levels of HERV-K, nor what effects, if any, there may be. However, it is known that retroviral reactivation is induced by both hormone exposure and stress signaling (13,29,30), and these signaling pathways could become more commonly activated in African-American men. Additionally, Macfarlane and Simmonds reported that the frequency of allelic variation in the various HERV-K germline loci is greater in African populations than populations from Europe and Asia (31). Furthermore, others reported significantly higher insertion frequencies of HERV-K113 (21%) and K115 (35%) in African-Americans compared with European-Americans (K113 9% and K115 6%) within the USA (32). Because African-Americans may have inherited a greater number of polymorphic HERV-K loci than European-Americans, this would increase the number of HERV-K loci that are potentially transcribed, leading to a higher baseline expression of HERV-K encoded genes from these loci.
Cigarette smoking is not associated with early disease development in prostate cancer (33,34), but it is associated with an increased risk of fatal prostate cancer (33,35,36). It is also associated with increased risk of biochemical recurrence and metastasis (37). Gabriel et al. (38) showed previously that exposure of both normal human dermal fibroblasts and benign human uroepithelium to urine from current smokers increased the transcription of HERV-K (HML-6), indicating that HERV-K may be induced by tobacco metabolites; however, the authors did not specifically test the HERV-K family. Because of these observations, we decided to evaluate the influence of smoking on the application of blood-based HERV-K expression as a disease biomarker. We had smoking status and pack-year information for a large subset of subjects in our case–control study and therefore assessed the impact of smoking status on the association of HERV-K with prostate cancer. This analysis led to the finding that HERV-K was more predictive of prostate cancer in current smokers, and the association increased in strength with increased pack-years smoked. The underlying mechanism for this modifying effect of smoking on the relationship between HERV-K and prostate cancer remains unclear, and we could not demonstrate a statistically significant interaction between smoking, HERV-K and risk of prostate cancer in this study. Yet, we think our observation could be of particular significance because those patients who are current smokers tend to develop a more aggressive disease than other patients, as already pointed out by us, and improved disease detection at an early stage for this patient group may have a significant impact in reducing disease mortality.
We also observed that elevated HERV-K mRNA in the PBMC of prostate cancer patients was associated with increased serum IFNγ and its effector IP10. It has been previously reported that patients with HERV-K-positive tumors exhibit a humoral response to HERV-K. Rakoff-Nahoum et al. (39) identified HERV-K-specific T cells in seminoma patients, which display elevated IFNγ secretion in response to HERV-K gag peptides. Wang-Johanning et al. (40) also showed that breast cancer patients mount anti-HERV-K responses, including production of anti-HERV-K(HML-2) env immunoglobulin G, production of IFNγ and a T-helper cell cytokine response signature including increased production of IL-2, IL-6, IL-8 and IP10 during in vitro stimulation of breast cancer PBMC with HERV-K antigen. In this context, the finding of an INF response in patients with an elevated blood-based HERV-K message is consistent with a humoral response to HERV-K that was observed by others, indicating aberrant HERV-K expression in prostate cancer patients not only affects tumor immunobiology but also triggers a systemic antiviral response. The relationship of this antiviral response with disease outcome is still largely unknown, but Reis et al. (5) observed that 6.8% of patients with prostate cancer had serum antibodies to the gag-HERV-K protein encoded by the ch22q11.23 locus, and these patients tended to have a more aggressive disease and a higher disease mortality. We previously reported differences in the prostate tumor immunobiology between African-American and European-American men (41). Prostate tumors from African-American men were characterized by the activation of immune response and host defense pathways. Johnston et al. (42) reported that activated macrophages display increased expression of HERV-K, HERV-W and HERV-H mRNA, indicating that HERV reactivation may occur as a consequence of an elevated immune activity. A frequent activation of immune response pathways in tumors from African-American men may therefore contribute to the increased expression of HERV-K env protein in these tumors that we observed in this study.
One of the limitations of this study is our inability to tell where exactly the elevated HERV-K expression arises from in our PBMC from prostate cancer. HERV-K has previously been reported as being expressed at basal levels in healthy PBMC, with aberrant expression occurring in leukemia cells (43–45). However, circulating tumor cells (CTCs) are detectable in the blood of prostate cancer patients (~2–8 per 7.5 ml of blood), with higher levels of CTCs correlating with bone and visceral metastasis (46). Prostate CTCs reflect the biology of the cancer with the CTCs gene signatures switching from AR-on to AR-off within the first month of androgen deprivation therapy, indicating their sensitivity as indicators of tumor response to treatment (47). When isolating PBMC from blood, CTCs may also be pulled down in the PBMC fraction (48). Therefore, PBMC with high levels of HERV-K in prostate cancer patients may potentially reflect HERV-K-positive CTCs. Another potential source of HERV-K is from exosomes secreted by tumors. Exosomes are nanoscale membrane vesicles that are secreted from cells and are thought to be important intercellular communicators or, in a cancer setting, drivers of metastatic spread (49). A recent study has now implicated HERVs in this process, with the finding that HERV-K mRNA is selectively packaged into tumor exosomes and that this genetic material can be transferred to normal cells (50). Exosomes can be found abundantly in the blood of patients and may contribute to tumor dissemination (51). Future work will focus on investigating these options and identifying the specific HERV-K loci responsible for elevated HERV-K in the PBMC of prostate cancer patients and will also evaluate the relative contribution of CTCs and exosomes to the HERV-K expression signal in PBMC from cancer patients.
In summary, we made the novel observation that blood-based HERV-K expression is a candidate early detection biomarker for prostate cancer that may specifically improve disease detection among older men. Furthermore, we obtained evidence that this test would perform similarly in African-Americans and European-Americans. Lastly, aberrant HERV-K expression seems to occur more frequently in both healthy African-Americans and African-Americans with prostate cancer than in their European-American counterparts. Although intriguing, these data need to be further studied.
Supplementary material
Supplementary Tables 1–4 and Figures 1–5 can be found at http://carcin.oxfordjournals.org/
Funding
The Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research , USA; the Galway University Foundation, Ireland (RNR1008 to F.J.S. and S.A.G.); the Health Research Board of Ireland Clinical Research Facility, Galway, Ireland (RSU004 to R.F.D. and F.J.G.); the Department of Defense Breast Cancer Research Program, USA (BC113114 to F.W.J.); the National Cancer Institute, National Institutes of Health, USA (U01CA171146 to R.G.); the Department of Defense Prostate Cancer Research Program, USA (PC081609 to R.G.); the Breast Cancer Campaign, UK (2013MayPR019 to S.A.G. and F.J.S.); the Irish Cancer Society, Ireland (PCT13MCD to S.A.G. and F.J.S.).
Conflict of Interest Statement: None declared.
Supplementary Material
Acknowledgement
We would like to thank Dr Aideen Ryan (National University of Ireland Galway) for her help with preparation of images for publication.
Glossary
Abbreviations:
- CTC
circulating tumor cell
- INF
interferon
- HERV
human endogenous retroviruses
- IHC
immunohistochemistry
- IL
interleukin
- IRB, Institutional Review Board; mRNA
messenger RNA
- NCI
National Cancer Institute
- OR
odds ratio
- PBMC
peripheral blood mononuclear cells
- PSA
prostate-specific antigen.
References
- 1. Siegel R., et al. (2013). Cancer statistics, 2013. CA. Cancer J. Clin., 63, 11–30 [DOI] [PubMed] [Google Scholar]
- 2. Grönberg H. (2003). Prostate cancer epidemiology. Lancet, 361, 859–864 [DOI] [PubMed] [Google Scholar]
- 3. De Marzo A.M., et al. (2007). Inflammation in prostate carcinogenesis. Nat. Rev. Cancer, 7, 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sutcliffe S., et al. (2008). Inflammation and prostate cancer: a focus on infections. Curr. Urol. Rep., 9, 243–249 [DOI] [PubMed] [Google Scholar]
- 5. Reis B.S., et al. (2013). Prostate cancer progression correlates with increased humoral immune response to a human endogenous retrovirus GAG protein. Clin. Cancer Res., 19, 6112–6125 [DOI] [PubMed] [Google Scholar]
- 6. Goering W., et al. (2011). Selective changes of retroelement expression in human prostate cancer. Carcinogenesis, 32, 1484–1492 [DOI] [PubMed] [Google Scholar]
- 7. Perot P., et al. (2013). Microarray-based Identification of Individual HERV Loci Expression: Application to Biomarker Discovery in Prostate Cancer. J Vis Exp, e50713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bannert N., et al. (2004). Retroelements and the human genome: new perspectives on an old relation. Proc. Natl Acad. Sci. USA, 101(suppl 2), 14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lander E.S., et al. ; International Human Genome Sequencing Consortium. (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 10. Schulz W.A., et al. (2006). Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol., 310, 211–250 [DOI] [PubMed] [Google Scholar]
- 11. Moyes D., et al. (2007). Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet., 23, 326–333 [DOI] [PubMed] [Google Scholar]
- 12. Bannert N., et al. (2006). The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet., 7, 149–173 [DOI] [PubMed] [Google Scholar]
- 13. Hohn O., et al. (2013). HERV-K(HML-2), the Best Preserved Family of HERVs: Endogenization, Expression, and Implications in Health and Disease. Front. Oncol., 3, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruprecht K., et al. (2008). Endogenous retroviruses and cancer. Cell. Mol. Life Sci., 65, 3366–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Downey R.F., et al. (2014). Human endogenous retrovirus K and cancer: innocent bystander or tumorigenic accomplice? International Journal of Cancer. :10.1002/ijc.29003. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agoni L., et al. (2013). Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines. Front. Oncol., 3, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liew C.C., et al. (2006). The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J. Lab. Clin. Med., 147, 126–132 [DOI] [PubMed] [Google Scholar]
- 18. Mohr S., et al. (2007). The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol. Med., 13, 422–432 [DOI] [PubMed] [Google Scholar]
- 19. Hudson R.S., et al. (2012). MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res., 40, 3689–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Contreras-Galindo R., et al. (2008). Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol., 82, 9329–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bookout A.L., et al. (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal., 1, e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang-Johanning F., et al. (2012). Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J. Natl Cancer Inst., 104, 189–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang-Johanning F., et al. (2014). Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int. J. Cancer, 134, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishida T., et al. (2008). Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun., 8, 15. [PMC free article] [PubMed] [Google Scholar]
- 25. You J., et al. (2010). Innovative biomarkers for prostate cancer early diagnosis and progression. Crit. Rev. Oncol. Hematol., 73, 10–22 [DOI] [PubMed] [Google Scholar]
- 26. Gulati R., et al. (2011). What if i don’t treat my PSA-detected prostate cancer? Answers from three natural history models. Cancer Epidemiol. Biomarkers Prev., 20, 740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carter H.B. (2013). American Urological Association (AUA) guideline on prostate cancer detection: process and rationale. BJU Int., 112, 543–547 [DOI] [PubMed] [Google Scholar]
- 28. Martin D.N., et al. (2013). Biological determinants of health disparities in prostate cancer. Curr. Opin. Oncol., 25, 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang-Johanning F., et al. (2003). Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene, 22, 1528–1535 [DOI] [PubMed] [Google Scholar]
- 30. Serafino A., et al. (2009). The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res., 315, 849–862 [DOI] [PubMed] [Google Scholar]
- 31. Macfarlane C., et al. (2004). Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol., 59, 642–656 [DOI] [PubMed] [Google Scholar]
- 32. Jha A.R., et al. (2009). Cross-sectional dating of novel haplotypes of HERV-K 113 and HERV-K 115 indicate these proviruses originated in Africa before Homo sapiens. Mol. Biol. Evol., 26, 2617–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giovannucci E., et al. (1999). Smoking and risk of total and fatal prostate cancer in United States health professionals. Cancer Epidemiol. Biomarkers Prev., 8(Pt 1), 277–282 [PubMed] [Google Scholar]
- 34. Hickey K., et al. (2001). Smoking and prostate cancer. Epidemiol. Rev., 23, 115–125 [DOI] [PubMed] [Google Scholar]
- 35. Huncharek M., et al. (2010). Smoking as a risk factor for prostate cancer: a meta-analysis of 24 prospective cohort studies. Am. J. Public Health, 100, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kenfield S.A., et al. (2011). Smoking and prostate cancer survival and recurrence. JAMA, 305, 2548–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moreira D.M., et al. (2013). Cigarette smoking is associated with an increased risk of biochemical disease recurrence, metastasis, castration-resistant prostate cancer, and mortality after radical prostatectomy: Results from the SEARCH database. Cancer, 120, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gabriel U., et al. (2010). Smoking increases transcription of human endogenous retroviruses in a newly established in vitro cell model and in normal urothelium. AIDS Res. Hum. Retroviruses, 26, 883–888 [DOI] [PubMed] [Google Scholar]
- 39. Rakoff-Nahoum S., et al. (2006). Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res. Hum. Retroviruses, 22, 52–56 [DOI] [PubMed] [Google Scholar]
- 40. Wang-Johanning F., et al. (2008). Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res., 68, 5869–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wallace T.A., et al. (2008). Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res., 68, 927–936 [DOI] [PubMed] [Google Scholar]
- 42. Johnston J.B., et al. (2001). Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann. Neurol., 50, 434–442 [DOI] [PubMed] [Google Scholar]
- 43. Medstrand P., et al. (1992). Expression of human endogenous retroviral sequences in peripheral blood mononuclear cells of healthy individuals. J. Gen. Virol., 73(Pt 9), 2463–2466 [DOI] [PubMed] [Google Scholar]
- 44. Brodsky I., et al. (1993). Expression of human endogenous retrovirus (HERV-K) in chronic myeloid leukemia. Leuk. Lymphoma, 11(suppl 1), 119–123 [DOI] [PubMed] [Google Scholar]
- 45. Depil S., et al. (2002). Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia, 16, 254–259 [DOI] [PubMed] [Google Scholar]
- 46. Thalgott M., et al. (2013). Detection of circulating tumor cells in different stages of prostate cancer. J. Cancer Res. Clin. Oncol., 139, 755–763 [DOI] [PubMed] [Google Scholar]
- 47. Miyamoto D.T., et al. (2012). Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov., 2, 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kallergi G., et al. (2007). Phosphorylation of FAK, PI-3K, and impaired actin organization in CK-positive micrometastatic breast cancer cells. Mol. Med., 13, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Théry C., et al. (2002). Exosomes: composition, biogenesis and function. Nat. Rev. Immunol., 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 50. Balaj L., et al. (2011). Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun., 2, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang H.G., et al. (2014). Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am. J. Pathol., 184, 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.