Summary
This study comprehensively assessed whether variation in prostaglandin synthesis and related pathways influences CRC risk by examining associations between 192 SNPs and two functional VNTRs within 17 candidate genes.
Abstract
Although use of non-steroidal anti-inflammatory drugs (NSAIDs) generally decreases colorectal cancer (CRC) risk, inherited genetic variation in inflammatory pathways may alter their potential as preventive agents. We investigated whether variation in prostaglandin synthesis and related pathways influences CRC risk in the Colon Cancer Family Registry by examining associations between 192 single nucleotide polymorphisms (SNPs) and two variable nucleotide tandem repeats (VNTRs) within 17 candidate genes and CRC risk. We further assessed interactions between these polymorphisms and NSAID use on CRC risk. Using a case-unaffected-sibling-control design, this study included 1621 primary invasive CRC cases and 2592 sibling controls among Caucasian men and women aged 18–90. After adjustment for multiple comparisons, two intronic SNPs were associated with rectal cancer risk: rs11571364 in ALOX12 [ORhet/hzv = 1.87, 95% confidence interval (CI) = 1.19–2.95, P = 0.03] and rs45525634 in PTGER2 (ORhet/hzv = 0.49, 95% CI = 0.29–0.82, P = 0.03). Additionally, there was an interaction between NSAID use and the intronic SNP rs2920421 in ALOX12 on risk of CRC (P = 0.03); among those with heterozygous genotypes, risk was reduced for current NSAID users compared with never or former users (ORhet = 0.60, 95% CI = 0.45–0.80), though not among those with homozygous wild-type or variant genotypes. The results of this study suggest that genetic variation in ALOX12 and PTGER2 may affect the risk of rectal cancer. In addition, this study suggests plausible interactions between NSAID use and variants in ALOX12 on CRC risk. These results may aid in the development of genetically targeted cancer prevention strategies with NSAIDs.
Introduction
Inflammation has long been recognized as an important contributor to colorectal cancer (CRC) risk (1–3). For example, chronic inflammatory conditions, such as inflammatory bowel disease, are associated with increased CRC risk and prior studies suggest that both inflammatory bowel disease-associated and sporadic CRC share mechanisms linked to inflammation (3–5). Although the exact mechanisms that contribute to this process are still unclear (3,4), inflammation may enable carcinogenesis by contributing to cell proliferation and survival, angiogenesis and genetic mutations (4,6,7). Use of non-steroidal anti-inflammatory drugs (NSAIDs), which inhibit inflammation, decreases CRC risk (8–12); however, inherited variation in genes that affect NSAID metabolism may alter their potential as preventive agents.
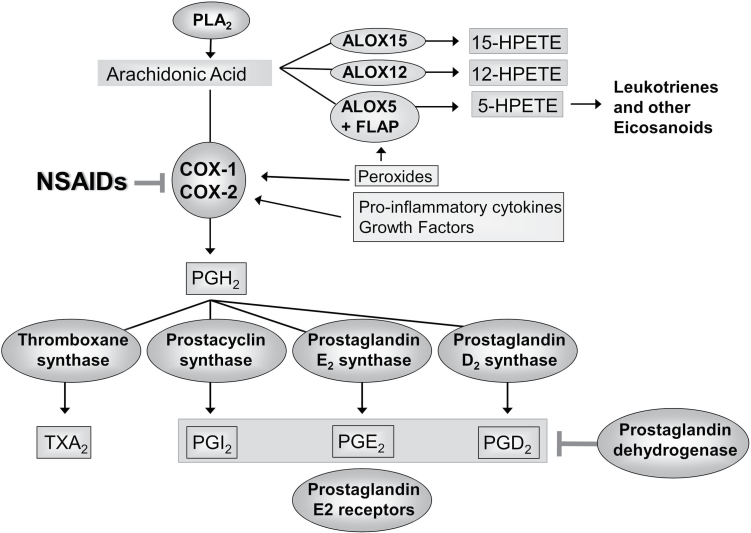
NSAIDs inhibit the cyclooxygenase (COX) enzymes, which control the conversion of arachidonic acid (AA) to prostaglandins and thromboxane (Figure 1) (2,12). COX1 and COX2 (encoded by PTGS1 and PTGS2) are the main targets of traditional NSAIDs (such as aspirin and ibuprofen) as well as COX2-specific inhibitors. They are the main enzymes responsible for reducing AA to prostaglandin H2, which is metabolized to a substrate for thromboxane synthase, prostacyclin synthase (PGI) and prostaglandin synthase (PG). Via these synthases, prostaglandin H2 is metabolized into thromboxane A2, PGI2, prostaglandin E2 (PGE2) and prostaglandin D2, which modulate important cellular functions by binding to their respective receptors: TP to thromboxane A2, IP to PGI2, EP (EP1-4) to PGE2 and DP to prostaglandin D2 (12–14). Arachidonate lipoxygenases (ALOXs) convert AA into leukotrienes and other eicosanoids that regulate inflammatory processes (15,16). Additionally, eicosanoid levels differ between normal and colon cancer tissues, with PGE2 levels increased and PGI2 levels decreased in colon cancer samples (17).
Fig. 1.
Prostaglandin synthesis pathway. The COX enzymes COX1 and COX2, which reduce arachidonic acid to prostaglandin H2 (PGH2), are the main targets of COX2-specific inhibitors and NSAIDs. PGH2 becomes a substrate for thromboxane, prostacyclin and prostaglandin synthases, which are converted into thromboxane A2 (TXA2), PGI2, PGE2 and prostaglandin D2 (PGD2). Arachidonic lipoxygenesases and hydroperoxides (HPETEs) convert arachidonic acid into leukotrienes and other inflammation-mediating eicosanoids.
Prior studies, including ours, reported associations between polymorphisms in genes involved in inflammation pathways and CRC risk and mortality (18–24). Likewise, numerous epidemiologic studies and randomized trials have shown that NSAID use reduces CRC risk and mortality (8–11,25–27). However, as we have shown previously, genetic variation in NSAID-metabolizing pathways (upstream and downstream) may affect the capacity of NSAIDs to serve as chemopreventive agents (28). Therefore, we investigated whether variation in genes involved in prostaglandin synthesis or related pathways might impact CRC risk. Using data from the population-based Colon Cancer Family Registry (CCFR) (29), we assessed whether single nucleotide polymorphisms (SNPs) and variable nucleotide tandem repeats (VNTRs) were independently associated with CRC risk and whether these associations were modified by pre-diagnostic use of NSAIDs.
Materials and methods
Study population
The study population has been described previously (29). Briefly, cases were men and women recruited from six registry centres and aged 18–90 when diagnosed with primary invasive CRC from 1998–2002. Cases included probands and any affected relatives, and were interviewed within 5 years of diagnosis. CRC included distal (left-sided), proximal (right-sided) and not otherwise specified colon cancers as well as rectal cancers. Controls were siblings not diagnosed with CRC at the time of ascertainment. Individuals whose self-reported sex did not match that determined by genotyping were excluded, as were cases that did not have at least one matched unaffected sibling control. Further, this study included only participants ascertained through population-based recruitment who self-reported as Caucasian, with no known familial adenomatous polyposis and no personal history of ulcerative colitis or Crohn’s disease. Other racial/ethnic groups and clinic-based populations were excluded as sample sizes were too small to examine associations.
Epidemiologic data were collected from CCFR study participants using standard questionnaires on demographic characteristics, medical history, NSAID use, family history of cancer, smoking history, diet, physical activity, height and weight. Blood and tissue samples were collected using standardized procedures across CCFR sites and DNA was extracted from peripheral blood leucocytes. Participants were provided a list of common aspirin and non-aspirin NSAIDs brands and reported their average weekly consumption. Current NSAID use was defined as use two or more times per week for at least 1 month in the 2 years prior to study enrollment. The Institutional Review Boards at each CCFR site approved this study and informed consent was obtained from all study participants.
Polymorphism selection
We selected a total of 220 candidate and tagging SNPs (tagSNPs) and three functional VNTRs in 17 candidate genes involved in prostaglandin synthesis and related pathways. The following genes were included: ALOX12, ALOX5, ALOX5AP, CRP, EGFR, GIPC1, HPGD, PARP12, PKN1, PLA2G1B, PTGER1, PTGER2, PTGER3, PTGER4, PTGES, PTGIS and TBXAS1. The procedure for tagSNP selection has previously been published (30). Briefly, using Haploview Tagger on publicly available HapMap 2 data (31), tagSNPs were selected based on a pairwise r 2 > 0.95, a minor allele frequency >5% and distance from the closest SNP of >60 base pairs (bp). Within linkage disequilibrium (LD) blocks for each gene, the 5′- and 3′-untranslated regions (UTR) were extended to include the most up- or downstream SNP (~10kb upstream and 5 kb downstream). In regions of no or low LD, SNPs with a minor allele frequency >5% at a density of ~1 per kb were selected from either HapMap (31) or dbSNP (32). When we designed the study using HapMap 2 as a reference panel, tagSNPs genotyped by the CCFR were estimated to cover at least 80% of the variation in these loci. With the HapMap 3 release, ~50% of these loci are covered.
Genotyping assays
SNPs were genotyped using the Illumina BeadXpress GoldenGate at Translational Genomics Research Institute (TGen, Phoenix, AZ). Replicate aliquots were included for 5% of samples to assess reliability. Concordance among duplicates was at least 99.8% for all SNPs. Monomorphic SNPs, SNPs with call rates <90% and SNPs with Hardy–Weinberg Equilibrium P values < 0.001 among unrelated Caucasian controls were excluded (N SNPs excluded = 28), resulting in 192 SNPs available for analysis.
Three VNTRs were also genotyped: the 9-base microsatellite polymorphism at nt –3 in the PTGIS (CYP8A1) 5′-untranslated region, which is a repeat of 4–7 copies of a putative Sp1 binding site (33), the ALOX5 VNTR promoter polymorphism [−176(GGGCGG)2–8], and the ALOX5AP (FLAP) poly(A) promoter repeat [−169poly(A)]. The PTGIS VNTR was genotyped by GeneScan assay as described previously (34). Briefly, the PTGIS (CYP8A1) promoter region was PCR amplified with a fluorescently tagged primer and size separated by capillary electrophoresis using an ABI 3130xl Sequence Analyzer (Life Technologies). Amplicon length was analysed using Genotyper software. The DNA input was 40 ng with a total reaction volume of 5 μl. The ALOX5 and ALOX5AP VNTRs were genotyped using GeneScan on an ABI 3130xl, based on previously published protocols (34,35). Briefly, multiplex PCR reactions consisted of 40 ng genomic template DNA, 1.5 mmol/l MgCl2, 1x PCR Gold Buffer (Applied Biosystems), 0.5 units of Amplitaq Gold Polymerase (Applied Biosystems), 200 nmol/l each of oligonucleotide primers ALOX5sp1-F 5′-6FAM-AGGAA CAGACACC TCGCTG AGGAGAG-3′, ALOX5sp1-R 5′GAGCAGCGAGCGCCGGGAGCCTCGGC-3′, -F: 5′ VIC-CGTGCTCCTCTGCCAAGCCCTGCTTC- 3′ and ALOX5AP-R02: 5′-GCTCT GCCTCCAGCTGCACAACCTG-3′, 150 µmol/l each of dATP, dCTP and dTTP, 75 µmol/l each of dGTP and 7-deaza-2V-dGTP (Roche Diagnostics GmbH, Mannheim, Germany) and 8% (vol/vol) DMSO (Sigma, St Louis, MO) in 5 μl. Cycling was as follows: 96°C for 7 min, 30 cycles of: 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, 72°C for 7 min. The observed 6FAM-labeled ALOX5 amplicon lengths ranged from 254 bp (26-bp repeats) to 290 bp (86-bp repeats) and VIC-labeled ALOX5AP amplicon was either 212 bp (19A) or 216 bp (23A). However, we excluded the ALOX5AP VNTR from analysis as it had a Hardy–Weinberg Equilibrium P value < 0.001 among Caucasian controls.
Statistical methods
Because colon and rectal cancer could have unique etiological mechanisms, we chose a priori to assess separately the association between each polymorphism and colon and rectal cancer risk as well as combined CRC risk using conditional logistic regression to compute odds ratios (OR) and 95% confidence intervals (CI). Conditional logistic regression was also used to examine interactions between polymorphisms and current NSAID use for CRC, colon and rectal cancer risk. For interaction analyses, ORs and 95% CI were computed within categories of NSAID use in order to compare genotypes. Likelihood ratio tests were used to calculate P values for polymorphism main effects and polymorphism–NSAID interactions. Statistical significance of main effects was determined using minP permutation tests (36) with 10 000 replications to obtain multiple comparison corrected P values. Statistical significance of interactions was determined using Bonferroni correction (37). VNTR genotypes were analysed categorically based on the number of repeats (≥6/≥6, <6/≥6 and <6/<6 for the PTGIS repeat and ≥5/≥5, <5/≥5 and <5/<5 for the ALOX5 repeat). We used co-dominant models for each polymorphism, except where genotype cell counts were <10 in either cases or controls in which case we used a dominant model. We did not estimate associations where genotype cell counts were less than five after combining heterozygotes and homozygous minor allele carriers. All models used sibling kinship as a matching variable and were adjusted for continuous linear age and sex. Polymorphism–NSAID interactions were further adjusted for body mass index (continuous), cigarette smoking in pack–years (continuous) and physical activity [categorized from average metabolic equivalent of task hours into inactive (0–6), less active (6.1–20), active (20.1–44) and very active (>44)]. All analyses were performed using SAS 9.3 or R version 2.13.2.
Results
Distributions of demographic characteristics were mostly similar by case–control status (Table I). Although slightly more cases than controls were male, there were no substantial differences by age, study center, NSAID use, physical activity, body mass index or cigarette smoking.
Table I.
Selected characteristics of colorectal cancer cases and unaffected sibling controls
| Controls | Cases | |||
|---|---|---|---|---|
| (N = 2592) | (N = 1621) | |||
| N | % | N | % | |
| Age | ||||
| Years (mean ± SD) | 53.9±11.7 | 53.6±10.8 | ||
| Sex | ||||
| Male | 1158 | 44.7 | 833 | 51.4 |
| Center | ||||
| Ontario | 499 | 19.3 | 301 | 18.6 |
| Los Angeles | 461 | 17.8 | 335 | 20.7 |
| Australia | 594 | 22.9 | 327 | 20.2 |
| Hawaii | 8 | 0.3 | 7 | 0.4 |
| Mayo | 474 | 18.3 | 251 | 15.5 |
| Seattle | 556 | 21.5 | 400 | 24.7 |
| NSAID use | ||||
| Never/former | 1974 | 76.8 | 1265 | 78.3 |
| Current | 595 | 23.2 | 351 | 21.7 |
| Physical activity | ||||
| Inactive | 598 | 24.5 | 383 | 24.8 |
| Less active | 702 | 28.8 | 433 | 28.0 |
| Active | 582 | 23.9 | 389 | 25.2 |
| Very active | 558 | 22.9 | 340 | 22.0 |
| BMI | ||||
| kg/m2 (mean ± SD) | 26.7±5.2 | 27.2±5.5 | ||
| Cigarette smoking | ||||
| Pack–years (mean ± SD) | 11.7±19.4 | 12.9±19.7 | ||
| Tumor site | ||||
| Colon | ||||
| Right | — | — | 542 | 33.4 |
| Left | — | — | 466 | 28.7 |
| NOS | — | — | 76 | 4.7 |
| Rectal | — | — | 537 | 33.1 |
BMI, body mass index; NOS, not otherwise specified.
After correcting for multiple comparisons, no polymorphisms reached statistical significance for associations with CRC or colon cancer risk. However, two SNPs were associated with rectal cancer risk: rs11571364 in ALOX12 and rs45525634 in PTGER2 (Table II). Individuals carrying the variant allele of rs11571364 (G>A) in ALOX12 were at an almost 90% increased risk of rectal cancer (ORhet/hzv = 1.87, 95% CI = 1.19–2.95). In contrast, those carrying the variant allele of rs45525634 (G>T) in PTGER2 had an ~50% decreased risk of rectal cancer (ORhet/hzv = 0.49, 95% CI = 0.29–0.82). All other associations with CRC, colon and rectal cancer risk are presented in Supplementary Table 1, available at Carcinogenesis Online.
Table II.
Statistically significanta associations between polymorphisms and rectal cancer risk
| Rectal cancer | ||||||||
|---|---|---|---|---|---|---|---|---|
| Controls | Cases | ORb | 95% CI | P c | ||||
| N | % | N | % | |||||
| ALOX12 | ||||||||
| rs11571364 | G/G | 762 | 87.8 | 429 | 83.3 | ref | — | 0.03 |
| Intron | G/A or A/A | 106 | 12.2 | 86 | 16.7 | 1.87 | 1.19–2.95 | |
| PTGER2 | ||||||||
| rs45525634 | G/G | 764 | 87.9 | 468 | 91.2 | ref | — | 0.03 |
| Intron | G/T or TT | 105 | 12.1 | 45 | 8.8 | 0.49 | 0.29–0.82 | |
aStatistical significance considered a minP Pvalue ≤ 0.05.
bAdjusted for continuous linear age and sex.
cMultiple comparisons corrected P value from a minP permutation test with 10 000 replications.
Although current NSAID use was not associated with a statistically significant reduced risk of CRC in this study population when compared with never or former use (OR = 0.86, 95% CI = 0.71–1.04, P = 0.11), after correcting for multiple comparisons we observed an interaction (P = 0.03) between NSAID use and the intronic SNP rs2920421 (G>A) in ALOX12 on the risk of CRC (Table III). Among those with heterozygous genotypes for rs2920421, current NSAID users had an ~40% decreased risk of CRC (ORhet = 0.60, 95% CI = 0.45–0.80) compared with never or former users, whereas this association was null among individuals with homozygous wild-type genotypes (ORhzw = 1.12, 95% CI = 0.86–1.46) and not significantly reduced among homozygous variant genotypes (ORhzv = 0.83, 95% CI = 0.45–1.51). We observed similar effect sizes for colon and rectal cancer risk separately, though these associations were not statistically significant after multiple comparison adjustment (Supplementary Table 2a and b, available at Carcinogenesis Online). Results for all other potential interactions for CRC, colon and rectal cancer risk are presented in Supplementary Table 2a and b, available at Carcinogenesis Online.
Table III.
Statistically significanta polymorphism–NSAID use interactions and colorectal cancer risk
| NSAID use | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Never/former | Currentb | P d | ||||||||
| Controls | Cases | ORc | 95% CI | Controls | Cases | ORc | 95% CI | |||
| ALOX12 | ||||||||||
| rs2920421 | G/G | 920 | 586 | ref | — | 257 | 183 | 1.12 | 0.86–1.46 | 0.03 |
| Intron | G/A | 818 | 541 | ref | — | 268 | 132 | 0.60 | 0.45–0.80 | |
| A/A | 203 | 124 | ref | — | 59 | 29 | 0.83 | 0.45–1.51 | ||
| rs2920421e | G/G | 920 | 586 | ref | — | 257 | 183 | 1.12 | 0.86–1.46 | 0.03 |
| Intron | G/A | 818 | 541 | 1.16 | 0.95–1.43 | 268 | 132 | 0.70 | 0.51–0.95 | |
| A/A | 203 | 124 | 1.09 | 0.77–1.53 | 59 | 29 | 0.90 | 0.49–1.65 | ||
BMI, body mass index.
aStatistical significance considered a Bonferroni-adjusted P value ≤ 0.05.
bCurrent NSAID use defined as use two or more times per week for at least 1 month in the 2 years prior to study enrollment.
cAdjusted for continuous linear age, BMI, and smoking as well as sex and physical activity (categorized from average MET hours into inactive, less active, active and very active).
dBonferroni-adjusted interaction P value.
eResults presented using a common reference group.
Discussion
We examined the association between CRC risk, NSAID use, and 192 candidate and tagSNPs and two VNTRs within 17 candidate genes involved in prostaglandin synthesis and related pathways. We observed that variants in two intronic SNPs, rs11571364 in ALOX12 and rs45525634 in PTGER2, were associated with rectal cancer risk. Our observation that these SNPs were associated with rectal cancer risk and not colon or combined colorectal cancer risk reflects the likelihood that colon and rectal cancer have unique etiological mechanisms (38,39). A post-hoc case–case comparison suggested a marginally significant difference in relative risk between colon and rectal cancers for rs11571364 in ALOX12 (P = 0.05), yet less so for rs45525634 in PTGER2 (P = 0.09) (data not shown). We observed an interaction between current NSAID use and rs2920421 in ALOX12 in relation to risk of CRC. A post-hoc case–case comparison of the interaction between this SNP and current NSAID use indicated there was no difference in the association between colon and rectal cancers (P = 0.20) (data not shown). The SNPs for which we observed statistically significant associations in ALOX12 (rs2920421 and rs11571364) and PTGER2 (rs45525634) were not in high LD with any of the other genotyped SNPs within these genes among controls (all pairwise r 2 < 0.65).
We observed that current NSAID users had a decreased risk of CRC compared with never or former users among those with heterozygous genotypes for rs2920421 in ALOX12. We conducted a post-hoc analysis with NSAID use categorized as never, former and current. When compared with never users the risk of CRC was reduced among current NSAID users (OR = 0.88, 95% CI = 0.72–1.07) and null among former users (OR = 1.07, 95% CI = 0.88–1.29), though the overall association between NSAID use and CRC risk was not statistically significant (global P = 0.22). When examining the interaction between NSAID use and rs2920421, we observed that current NSAID users with heterozygous genotypes had an ~40% decreased risk of CRC (ORhet = 0.61, 95% CI = 0.45–0.83) while there was no association for former NSAID users with heterozygous genotypes (ORhet = 1.07, 95% CI = 0.79–1.44) when compared with never users. Among individuals with homozygous wild-type genotypes, this association was null when comparing former users (ORhzw = 1.14, 95% CI = 0.86–1.50) and current users (ORhzw = 1.17, 95% CI = 0.89–1.54) to never users. Among individuals with homozygous variant genotypes, this association was modestly, although not statistically significantly, reduced when comparing former users (ORhzv = 0.84, 95% CI = 0.46–1.52) and current users (ORhzv = 0.79, 95% CI = 0.43–1.47) to never users. Overall, our results suggest that the expected protective association with NSAIDs is present only among subjects with variant genotypes. Variants in ALOX12 could modify the effect of NSAIDs on CRC risk, with this association likely being driven by current NSAID use. However, larger sample sizes are needed to verify these results.
ALOX12 is one of three main lipoxygenases responsible for mediating inflammatory responses and converting AA into leukotrienes and other eicosanoids (16,40–42). Further, ALOX12 is up-regulated in CRC tumour tissues and promotes vascular endothelial growth factor production in tumour stroma, which contributes to angiogenesis and the motility of cancer cells (43,44). Of interest, the SNP in ALOX12 for which we observed an interaction between current NSAID use and CRC risk (rs2920421) is in high LD (r 2 = 1.0) with another SNP (rs2440129) that has been associated with microRNA expression levels in adipose tissue and body mass index in genome-wide association studies (45). Although genome-wide association studies have not observed associations between CRC risk and variants in ALOX12, results from previous candidate-gene studies suggest plausible associations. In a recent case–control study of colon and rectal cancer risk (N colon cases = 1574, N rectal cases = 791, N controls = 2,969), there was evidence that fatty acid intake and aspirin use modified the associations between two variants in ALOX12, the intronic SNP rs11571339 and the coding SNP rs312462 (L634L), and the risk of rectal cancer (46). Another study of 2300 Chinese CRC cases and controls observed that the coding SNP rs1126667 (G261A) in ALOX12 was associated with an increased risk of CRC, and further observed an interaction between this SNP and rs689466 in PTGS2 on CRC risk (47). A study assessing the association of variants in ALOX12 and ALOX5 with CRC risk among 162 cases and 211 controls reported a borderline association between rs1126667 and CRC risk (48). However, two other studies reported no association between this SNP and CRC risk (15,49). In the current study, rs1126667 was associated with a decreased risk of CRC (ORAA versus wtGG = 0.71, 95% CI 0.55–0.93); however, this association was not statistically significant after correcting for multiple comparisons. Nonetheless, our study presents novel results for variants in ALOX12, as no prior studies have reported an association between rs11571364 and rectal cancer risk or an interaction between NSAID use and rs2920421 on the risk of CRC.
This study is also the first to report an association between rectal cancer risk and the intronic SNP rs45525634 in PTGER2. Based on data from the Encyclopedia of DNA Elements (ENCODE) Consortium (50), rs45525634 may regulate transcription. PTGER2 encodes the receptor EP2 for PGE2 (21,51–55), which is thought to be critical in CRC development as it promotes angiogenesis as well as the survival, motility and invasiveness of cancer cells (52–57). In a recent study, overexpression of PTGER2 was associated with high microsatellite instability in CRC tumours (58). EP2 also plays an important role in colorectal carcinogenesis, having been shown to contribute to intestinal polyps and to affect tumour growth in mice (59–61). Genome-wide association studies have not identified SNPs in PTGER2 as associated with CRC risk, though some candidate-gene studies have reported associations. In a study of 1225 CRC cases and 2032 controls, the intergenic SNP rs17831718 was associated with a roughly 30% decreased risk of CRC (OR = 0.73, 95% CI: 0.58–0.91) (62). Moreover, this study observed a statistically significant inverse association (P = 0.006) between CRC risk and the PTGER2 GGG haplotype based on the SNPs rs17831718, rs17125362 and rs1254580. Another recent large study observed an increased polyp risk for rs17125318, an intergenic SNP close to PTGER2 (P = 0.008) (63). In our prior study examining adenoma cases and polyp-free controls, we observed the intronic SNP rs33993630 in PTGER2 to be associated with a reduced risk of colorectal adenoma (OR = 0.71, 95% CI: 0.52–0.99) (21). In the present study, those carrying the variant allele for rs33993630 had a reduced risk of rectal cancer (OR = 0.49, 95% CI: 0.29–0.82, P = 0.02); however, this association was not statistically significant after correcting for multiple comparisons (P = 0.07). Likewise, we observed no evidence of an interaction between rs33993630 and NSAID use on CRC, colon or rectal cancer risk.
This study is the first to examine associations and NSAID use interactions between CRC risk and the functional VNTRs in ALOX5 and PTGIS. Our previous study observed an increased risk of colorectal polyps associated with the PTGIS VNTR (OR<6/<6 versus 6/6 = 1.90, 95% CI: 1.09–3.30), though not with the ALOX5 VNTR (34). In the current study, neither the ALOX5 nor PTGIS VNTR was associated with CRC risk, nor did we observe evidence of interactions with NSAID use.
Although we observed an association only between rectal cancer risk and variants in ALOX12 and PTGER2, previous studies have shown variants in other genes involved in prostaglandin synthesis and related pathways to be associated with CRC risk. For example, prior studies have suggested associations between CRC risk and variants in the cytokines IL6 and TNF (64–66) as well as PTGS2 (COX2), including rs689466, rs20417 and rs4648298 (47,67). Further, a prior study of variants in ALOX5, ALOX12, PTGS1 and PTGS2 observed two SNPs both in the flanking 5′-untranslated region of ALOX5, rs6413416 and rs4986832, to be associated with colon cancer risk among Caucasians (15). We assessed 21 SNPs in ALOX5 in this study, including rs4986832, but none was significantly associated with CRC, colon or rectal cancer risk after adjustment for multiple comparisons.
There were some potential limitations to this study. We observed that current NSAID use was associated with a modestly and non-significantly reduced risk of CRC compared with never or former use (OR = 0.86, 95% CI: 0.71–1.04; P = 0.11); therefore, we may have been limited in our ability to detect interactions between current NSAID use and the polymorphisms analysed. It is possible that we missed relevant variation in the genes we examined due to incomplete coverage, though this is unlikely given our thorough candidate and tagSNP approach. We may also have missed variation by not examining deletions, copy number variation or additional variants in repeat regions; however, we were unable to examine some of this variation given our genotyping methods. Finally, although assays may have misclassified or failed to detect variation in the genes examined for this study, this was not likely an issue since we had high concordance among duplicate samples.
There were several strengths to this study. Our use of a combined candidate and tagSNP approach enabled comprehensive coverage of most of our genes of interest, including an assessment of VNTRs in ALOX5 and PTGIS. By using data from the CCFR, this study had large numbers of cases and controls as well as access to extensive data on demographic and lifestyle factors, including NSAID use. These design features allowed us to examine the association between polymorphism–NSAID interactions and CRC, colon and rectal cancer risk. Further, although this study made a number of comparisons, all results were interpreted after correcting for multiple comparisons.
Results from this study suggest that genetic variation in ALOX12 and PTGER2 may be associated with the risk of rectal cancer. In addition, this study suggests that current use of NSAIDs may interact with variants in ALOX12 to modify CRC risk. As these results may aid in the development of genetically targeted cancer prevention strategies with NSAIDs, further studies are warranted that examine the association between CRC risk, NSAID use, and polymorphisms in genes involved in prostaglandin synthesis and related pathways.
Supplementary material
Supplementary Tables 1 and 2 can be found at http://carcin. oxfordjournals.org/
Funding
National Cancer Institute ; National Institutes of Health (R01 CA129063, T32 CA09168, R01 CA112516, R01 CA114467-05).
Supplementary Material
Acknowledgements
We would like to acknowledge the support this study received from the National Cancer Institute and the National Institutes of Health grants R01 CA129063 (Inflammation and Innate Immunity Genes and Colorectal Cancer Risk) and T32 CA09168 (Cancer Epidemiology and Biostatistics Training Grant, for A.J.R. and M.N.P.), as well as through cooperative agreements with members of the Colon Cancer Family Registry (CCFR) and Principal Investigators. The contents of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the CCFRs, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the CCFR. CCFR centers providing data for the analysis include: Australasian Colorectal Cancer Family Registry (U01 CA097735); Familial Colorectal Neoplasia Collaborative Group (U01 CA074799); Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800); Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); Seattle Colorectal Cancer Family Registry (U01 CA074794); University of Hawaii Colorectal Cancer Family Registry (U01 CA074806) and University of California, Irvine Informatics Center (U01 CA078296). We would like to thank S.Thomas, C.Rimorin and J.DaGloria of the FHCRC Molecular Epidemiology Laboratory for genotyping assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AA
arachidonic acid
- bp
base pair
- ALOX
arachidonate lipoxygenase
- CI
confidence interval
- COX
cyclooxygenase
- CRC
colorectal cancer
- LD
linkage disequilibrium
- NSAID
non-steroidal anti-inflammatory drug
- OR
odds ratio
- PGE2
prostaglandin E2
- PGI
prostacyclin synthase
- SNP
single nucleotide polymorphism
- VNTR
variable nucleotide tandem repeat.
References
- 1. Terzić J., et al. (2010). Inflammation and colon cancer. Gastroenterology, 138, 2101–2114.e5 [DOI] [PubMed] [Google Scholar]
- 2. Ulrich C.M., et al. (2006). Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat. Rev. Cancer, 6, 130–140 [DOI] [PubMed] [Google Scholar]
- 3. Rizzo A., et al. (2011). Intestinal inflammation and colorectal cancer: a double-edged sword? World J. Gastroenterol., 17, 3092–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kraus S., et al. (2009). Inflammation and colorectal cancer. Curr. Opin. Pharmacol., 9, 405–410 [DOI] [PubMed] [Google Scholar]
- 5. Rhodes J.M., et al. (2002). Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol. Med., 8, 10–16 [DOI] [PubMed] [Google Scholar]
- 6. Hanahan D., et al. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 7. Mantovani A. (2009). Cancer: inflaming metastasis. Nature, 457, 36–37 [DOI] [PubMed] [Google Scholar]
- 8. Sandler R.S., et al. (2003). A randomized trial of aspirin to prevent colorectal adenomas in patients with previous colorectal cancer. N. Engl. J. Med., 348, 883–890 [DOI] [PubMed] [Google Scholar]
- 9. Baron J.A., et al. (2003). A randomized trial of aspirin to prevent colorectal adenomas. N. Engl. J. Med., 348, 891–899 [DOI] [PubMed] [Google Scholar]
- 10. Baron J.A., et al. ; APPROVe Trial Investigators. (2006). A randomized trial of rofecoxib for the chemoprevention of colorectal adenomas. Gastroenterology, 131, 1674–1682 [DOI] [PubMed] [Google Scholar]
- 11. Baron J.A. (2009). Aspirin and NSAIDs for the prevention of colorectal cancer. Recent Results Cancer Res., 181, 223–229 [DOI] [PubMed] [Google Scholar]
- 12. Wang D., et al. (2010). The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene, 29, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenhough A., et al. (2009). The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis, 30, 377–386 [DOI] [PubMed] [Google Scholar]
- 14. Cebola I., et al. (2012). Epigenetic deregulation of the COX pathway in cancer. Prog. Lipid Res., 51, 301–313 [DOI] [PubMed] [Google Scholar]
- 15. Goodman J.E., et al. (2004). Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis, 25, 2467–2472 [DOI] [PubMed] [Google Scholar]
- 16. Shureiqi I., et al. (2001). Lipoxygenase modulation to reverse carcinogenesis. Cancer Res., 61, 6307–6312 [PubMed] [Google Scholar]
- 17. Rigas B., et al. (1993). Altered eicosanoid levels in human colon cancer. J. Lab. Clin. Med., 122, 518–523 [PubMed] [Google Scholar]
- 18. Boland C.R. (2010). Chronic inflammation, colorectal cancer and gene polymorphisms. Dig. Dis., 28, 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coghill A.E., et al. (2011). Genetic variation in inflammatory pathways is related to colorectal cancer survival. Clin. Cancer Res., 17, 7139–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poole E.M., et al. (2007). Genetic variability in prostaglandin synthesis, fish intake and risk of colorectal polyps. Carcinogenesis, 28, 1259–1263 [DOI] [PubMed] [Google Scholar]
- 21. Poole E.M., et al. (2010). Genetic variation in prostaglandin E2 synthesis and signaling, prostaglandin dehydrogenase, and the risk of colorectal adenoma. Cancer Epidemiol. Biomarkers Prev., 19, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poole E.M., et al. (2012). Genetic variability in IL23R and risk of colorectal adenoma and colorectal cancer. Cancer Epidemiol., 36, e104–e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ulrich C.M., et al. (2004). Polymorphisms in PTGS1 (=COX-1) and risk of colorectal polyps. Cancer Epidemiol. Biomarkers Prev., 13, 889–893 [PubMed] [Google Scholar]
- 24. Ulrich C.M., et al. (2005). PTGS2 (COX-2) -765G > C promoter variant reduces risk of colorectal adenoma among nonusers of nonsteroidal anti-inflammatory drugs. Cancer Epidemiol. Biomarkers Prev., 14, 616–619 [DOI] [PubMed] [Google Scholar]
- 25. Algra A.M., et al. (2012). Effects of regular aspirin on long-term cancer incidence and metastasis: a systematic comparison of evidence from observational studies versus randomised trials. Lancet Oncol., 13, 518–527 [DOI] [PubMed] [Google Scholar]
- 26. Flossmann E., et al. ; British Doctors Aspirin Trial and the UK-TIA Aspirin Trial. (2007). Effect of aspirin on long-term risk of colorectal cancer: consistent evidence from randomised and observational studies. Lancet, 369, 1603–1613 [DOI] [PubMed] [Google Scholar]
- 27. Dubé C., et al. ; U.S. Preventive Services Task Force. (2007). The use of aspirin for primary prevention of colorectal cancer: a systematic review prepared for the U.S. Preventive Services Task Force. Ann. Intern. Med., 146, 365–375 [DOI] [PubMed] [Google Scholar]
- 28. Bigler J., et al. (2001). CYP2C9 and UGT1A6 genotypes modulate the protective effect of aspirin on colon adenoma risk. Cancer Res., 61, 3566–3569 [PubMed] [Google Scholar]
- 29. Newcomb P.A., et al. ; Colon Cancer Family Registry. (2007). Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol. Biomarkers Prev., 16, 2331–2343 [DOI] [PubMed] [Google Scholar]
- 30. Levine A.J., et al. (2010). A candidate gene study of folate-associated one carbon metabolism genes and colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev., 19, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thorisson G.A., et al. (2005). The International HapMap Project Web site. Genome Res., 15, 1592–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sherry S.T., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chevalier D., et al. (2001). Characterization of new mutations in the coding sequence and 5’-untranslated region of the human prostacylcin synthase gene (CYP8A1). Hum. Genet., 108, 148–155 [DOI] [PubMed] [Google Scholar]
- 34. Poole E.M., et al. (2006). Prostacyclin synthase and arachidonate 5-lipoxygenase polymorphisms and risk of colorectal polyps. Cancer Epidemiol. Biomarkers Prev., 15, 502–508 [DOI] [PubMed] [Google Scholar]
- 35. Sayers I., et al. (2003). Promoter polymorphism in the 5-lipoxygenase (ALOX5) and 5-lipoxygenase-activating protein (ALOX5AP) genes and asthma susceptibility in a Caucasian population. Clin. Exp. Allergy, 33, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 36. Dudoit S., et al. (2004). Multiple testing. Part I. Single-step procedures for control of general type I error rates. Stat. Appl. Genet. Mol. Biol., 3, Article13. [DOI] [PubMed] [Google Scholar]
- 37. Murcray C.E., et al. (2009). Gene-environment interaction in genome-wide association studies. Am. J. Epidemiol., 169, 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Slattery M.L., et al. (2005). Associations between apoE genotype and colon and rectal cancer. Carcinogenesis, 26, 1422–1429 [DOI] [PubMed] [Google Scholar]
- 39. Slattery M.L., et al. (2007). IL6 genotypes and colon and rectal cancer. Cancer Causes Control, 18, 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wecksler A.T., et al. (2009). Mechanistic investigations of human reticulocyte 15- and platelet 12-lipoxygenases with arachidonic acid. Biochemistry, 48, 6259–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sears D.D., et al. (2009). 12/15-lipoxygenase is required for the early onset of high fat diet-induced adipose tissue inflammation and insulin resistance in mice. PLoS One, 4, e7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cole B.K., et al. (2012). 12/15-Lipoxygenase signaling in the endoplasmic reticulum stress response. Am. J. Physiol. Endocrinol. Metab., 302, E654–E665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szefel J., et al. (2011). Eicosanoids in prevention and management of diseases. Curr. Mol. Med., 11, 13–25 [DOI] [PubMed] [Google Scholar]
- 44. Klampfl T., et al. (2012). Up-regulation of 12(S)-lipoxygenase induces a migratory phenotype in colorectal cancer cells. Exp. Cell Res., 318, 768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parts L., et al. ; MuTHER Consortium. (2012). Extent, causes, and consequences of small RNA expression variation in human adipose tissue. PLoS Genet., 8, e1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Habermann N., et al. (2013). PTGS1, PTGS2, ALOX5, ALOX12, ALOX15, and FLAP SNPs: interaction with fatty acids in colon cancer and rectal cancer. Genes Nutr., 8, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tan W., et al. (2007). Associations of functional polymorphisms in cyclooxygenase-2 and platelet 12-lipoxygenase with risk of occurrence and advanced disease status of colorectal cancer. Carcinogenesis, 28, 1197–1201 [DOI] [PubMed] [Google Scholar]
- 48. Gong Z., et al. (2007). Common polymorphisms in 5-lipoxygenase and 12-lipoxygenase genes and the risk of incident, sporadic colorectal adenoma. Cancer, 109, 849–857 [DOI] [PubMed] [Google Scholar]
- 49. Küry S., et al. (2008). Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer, 8, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosenbloom K.R., et al. (2012). ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res., 40(Database issue), D912–D917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Obermajer N., et al. (2011). Positive feedback between PGE2 and COX2 redirects the differentiation of human dendritic cells toward stable myeloid-derived suppressor cells. Blood, 118, 5498–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kamei D., et al. (2003). Potential role of microsomal prostaglandin E synthase-1 in tumorigenesis. J. Biol. Chem., 278, 19396–19405 [DOI] [PubMed] [Google Scholar]
- 53. Amano H., et al. (2009). Roles of a prostaglandin E-type receptor, EP3, in upregulation of matrix metalloproteinase-9 and vascular endothelial growth factor during enhancement of tumor metastasis. Cancer Sci., 100, 2318–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kopp K.L., et al. (2010). COX-2-dependent PGE(2) acts as a growth factor in mycosis fungoides (MF). Leukemia, 24, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 55. Wang D., et al. (2010). Eicosanoids and cancer. Nat. Rev. Cancer, 10, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheng H., et al. (2001). Prostaglandin E2 increases growth and motility of colorectal carcinoma cells. J. Biol. Chem., 276, 18075–18081 [DOI] [PubMed] [Google Scholar]
- 57. Wang D., et al. (2004). Prostaglandin E(2) promotes colorectal adenoma growth via transactivation of the nuclear peroxisome proliferator-activated receptor delta. Cancer Cell, 6, 285–295 [DOI] [PubMed] [Google Scholar]
- 58. Baba Y., et al. (2010). PTGER2 overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Cancer Epidemiol. Biomarkers Prev., 19, 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sonoshita M., et al. (2001). Acceleration of intestinal polyposis through prostaglandin receptor EP2 in Apc(Delta 716) knockout mice. Nat. Med., 7, 1048–1051 [DOI] [PubMed] [Google Scholar]
- 60. Seno H., et al. (2002). Cyclooxygenase 2- and prostaglandin E(2) receptor EP(2)-dependent angiogenesis in Apc(Delta716) mouse intestinal polyps. Cancer Res., 62, 506–511 [PubMed] [Google Scholar]
- 61. Yang L., et al. (2003). Cancer-associated immunodeficiency and dendritic cell abnormalities mediated by the prostaglandin EP2 receptor. J. Clin. Invest., 111, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hoeft B., et al. (2010). Polymorphisms in fatty-acid-metabolism-related genes are associated with colorectal cancer risk. Carcinogenesis, 31, 466–472 [DOI] [PubMed] [Google Scholar]
- 63. Edwards T.L., et al. (2012). A study of prostaglandin pathway genes and interactions with current nonsteroidal anti-inflammatory drug use in colorectal adenoma. Cancer Prev. Res. (Phila)., 5, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Landi S., et al. ; Bellvitge Colorectal Cancer Study Group. (2003). Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res., 63, 3560–3566 [PubMed] [Google Scholar]
- 65. Gunter M.J., et al. (2006). Inflammation-related gene polymorphisms and colorectal adenoma. Cancer Epidemiol. Biomarkers Prev., 15, 1126–1131 [DOI] [PubMed] [Google Scholar]
- 66. Theodoropoulos G., et al. (2006). Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J. Gastroenterol., 12, 5037–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cox D.G., et al. ; Bellvitge Colorectal Cancer Study Group. (2004). Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br. J. Cancer, 91, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.