Summary
We performed a genome-wide association study and identified single-nucleotide polymorphism rs4698934 located in the intron of the TET2 gene on chromosome 4q24 nominally significantly associated with melanoma risk, and a novel somatic mutation of TET2 was identified in melanoma case.
Abstract
Although genetic studies have reported a number of loci associated with melanoma risk, the complex genetic architecture of the disease is not yet fully understood. We sought to identify common genetic variants associated with melanoma risk in a genome-wide association study (GWAS) of 2298 cases and 6654 controls. Thirteen of 15 known loci were replicated with nominal significance. A total of 69 single-nucleotide polymorphisms (SNPs) were selected for in silico replication in two independent melanoma GWAS datasets (a total of 5149 cases and 12 795 controls). Seven novel loci were nominally significantly associated with melanoma risk. These seven SNPs were further genotyped in 234 melanoma cases and 238 controls. The SNP rs4698934 was nominally significantly associated with melanoma risk. The combined odds ratio per T allele = 1.18; 95% confidence interval (1.10–1.25); combined P = 7.70 × 10− 7. This SNP is located in the intron of the TET2 gene on chromosome 4q24. In addition, a novel somatic mutation of TET2 was identified by next-generation sequencing in 1 of 22 sporadic melanoma cases. TET2 encodes a member of TET family enzymes that oxidizes 5-methylcytosine to 5-hydroxymethylcytosine (5hmC). It is a putative epigenetic biomarker of melanoma as we previously reported, with observation of reduced TET2 transcriptional expression. This study is the first to implicate TET2 genetic variation and mutation in melanoma.
Introduction
Melanoma is the leading cause of skin cancer-related mortality and, in advanced stages, is characterized by poor prognosis. The incidence of melanoma has steadily increased worldwide (1,2). Much of this increase has been seen in relatively young adults, and consequently, the number of life years lost per melanoma death is higher than that of most other solid tumors (3). Susceptibility to melanoma is determined by both environmental risk factors, including excessive exposure to ultraviolet radiation and genetically controlled phenotypic traits such as nevus propensity, red or blonde hair, light-colored eyes, fair skin, and limited tanning ability (4–6). Common low-risk predisposing alleles probably account for most of the melanoma burden (7,8). Many putative risk alleles involved in cellular pathways such as pigmentation, DNA repair, telomere maintenance, oxidation stress, apoptosis, cell growth, and melanocyte differentiation and migration have been implicated in melanoma susceptibility (9–11). Genome-wide association studies (GWASs) have propelled this field forward by identifying many novel low- to moderate-risk loci that may cause sporadic melanoma. Within the last several years, several GWASs have reported their findings on melanoma risk (11–18), and most of the loci identified in earlier GWASs were within pigmentation genes (19,20), such as MC1R and TYR. Recent melanoma GWASs identified new loci not related to pigmentation, including ATM, MX2, CASP8, PARP1, CCND1, and a locus (probably SETDB1 and ARNT) at 1q21.3 (15–17). However, all currently identified loci are believed to account for only a small proportion of the genetic susceptibility to melanoma (21,22). To identify additional melanoma risk alleles, we conducted a multistage melanoma GWAS in Caucasians. In addition, somatic gene mutational status was detected by next-generation sequencing in sporadic melanoma samples.
Materials and methods
Harvard cohorts
Description of study populations.
Nurses’ Health Study. The Nurses’ Health Study (NHS) was established in 1976, when 121700 female registered nurses between the ages of 30 and 55 years residing in 11 larger US states completed and returned an initial self-administered questionnaire on their medical histories and baseline health-related exposures. Biennial questionnaires with collection of exposure information on risk factors have been collected prospectively. Every 2 years, along with exposures, outcome data with appropriate follow-up of reported disease events are collected. Overall, follow-up has been high; after more than 20 years, ~90% of participants continue to complete questionnaires. From May 1989 through September 1990, we collected blood samples from 32 826 participants in the NHS. Information on melanoma development was first collected in the 1984 questionnaire.
Health Professionals Follow-up Study. In 1986, 51 529 men from all 50 US states in health professions (dentists, pharmacists, optometrists, osteopath physicians, podiatrists, and veterinarians) aged 40–75 years answered a detailed mailed questionnaire, forming the basis of the study. The average follow-up rate for this cohort over 10 years is >90%. On each biennial questionnaire, we obtained disease- and health-related information. Between 1993 and 1994, 18 159 study participants provided blood samples by overnight courier. Information on melanoma development was first collected in the 1986 questionnaire. Description of the study population can be found elsewhere (23,24).
Melanoma cases and controls in the discovery set.
Eligible cases in the NHS and the Health Professionals Follow-up Study (HPFS) consisted of participants with pathologically confirmed invasive melanoma, diagnosed any time after baseline up to the 2008 follow-up cycle for both cohorts. All subjects were United States non-Hispanic Caucasians.
We have previously conducted several GWASs on different disease outcomes (NHS: breast cancer, coronary heart disease, type 2 diabetes, kidney stone, pancreatic cancer, and glaucoma; HPFS: coronary heart disease, type 2 diabetes, kidney stone, advanced prostate cancer, and glaucoma). The study description for eight GWAS sets of the discovery set is presented in Supplementary Methods, available at Carcinogenesis Online. We included only controls in each GWAS, except for the kidney stone GWAS, in which we used both cases and controls. Participants without melanoma diagnosis were the controls in the current study, and those with melanoma diagnosis were the cases. In addition, we genotyped the rest of the melanoma cases in both cohorts who were not included in these previous GWASs. A detailed description of the numbers of cases and controls included and the platform for each study are shown in Table I. Finally, we included 494 melanoma cases and 5628 controls.
Table I.
Melanoma cases and controls (SNPs genotyped by studies in the NHS and HPFS of the discovery phase)
| NHS | Control | Case | Genotyped SNPs | Platform | HPFS | Control | Case | Genotyped SNPs | Platform |
|---|---|---|---|---|---|---|---|---|---|
| Melanoma | 0 | 264 | 620901 | Illumina 610 | Melanoma | 0 | 136 | 620901 | Illumina 610 |
| Breast cancer | 840 | 5 | 546646 | Illumina 550 | |||||
| Coronary heart disease | 557 | 13 | 721316 | Affymetrix 6.0 | Coronary heart disease | 636 | 13 | 724881 | Affymetrix 6.0 |
| Glaucoma | 337 | 5 | 495161 | Illumina 660 | Glaucoma | 197 | 1 | 495161 | Illumina 660 |
| Kidney stone | 359 | 5 | 620901 | Illumina 610 | Kidney stone | 410 | 2 | 620901 | Illumina 610 |
| Type 2 diabetes | 1220 | 23 | 704409 | Affymetrix 6.0 | Type 2 diabetes | 867 | 23 | 706040 | Affymetrix 6.0 |
| Pancreatic cancer | 64 | 2 | 559865 | Illumina 550 | |||||
| Advanced prostate cancer | 141 | 2 | 573612 | Illumina 610 | |||||
| Total | 3377 | 317 | Total | 2251 | 177 |
Laboratory assays.
Genotyping in eight GWASs of the discovery set. We performed genotyping in the breast cancer GWAS in NHS using the Illumina HumanHap550 array, as part of the National Cancer Institute’s Cancer Genetic Markers of Susceptibility Project. For the coronary heart disease and type 2 diabetes GWASs of the discovery set, we performed genotyping using the Affymetrix 6.0 array. For the glaucoma GWAS, we performed genotyping using the Illumina HumanHap660 array. For the kidney stone, advanced prostate cancer, and melanoma GWASs, we performed genotyping using the Illumina HumanHap610 array. The quality control procedures for eight GWAS sets of the discovery set are presented in Supplementary Methods, available at Carcinogenesis Online.
Imputation and meta-analysis.
On the basis of the genotyped single-nucleotide polymorphisms (SNPs) and haplotype information in the National Center for Biotechnology Information build 35 of phase II Hapmap CEU data, we imputed genotypes for >2.5 million SNPs using the program MACH (25).Only SNPs with imputation quality R 2 > 0.95 in each study were included in the final analysis. A total of 1 579 307 SNPs were included in the final meta-analysis of the NHS and HPFS (Table I). Betas from each study of the discovery set were combined in a meta-analysis with weights proportional to the inverse variance of the beta in each study.
MD Anderson Cancer Center
The study participants for the discovery analysis were from a hospital-based case–control study of melanoma, for which cases were recruited from among non-Hispanic white patients at MD Anderson between March 1998 and August 2008. Samples and data were available from 931 melanoma patients and 1026 cancer-free controls (friends of other patients reporting to clinics), who were frequency matched on age and sex, completed a comprehensive skin lifestyle questionnaire, and passed quality control filters for genotyping. This questionnaire was administered by an interviewer to 70% of patients and controls and was self-administered for the remaining 30%. An additional case series comprising 873 individuals presenting for treatment for melanoma at MD Anderson was also included, bringing the total number of melanoma patients to 1804. The study protocols were approved by the Institutional Review Board at MD Anderson, and informed consent was obtained from all participants. Tissue samples were collected as whole blood, with various DNA extraction methods (including Gentra, Qiagen, and phenol–chloroform). DNA samples for the first-stage GWAS were genotyped using the Illumina Omnil-Quad array and were called using the Bead Studio algorithm at the John Hopkins University Center for Inherited Disease Research. Detailed information about the MD Anderson melanoma GWAS can be found elsewhere (17).
Melanoma GWASs in the replication set
The study description and quality control procedures for the replication sets from GenoMEL (15), Australia (16), and the NHS II are presented in Supplementary Methods, available at Carcinogenesis Online.
Statistical methods
We regressed a binary coding for melanoma case or control (0 or 1) on each SNP (dosage file) that passed quality control filters. The five largest principal components of genetic variation were nominally significantly associated with melanoma (P < 0.05), and we adjusted for them along with age. These principal components were calculated for all individuals on the basis of ~10,000 unlinked markers using the EIGENSTRAT software. We performed a meta-analysis to combine the replication sets and the discovery set. From the discovery stage, SNPs with P values <1×10−4 were selected for replication, and in the replication stages, P values < .05 were considered nominally statistically significant.
Somatic mutation detection by targeted next-generation sequencing in sporadic melanoma cases
DNA was isolated from formalin-fixed melanoma samples using standard methods. In brief, samples were incubated in proteinase K overnight, followed by subsequent purification of the DNA according to manufacturer’s instructions (QIAamp DNA Mini Kit, QIAGEN, Gaithersburg, MD). Then, DNA concentration was assessed using PicoGreen dsDNA detection (Life Technologies, Carlsbad, CA). Targeted next-generation sequencing was performed using cancer genomic assay to detect mutations in 275 cancer target genes. The complete coding sequence of the target genes, plus selected introns for 30 of the genes, was captured using a custom-designed Agilent SureSelect hybrid capture kit (Agilent Technologies, Santa Clara, CA) and massively parallel sequencing was performed on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA). Data analysis was performed with a bioinformatics pipeline that included open-sourced software (Mutect and GATK, Broad Institute, Cambridge, MA) as well as internally developed software (VisCap Cancer, Breakmer).
Results
We conducted a GWAS to identify common genetic variants associated with melanoma risk. The GWAS discovery set was a meta-analysis of the GWAS on melanoma risk in Harvard cohorts (NHS and Health Professionals Follow-up Study) and the MD Anderson GWAS on melanoma risk, including data for 2 449 178 SNPs that passed quality control procedures on a total of 8952 individuals (2298 cases and 6654 controls; (Table I).
In the discovery set of the GWAS, we further selected the 69 SNPs most significantly associated with melanoma risk (P < 5.0 × 10−5) and not in high linkage disequilibrium (R 2 < 0.8) with each other or with published melanoma risk SNPs for in silico replication in two independent GWAS datasets (Australian GWAS on melanoma risk and GenoMEL GWAS on melanoma risk; 5149 cases and 12 795 controls in total). Seven SNPs were nominally significantly associated with melanoma risk in the combined replication set (Supplementary Table 1, available at Carcinogenesis Online). The most significant SNP identified was on chromosome 3, rs13097028, P = 9.99 × 10−7 in the combined set. Another SNP identified on chromosome 3 was rs1031925, P = 4.27 × 10−6. Two SNPs on chromosome 2 were identified, rs11901831 (P = 1.48 × 10−5) and rs13404035 (P = 4.15 × 10−6). One SNP on chromosome 4 was identified, rs4698934 (P = 5.21 × 10−6). One SNP on chromosome 6 was identified, rs1889497 (P = 2.45×10−6). One SNP on chromosome 13 was also identified, rs4773180 (P = 5.69 × 10−5). None of the other 62 SNPs reached nominal significance in the combined replication set (Supplementary Table 1, available at Carcinogenesis Online).
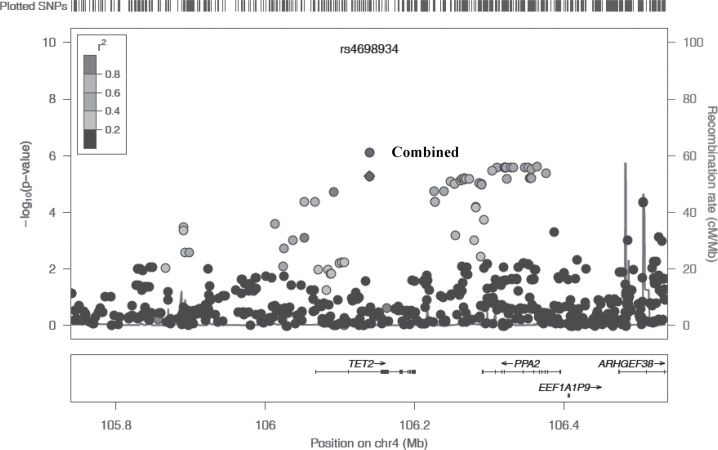
We further genotyped these seven SNPs in a nested case–control study within the NHS II of 234 melanoma cases and 238 controls. The SNP rs4698934 was replicated with a P value of 0.009. The OR per T allele was 1.18 (95% CI: 1.10–1.25, combined P = 7.70 × 10−7) (Table II). Regional association plot of SNPs at the TET2 locus is displayed in Figure 1.
Table II.
Association between seven SNPs and melanoma risk
| SNP information | Discovery set (Harvard and MD Anderson) | Replication 1 (GenoMEL and Australia) | Replication 2 (NHS II) | Meta-analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | Position | Gene | A1 | A2 | R 2 | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P |
| rs11901831 | 2 | 212225520 | ERBB4 | T | C | 1.96 | 1.39 (1.20–1.61) | 1.1×10−5 | 1.12 (1.01–1.25) | 0.030 | 1.13 (0.74–1.74) | 0.563 | 1.21 (1.11–1.31) | 1.2×10−5 |
| rs13404035 | 2 | 215121966 | SPAG16 | T | C | 1.99 | 0.84 (0.78–0.92) | 3.7×10−5 | 0.92 (0.86–0.98) | 0.008 | 1.22 (0.91–1.63) | 0.182 | 0.90 (0.86–0.94) | 2.0×10−5 |
| rs1031925 | 3 | 51379274 | DOCK3 | T | C | 1.97 | 1.29 (1.15–1.44) | 2.1×10−5 | 1.11 (1.03–1.19) | 0.005 | 0.92 (0.64–1.33) | 0.662 | 1.15 (1.08–1.22) | 8.1×10−6 |
| rs13097028 | 3 | 169464942 | ACTRT3 | T | C | 1.99 | 0.85 (0.78–0.92) | 4.9×10−5 | 0.92 (0.87–0.97) | 0.002 | 0.88 (0.67–1.16) | 0.376 | 0.89 (0.85–0.93) | 6.7×10−7 |
| rs4698934 | 4 | 106139387 | TET2 | T | C | 1.98 | 1.30 (1.16–1.45) | 5.2×10−6 | 1.10 (1.02–1.19) | 0.018 | 1.67 (1.13–2.45) | 0.009 | 1.18 (1.10–1.25) | 7.7×10−7 |
| rs1889497 | 6 | 65432283 | EYS | A | T | 1.99 | 0.80 (0.72–0.88) | 7.0×10−6 | 0.91 (0.84–0.98) | 0.011 | 0.82 (0.59–1.14) | 0.245 | 0.87 (0.82–0.92) | 1.6×10−6 |
| rs4773180 | 13 | 111062015 | COL4A2 | T | G | 1.96 | 1.32 (1.15–1.50) | 3.9×10−5 | 1.10 (1.00–1.21) | 0.044 | 1.31 (0.82–2.10) | 0.260 | 1.17 (1.09–1.26) | 3.2×10−5 |
A1, allele1, test allele; A2, allele2, reference allele; CHR, chromosome; CI, confidence interval; OR, odds ratio; Position, genome build 37.3; R 2, imputation R 2 in Harvard data.
Fig. 1.
Regional association plot of SNPs with melanoma risk. The vertical axis represents the −log10 P values. Recombination rates in this region are plotted in the background. LD is represented by R2 as five levels: 0–0.2, 0.2–0.4, 0.4–0.6, 0.6–0.8, and 0.8–1.0. The significance level of the SNP rs4698934 was shown for both discovery set and discovery and replication combined. The plot was generated based on Hapmap Build 37.3
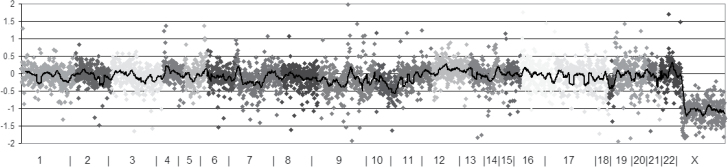
We examined the results of the previously identified melanoma susceptibility loci in the discovery set, and they were comparable with previous findings (Table III). All but two (CASP8 and CCND1) of the known loci were nominally replicated. For the CASP8 locus, the direction of association was consistent with prior publications. Among the novel SNPs we indentified in this study, SNPs at TET2 locus is the most significantly associated with melanoma susceptibility. As we reported previously, TET2-mediated global loss of 5-hmC in melanoma genome is an epigenetic hallmark event (26). Then, we sequenced 22 melanoma samples to a depth of 110 reads on average for each melanoma sample. A c.650C>T nucleotide mutation was identified in one melanoma sample at an allelic fraction of 15.5%. The variant affects Serine nucleotide 217 to Phenylalanine, resulting from the substitution to missense of amino acid p.S217F (Figure 2).
Table III.
Effect sizes for previously published melanoma risk loci in our melanoma GWAS
| Ref | Gene | CHR | SNP | MA | Other studies | Our study | ||
|---|---|---|---|---|---|---|---|---|
| P | OR | P | OR | |||||
| Barrett et al. (15) | — | 1 | rs7412746 | C | 9.0×10−11 | 0.89 (0.85–0.95) | 1.9×10−4 | 0.86 (0.79–0.93) |
| Barrett et al. (15) | PARP1 | 1 | rs3219090 | T | 9.3×10−8 | 0.82 (NA) | 0.005 | 0.89 (0.82–0.97) |
| Falchi. et al. (14) | CASP8 | 2 | rs13016963 | A | 8.6×10−10 | 1.14 (1.09–1.19) | 0.15 | 1.06 (0.98–1.15) |
| Amos et al. (17) | CLPTM1L/TERT | 5 | rs401681 | C | 0.004 | 0.73 (0.59–0.91) | 0.05 | 0.93 (0.85–1.00) |
| Brown et al. (12) | MTAP/CDKN2A | 9 | rs7023329 | G | 4.0×10−7 | 0.85 (0.80–0.91) | 2.3×10−5 | 0.85 (0.78–0.91) |
| Bishop. et al. (13) | MTAP | 9 | rs10757257 | G | 3.4×10−8 | 1.23 (1.15–1.30) | 3.9×10−5 | 1.18 (1.09–1.28) |
| Nan et al. (10) | TYR | 11 | rs1126809* | A | 2.8×10−7 | 1.21 (1.13–1.30) | 5.6×10−8 | 1.25 (1.15–1.35) |
| Brown et al. (12) | TYR | 11 | rs1393350 | A | 2.4×10−14 | 1.29 (1.21–1.38) | 6.7×10−5 | 1.20 (1.10–1.32) |
| Falchi. et al. (14) | CCND1 | 11 | rs1485933 | A | 0.0012 | 1.11 (1.04–1.18) | / | / |
| Falchi. et al. (14) | ATM | 11 | rs1801516 | A | 3.4×10−9 | 0.84 (0.78–0.90) | 0.03 | 0.88 (0.78–0.99) |
| Brown et al. (12) | MC1R | 16 | rs258322 | A | 2.5×10−27 | 1.67 (1.52–1.83) | 1.6×10−9 | 1.50 (1.31–1.71) |
| Gudbjartsson et al. (11) | PIGU | 20 | rs910873 | C | 9.9×10−16 | 1.75 (1.53–2.01) | 9.8×10−5 | 1.42 (1.19–1.69) |
| Gudbjartsson et al. (11) | MYH7B | 20 | rs1885120 | G | 9.8×10−16 | 1.78 (1.54–2.04) | 7.1×10−5 | 1.43 (1.20–1.69) |
| Falchi. et al. (14) | MX2 | 21 | rs45430 | G | 2.9×10−9 | 0.88 (0.85–0.92) | 0.035 | 0.90 (0.81–0.99) |
| Bishop. et al. (13) | PLA2G6 | 22 | rs132985 | C | 2.6×10−7 | 1.23 (1.15–1.30) | 2.8×10−5 | 1.18 (1.10–1.28) |
CHR, chromosome; MA, minor allele and also test allele; NA, not available; OR, odds ratio; our study, Harvard and MD Anderson melanoma GWAS; Ref, reference.
*SNP rs1126809 was not included in our GWAS, and we used SNP rs1847134 as substitution, the LD between the two SNPs is 0.96. SNP rs1485933 was not included in our GWAS, nor any SNPs in high linkage disequilibrium with it.
Fig. 2.
Somatic mutation of TET2 detection by next-generation sequencing in sporadic melanoma. Plot of copy number variation by chromosomes, sex chromosomes are excluded from the analysis. The vertical axis is the ratio of number of reads for this specimen and a panel of normal in log base 2 scales. A value of 0 denotes no difference from normal (diploid). When the sample contains 100% tumor cells, a value of −1 equals to 1 copy loss and 0.58 is 1 copy gain.
Discussion
We replicated most of the known loci, and we identified several novel loci predisposing to melanoma. The SNP rs13097028 on chromosome 3q26 locus was the most significant, but was not replicated in the NHS II. The SNP rs4698934 locus on chromosome 4q24 was replicated. The NHS II has a modest sample size, and the statistical power for this replication is limited. The SNP rs13097028 is not located in gene region, and genes near this locus are ACTRT3, MYNN, LRRC34, LRRIQ4, LRRC31, and MECOM.
The SNP rs4698934 is within the intron of TET2 gene at position of 106139387. Interestingly, the somatic mutation of exon 3 at position of 106155749 is identified in our study. TET2 (tet oncogene family member 2, or Ten-Eleven Translocation 2) is a family member of genes with enzymatic activities and plays important roles in epigenetic DNA demethylation (27). TET proteins have enzymatic activity for the conversion from 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC), which is a key intermediate of DNA demethylation. Mutations to the TET gene have been reported to be the most frequently mutated gene in myelodysplastic syndrome and tightly associated with reduced overall survival in hematologic malignancies (28,29). TET-mediated 5-hmC loss recently also has been reported in various solid malignancies, including breast cancer (30,31), oral squamous cell carcinoma (32), gastrointestinal stromal tumor (33,34), hepatocellular carcinoma (35), and brain tumor (36). In melanoma, several groups and we reported that the TET2-mediated 5-hmC loss is associated with prognosis and as a putative biomarker (26,37–39). In addition, we reported that increasing 5-hmC levels via overexpressing TET2 reversed the genome-wide 5-hmC loss toward a benign nevus-like pattern (26). We functionally characterized the significant impact of TET2-mediated 5-hmC loss in melanoma progression with diminished TET2 gene expression as a possible molecular mechanisms underlying global loss of 5-hmC previously (26). The findings of germline and somatic mutations in this study provide further evidence for a possible novel mechanism of 5-hmC loss is due to inactivation of TET2 enzymatic function due to TET2 gene mutation.
The major limitation of this study is its modest statistical power, which prevents detection of the modest effects of some genetic variants. With the current sample size, we have 80% power to detect an effect of 1.44. Furthermore, constitutional and environmental risk factors for melanoma, such as pigmentation and sun exposure, vary among European populations. These may underlie previously noted differences between the US and Australian or GenoMEL study findings (17). Further meta-analysis of the existing GWASs is warranted to identify additional melanoma susceptibility loci. Although the identification of low-risk alleles has stimulated much enthusiasm in the field, gaining a deeper understanding of the biological consequences and clinical utility of these findings remains challenging. Future prospective studies to elucidate whether germline TET2 SNP correlates with somatic TET2 mutation in melanoma patients will provide critical insight, which will require larger clinical study with multiple institutions.
Supplementary material
Supplementary Methods and Supplementary Table 1 can be found at http://carcin.oxfordjournals.org/.
Funding
National Institutes of Health (CA122838, CA87969, CA49449, CA176726, CA67262, CA167552 and 5P50CA093459-10).
Supplementary Material
Acknowledgements
We thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. The authors assume full responsibility for analyses and interpretation of these data.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- GWAS
genome-wide association study
- HPFS
Health Professionals Follow-up Study
- NHS
Nurses’ Health Study
- SNP
single-nucleotide polymorphism.
References
- 1. Lian C.G., et al. (2014). Skin cancer. In Stewart B.W. et al. (eds) World Cancer Report 2014. World Health Organization, Geneva, Switzerland, pp. 495–503 [Google Scholar]
- 2. Jemal A., et al. (2011). Global cancer statistics. CA Cancer J. Clin., 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 3. MacKie R.M., et al. (2009). Epidemiology of invasive cutaneous melanoma. Ann. Oncol., 20 (suppl. 6, vi1–vi7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tucker M.A., et al. (2003). Melanoma etiology: where are we? Oncogene, 22, 3042–3052 [DOI] [PubMed] [Google Scholar]
- 5. Gandini S., et al. (2005). Meta-analysis of risk factors for cutaneous melanoma. III. Family history, actinic damage and phenotypic factors. Eur. J. Cancer, 41, 2040–2059 [DOI] [PubMed] [Google Scholar]
- 6. Gandini S., et al. (2005). Meta-analysis of risk factors for cutaneous melanoma. I. Common and atypical naevi. Eur. J. Cancer, 41, 28–44 [DOI] [PubMed] [Google Scholar]
- 7. Meyle K.D., et al. (2009). Genetic risk factors for melanoma. Hum. Genet., 126, 499–510 [DOI] [PubMed] [Google Scholar]
- 8. Lin J., et al. (2008). Genetics of melanoma predisposition. Br. J. Dermatol., 159, 286–291 [DOI] [PubMed] [Google Scholar]
- 9. Fargnoli M.C., et al. (2006). High- and low-penetrance cutaneous melanoma susceptibility genes. Expert Rev. Anticancer Ther., 6, 657–670 [DOI] [PubMed] [Google Scholar]
- 10. Nan H., et al. (2011). Genetic variants in telomere-maintaining genes and skin cancer risk. Hum. Genet., 129, 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gudbjartsson D.F., et al. (2008). ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet., 40, 886–891 [DOI] [PubMed] [Google Scholar]
- 12. Brown K.M., et al. (2008). Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet., 40, 838–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bishop D.T., et al. (2009). Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet., 41, 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falchi M., et al. (2009). Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet., 41, 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrett J.H., et al. ; GenoMEL Consortium. (2011). Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet., 43, 1108–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macgregor S., et al. (2011). Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat. Genet., 43, 1114–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amos C.I., et al. ; GenoMEL Investigators; Q-Mega Investigators; AMFS Investigators. (2011). Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet., 20, 5012–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rafnar T., et al. (2009). Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet., 41, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Udayakumar D., et al. (2009). Moderate- to low-risk variant alleles of cutaneous malignancies and nevi: lessons from genome-wide association studies. Genome Med., 1, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saint-Martin C., et al. ; French Group of Familial Myeloproliferative Disorders. (2009). Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood, 114, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 21. Stacey S.N., et al. (2008). Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat. Genet., 40, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 22. Duffy D.L., et al. (2010). Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J. Invest. Dermatol., 130, 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nan H., et al. (2011). Genome-wide association study identifies novel alleles associated with risk of cutaneous basal cell carcinoma and squamous cell carcinoma. Hum. Mol. Genet., 20, 3718–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nan H., et al. (2011). Genome-wide association study identifies nidogen 1 (NID1) as a susceptibility locus to cutaneous nevi and melanoma risk. Hum. Mol. Genet., 20, 2673–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Biernacka J.M., et al. (2009). Assessment of genotype imputation methods. BMC Proc., 3 (suppl. 7, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lian C.G., et al. (2012). Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell, 150, 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ito S., et al. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature, 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saint-Martin C., et al. ; French Group of Familial Myeloproliferative Disorders. (2009). Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood, 114, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 29. Abdel-Wahab O., et al. (2009). Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood, 114, 144–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haffner M.C., et al. (2011). Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget, 2, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun M., et al. (2013). HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl Acad. Sci. USA, 110, 9920–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jäwert F., et al. (2013). Loss of 5-hydroxymethylcytosine and TET2 in oral squamous cell carcinoma. Anticancer Res., 33, 4325–4328 [PubMed] [Google Scholar]
- 33. Mason E.F., et al. (2013). Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in gastrointestinal stromal tumors: implications for mechanisms of tumorigenesis. Mod. Pathol., 26, 1492–1497 [DOI] [PubMed] [Google Scholar]
- 34. Zhang L.T., et al. (2013). Quantification of the sixth DNA base 5-hydroxymethylcytosine in colorectal cancer tissue and C-26 cell line. Bioanalysis, 5, 839–845 [DOI] [PubMed] [Google Scholar]
- 35. Liu C., et al. (2013). Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One, 8, e62828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kraus T.F., et al. (2012). Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int. J. Cancer, 131, 1577–1590 [DOI] [PubMed] [Google Scholar]
- 37. Gambichler T., et al. (2013). Loss of 5-hydroxymethylcytosine and ten-eleven translocation 2 protein expression in malignant melanoma. Melanoma Res., 23, 218–220 [DOI] [PubMed] [Google Scholar]
- 38. Larson A.R., et al. (2014). Loss of 5-hydroxymethylcytosine correlates with increasing morphologic dysplasia in melanocytic tumors. Mod. Pathol., 27, 936–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uchiyama R., et al. (2014). 5-Hydroxymethylcytosine as a useful marker to differentiate between malignant melanomas and benign melanocytic nevi. J. Dermatol. Sci., 73, 161–163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.