Abstract
HMGB1 is a highly conserved nuclear protein that displays important biological activities inside as well as outside the cell and serves as a prototypic alarmin to activate innate immunity. The translocation of HMGB1 from inside to outside the cell occurs with cell activation as well as cell death, including apoptosis. Apoptosis is also a setting for the release of cellular microparticles (MPs) which are small-membrane bound vesicles that represent an important source of extracellular nuclear molecules. To investigate whether HMGB1 released from cells during apoptosis is also present on MPs, we determined the presence of HMGB1 on particles released from Jurkat and HL-60 cells induced to undergo apoptosis in vitro by treatment with either etoposide or staurosporine; MPs released from cells undergoing necrosis by freeze-thaw were also characterized. As shown by both Western blot analysis and flow cytometry, MPs from apoptotic cells contain HMGB1, with binding by antibodies indicating an accessible location in the particle structure. These results indicate that HMGB1, like other nuclear molecules, can translocate into MPs during apoptosis and demonstrate another biochemical form of this molecule that may be immunologically active.
Introduction
HMGB1 is a highly conserved non-histone nuclear protein that displays important biological activities inside as well as outside the cell [1,2]. Inside the cell, HMGB1 can bind DNA and regulate chromosome architecture and regulate transcription [3,4]. Outside the cell, HMGB1 can serve as an alarmin to activate innate immunity and mediate inflammation in both normal and aberrant immunity. As shown in studies in both in vivo and in vitro systems, HMGB1 can translocate from the nucleus to the cytoplasm of cells with eventual release during activation as well as cell death [5,6]. Depending on the setting for its release, HMGB1 can undergo post-translational modification and redox reactions that modulate its immunological properties [7–9].
Once in an extracellular locale, HMGB1 can trigger innate immune responses by binding to receptors including RAGE (receptor for advanced glycation end-products), TLR 2 and TLR4 [1,2,10,11]. Furthermore, HMGB1 can bind to other mediators such as cytokines (e.g., IL-1) or LPS to create novel structures that can drive responses via the receptor for the bound mediator [12–14]. An important contribution of HMGB1 to disease pathogenesis is supported by observations of increased levels of HMGB1 in the blood and tissue in disease settings as well as the efficacy of targeting HMGB1 in animal models such as collagen-induced arthritis, shock and liver cell injury [1,2].
As an alarmin or DAMP (damage-associated molecular pattern), HMGB1 is released from cells in conjunction with many nuclear, cytoplasmic and membrane constituents, some of which also have immune activity [15–17]. This release can occur during immune cell activation as well as cell death, whether by apoptosis, necrosis, NETosis or pyroptosis; pyroptosis is an inflammatory form of cell death that results from triggering of the inflammasome [18–21]. Importantly, HMGB1 release occurs in the same settings as the release of microparticles. Microparticles are small membrane-bound vesicles that emanate from cells by a blebbing process. Particles range in size from 0.1 to 1.0 µm and include, among their constituents, nuclear molecules such as DNA and histones. Like HMGB1, microparticles have potent biological activities and can induce inflammation and promote thrombosis [22,23].
In the current studies, we have investigated the presence of HMGB1 in microparticles derived from apoptotic cells, extending findings of other studies indicating its translocation during death processes. While original studies indicated nuclear retention of HMGB1 during apoptosis, subsequent studies demonstrated HMGB1 release from cells undergoing apoptosis [7,18]. The magnitude of HMGB1 release during apoptosis may be less than that observed during necrosis although models for necrosis vary significantly in the process of HMGB1 release [21]. To characterize further the expression of HMGB1 in a particulate form, we analyzed the content of HMGB1 on MPs from Jurkat and HL-60 cells undergoing apoptosis in vitro, using immunoblotting and flow cytometry. As results presented herein indicate, HMGB1 is a component of MPs derived from cells undergoing apoptosis in vitro, defining another form of HMGB1 that may have biological activity as well as utility as a biomarker.
Materials and Methods
Cell Culture
HL-60 and Jurkat cell lines were obtained from the Duke University Cell Culture Facility and were maintained in RPMI 1640 (Life Technologies, New York, NY) supplemented with 10% fetal calf serum (Atlanta Biologicals, Atlanta, GA) and 20 µg/ml gentamicin (Life Technologies). The cells were cultured at 37°C, 5% CO2, and divided every 2–3 days to maintain logarithmic growth.
Induction of cell death
HL-60 and Jurkat cell lines were induced to undergo apoptosis or necrosis as described below. The cells were first pelleted by centrifugation at 500×g and resuspended with fresh culture media to a concentration of 3×106/ml. Depending on the experiment, the cells were cultured in 6 well plates (Greiner, Monroe, NC) or T175 tissue culture flasks (Greiner). To induce apoptosis, the cells were incubated for 24h at 37°C, 5% CO2 with either 1µM staurosporine (STS) or 30µg/ml etoposide. Both compounds were purchased from Sigma-Aldrich, St. Louis, MO.
For these experiments, we employed conditions previously used to demonstrate MP release during apoptosis [24,25]. To verify induction of apoptosis, cells were collected after 5h and assessed for staining with annexin V and propidium iodide (PI). Briefly, HL-60 and Jurkat cells in culture media were treated with etoposide and STS at the doses indicated above for 5h at 37°C, 5% CO2. The 5h time point was used instead of 24h to provide a time frame to allow observation of early phases of apoptosis; at 24 hours, progression to late apoptosis or secondary necrosis occurs.
HL-60 and Jurkat cells from untreated cultures were included as negative controls for PI and annexin V staining. Heat treated HL-60 and Jurkat cells (56°C, 30min) were included as positive controls for PI staining. Approximately 6×106 cells were pelleted and washed with 1X phosphate buffered saline (PBS, Life Technologies). The cells were pelleted again and resuspended in 2mls of annexin V binding buffer [10mM Hepes, pH7.4 (Life Technologies), 140 mM NaCl (Sigma-Aldrich), 2.5mM CaCl2]. FACS tubes were then prepared with100 uls of cell suspension per tube. For each treatment, samples were stained with PI alone (5µl of a 1mg/ml stock/tube; Sigma-Aldrich); annexin V alone (3µl/tube; Becton Dickinson, Mansfield, MA); and PI and annexin V together. An unstained sample for each treatment was prepared to provide background levels of staining. The staining reactions proceeded for 30 min at room temperature (RT). On completion, the volume for each tube was increased to 400 µls with annexin V binding buffer. Cell death was verified by flow cytometry using a FACScan (Becton Dickinson) instrument with settings previously established to detect cells staining for annexin V and propidium iodide.
For necrosis, HL-60 and Jurkat cells were subjected to repetitive freeze-thaw. Briefly, 3×106/ml cells were distributed into 15 ml tubes (no more than 3ml/tube). The cells then underwent three cycles of freeze-thawing (2min in liquid nitrogen, directly followed by 4min in a 37°C water bath). Immediately after the freeze-thawing process, EDTA was added to a final concentration of 25mM to inhibit any DNase activity.
Quantitation of MPs by flow cytometry
For quantification of MPs, the treated cultures were first pipetted five times to assure suspension of MPs which may have become attached to cells or culture vessels. Following this step, cells and larger debris were removed by centrifugation at 500xg for 5min at 20°C. The resulting supernatant, containing the MPs, was then carefully removed and stored on ice. The numbers of MPs/µl in the supernatant were measured by flow cytometry using a FACScan instrument. Data were collected using FlowJo Collector’s Edition software (Tree Star, Inc., Ashland, OR) and analyzed using FlowJo analysis software (Tree Star). The supernatants were diluted with phosphate buffered saline to produce samples where detectable events/second were no more than 1000 MPs/second; dilutions ranged from 1:10–1:100 depending on the cellular treatment.
Events (MPs) were measured using side scatter (SSC) detection settings, and, using pre-determined parameters, defined by the use of sizing beads (sizes 0.1µm, 0.5µm and 1µm; Polysciences, Inc., Warrington, PA) events above a given size threshold were collected. Data were collected until 10000 events were counted or 30 seconds of counting had elapsed. Background events for buffer alone were subtracted and the counts/µl of the samples were calculated using the flow rate of the machine. Background events contributed between 7–9% of total counts depending on the culture treatment.
Western blotting of HMGB1 in MPs
Briefly, Jurkat and HL-60 cells were counted and resuspended at 3×106/ml and treated to induce apoptosis (etoposide or STS) or necrosis (freeze-thaw). To obtain an adequate number of MPs to allow detection of HMGB1 by Western blotting, usually 40 mls of supernatant were used. The resulting sample was centrifuged twice for 5min at 500×g at 20°C to remove cells and large sized debris. The supernatant was collected and filtered through a 1.2µm syringe filter (Pall Corporation, Cornwall, UK). The MPs were then isolated from the supernatant by ultracentrifugation (Beckman L7-65 Ultracentrifuge, SW 41 Ti swinging rotor) for 25min at 150000g at 10°C.
Following this procedure, the supernatant was carefully removed and the remaining MP pellet was resuspended in 1X PBS at a volume of 2mls/pellet. This stock was then transferred to a 2ml tube and the MPs were pelleted for a second time at 30 min at 16000g at RT using a microcentrifuge (Denville, South Plainfield, NJ). On completion, the MP pellet was suspended in the lowest volume of 1X PBS possible (25–30 µl) to try to maintain a concentrated MP sample. This MP stock was then stored at −20°C until further analysis.
Quantitation of the amount of protein in the MP suspension was performed using the BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). For Western blotting, samples were loaded at an equal amount of protein per lane (10ug/lane) and were resolved using 4–12% NuPAGE Tris-Bis SDS polyacrylamide gel (Life Technologies). Recombinant his-tagged human HMGB1 (purchased from MyBioSource, San Diego, CA) was also included on the gel at 10 ng/lane as a positive control for the antibody. Protein was transferred to polyvinylidene difluoride membranes and blocked with 5% dry milk in PBS-Tween (0.1%), and blotted with a 1:10000 dilution of rabbit anti-mouse HMGB1 antibody (polyclonal preparation developed to residues 150 of the c-terminus; Abcam, Cambridge, MA). The membrane was then incubated with an anti-rabbit HRP-conjugated antibody (Thermo Fisher Scientific), followed by incubation with SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher Scientific). The resulting signal was captured using charge-coupled device camera (FluorChem8900; Alpha Innotech, Santa Clara, CA).
Determination of HMGB1 levels on MPs after cell death
To determine the amount of HMGB1 present on MPs in the supernatant, cells were treated as described above to induce apoptosis (etoposide or STS) or necrosis (freeze-thaw method). After treatment, the culture was pipetted to assure particle suspension and then centrifuged as described above to remove cells. The resulting supernatant was divided into two portions, with one portion remaining on ice while the other was centrifuged at 16,000×g at 20°C for 30min. The number of MPs/µl of supernatant were then quantitated by flow cytometry (as described above) to verify MP removal. The samples were then stored at −20°C until further analysis. The two samples were then thawed on ice and concentrated using 10 kDa NWCO ultra centrifugal filters (Millepore, Billerica, MA) to the same final volume. The concentrated samples were analyzed by Western blot for HMGB1 content; an equal volume of sample was loaded per well.
Measurement of MP HMGB1 by flow cytometry
MPs were purified from treated cell supernatants as described above. After suspension of MPs in PBS, the MP numbers were quantitated by flow cytometry as described above. For experiments to digest DNA, MPs were treated with recombinant DNase I (rDNase; 10U rDNase / 4×106 MPs; Life Technologies) for 60 min at 37°C. After this treatment, the MPs were transferred to flow cytometry tubes at an MP density of 2×105/tube. To assess HMGB1 content, samples were stained with a Phycoerythrin (PE)-conjugated mouse anti-human HMGB1 antibody (2ul/tube; monoclonal antibody directed to residues 2–215, R&D systems, Minneapolis, MN) or a PE-conjugated mouse IgG2b (R&D Systems) for 60min at 20°C. To assess DNA, MPs were stained with a mouse monoclonal anti-dsDNA antibody (clone 163p.132; 4ug/tube; a kind gift from Dr. Tony Marion) or a mouse IgG2b (isotype control; 4ug/tube; Sigma-Aldrich). After this step, MPs treated with either anti-DNA or isotype control were incubated with a F(ab')2 sheep anti-mouse IgG PE (Sigma-Aldrich) for 30min at 20°C in the dark. The samples were then analyzed by flow cytometry.
Statistics
All data are expressed as mean and standard deviation or standard error as described in the figure legends. Statistical analysis was performed using Microsoft Excel 2010 (Redmond, WA). For analysis between different groups, the Student t test was used, and a value of p<0.05 was considered statistically significant.
Results
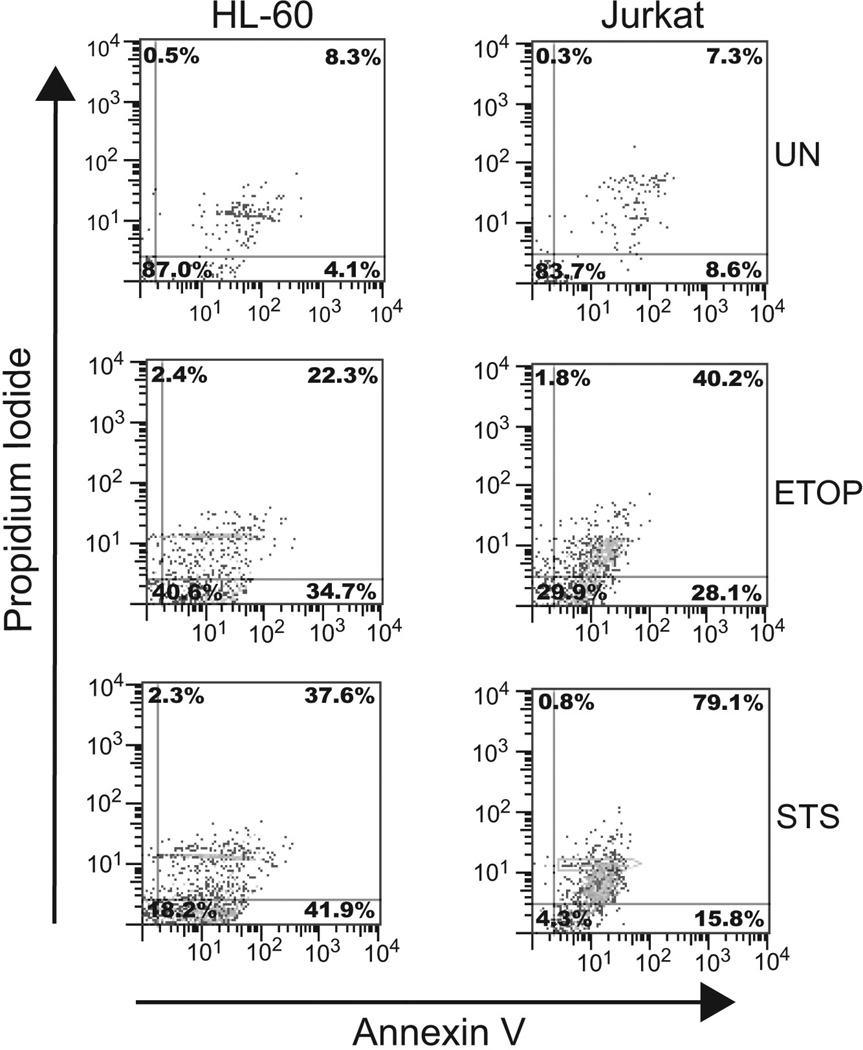
In this study, we used Jurkat and HL-60 cell lines as models to assess the expression of HMGB1 on microparticles released from cells undergoing apoptosis or necrosis in vitro. Jurkat cells were derived from a human T cell leukemia while HL-60 cells were derived from a patient with acute myelogeneous leukemia. To induce apoptosis, we used etoposide or staurosporine at concentrations previously shown to produce a high level of apoptosis as determined by staining with annexin V and propidium iodide [24,25]. In a representative experiment (Figure 1), Jurkat cells showed 78% annexin V positivity with etoposide treatment and 79% with staurosporine at 5 hours. Similarly, HL-60 cells showed 56% annexin V positivity with etoposide and 79% with staurosporine. Thus, the majority of cells were undergoing apoptosis under the conditions of these experiments.
Figure 1. Induction of apoptosis in HL-60 and Jurkat cells.
Cells were induced to undergo apoptosis by incubation with etoposide (ETOP; 30µg/ml) or STS (1µM). Untreated cells were included to show background levels of apoptosis induction in healthy cells. The treated cells were isolated, stained with annexin V and PI and examined by flow cytometry. The data collected are presented in the form of quadrant plots. The percentages of cells staining positive within a given quadrant are shown.
As a model of necrosis, we selected freeze-thaw in view of previous studies indicating that this method of necrosis induction leads to the most abundant release of both HMGB1 and MPs [21].
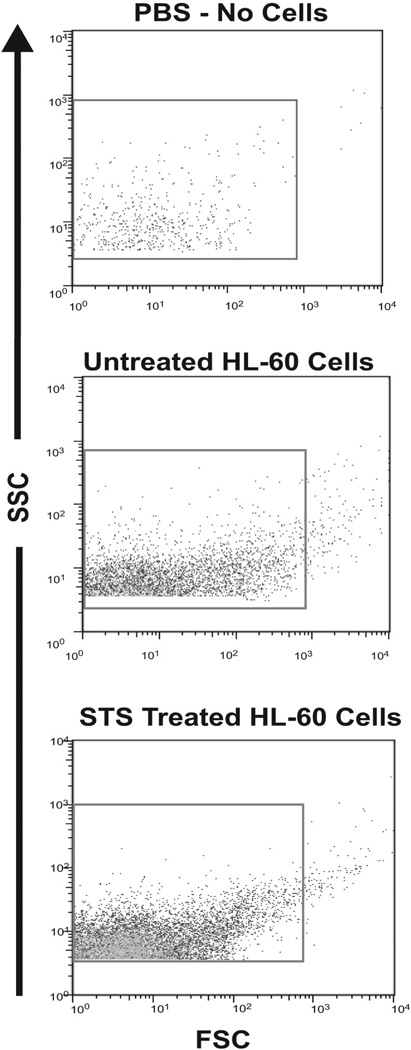
Flow cytometry was used to study the number of MPs released by the different treatments and their molecular content. To quantitate the numbers of MPs released from dying cells, the cells were treated to induce cell death. Supernatants from the treated cells were collected and centrifuged to remove of cells and large debris. The supernatants were diluted and the number of MPs present were counted. Figure 2 shows data in the form of density dot plots. Sizing beads were used to determine gates to select events falling within the size range of microparticles (0.1µm–1µm). The gate is indicated by a box within each of the presented plots.
Figure 2. Microparticle detection by flow cytometry.
Samples were analyzed by flow cytometry to assess microparticle production by cells. The data are represented as dot plots and show microparticle detection in PBS alone, untreated HL-60 cells and STS treated HL-60 cells. The box indicates the location of the gate used to select detected events that represent molecules of sizes appropriate to MPs. The gate was determined by the use of sizing beads.
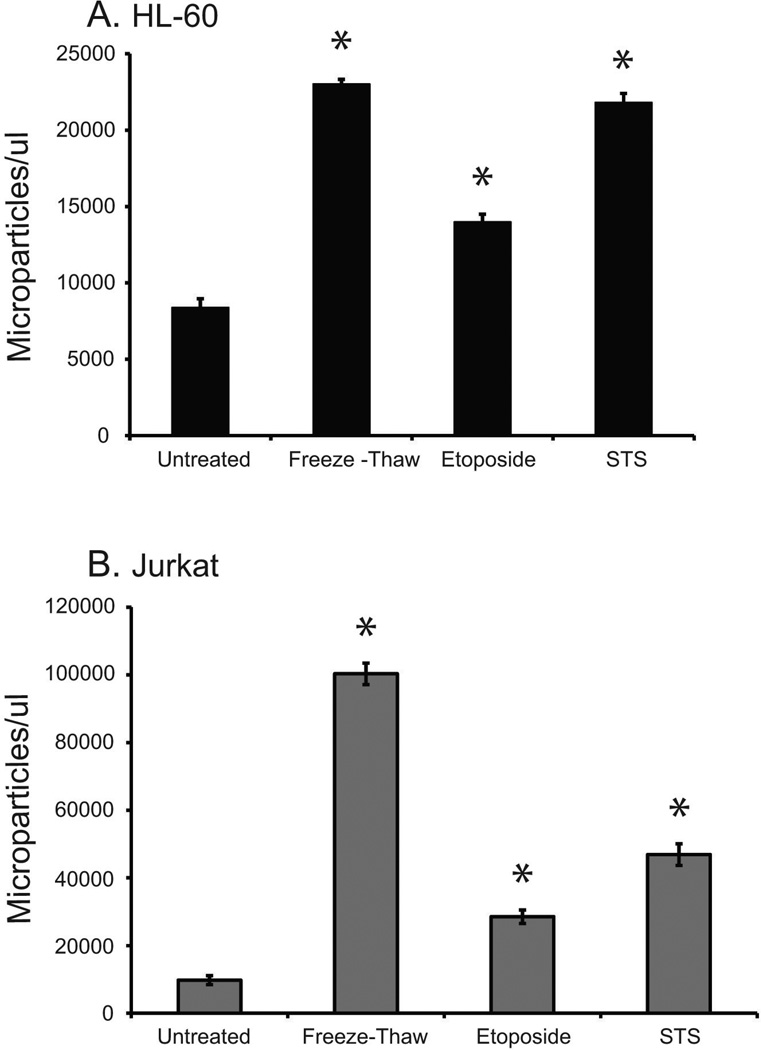
Figure 3 presents results of an analysis of particle release by the cell lines as assessed by flow cytometry. As the results indicate, the treatments led to increased production of MPs compared to untreated control cultures although levels differed between cell lines and inducing condition. The data shown in Table 1 indicate that Jurkat cells produced more MPs per treatment when compared to HL-60 cells. Also, of the treatments used in these experiments, MP production in HL-60 and Jurkat cells was greatest in response to staurosporine (20932 MPs/µl ± 906) and freeze-thaw (72782 MPs/µl ± 7777), respectively.
Figure 3. Microparticle release by Jurkat and HL-60 cell lines.
HL-60 (A) and Jurkat (B) cell lines were treated to induce necrosis (freeze-thaw) or apoptosis (etoposide (30ug/ml) or STS (1uM)). Data for untreated cells are also shown. Microparticle release by the cells in post-treatment supernatant was quantitated by flow cytometry analysis using SSC detection settings determined by sizing beads. The data shown are representative of three separate experiments. All treatments were performed in triplicate; standard deviations are shown. Microparticle release at a level that is statistically significant above untreated cells is denoted with an asterisk (*).
Table 1.
Microparticle production by untreated cells and cells induced to undergo apoptosis or necrosis.
| Mean MPs/ul ± SE | (range of values) | |
|---|---|---|
| HL-60 | ||
| Untreated | 7297 ± 125.3 | (5770–9431) |
| Etoposide | 14946 ± 357.4 | (12470–21200) |
| Staurosporine | 29032 ± 906.6 | (20653–40086) |
| Freeze-Thaw | 17727 ± 777.7 | (10572–22828) |
| Jurkat | ||
| Untreated | 10894 ± 311.1 | (7806–14970) |
| Etoposide | 41458 ± 1701.3 | (26226–73656) |
| Staurosporine | 62617 ± 1909.1 | (41160–87810) |
| Freeze-Thaw | 72782 ± 3786.1 | (43727–105800) |
For treatments shown, n=7–9. SE, standard error.
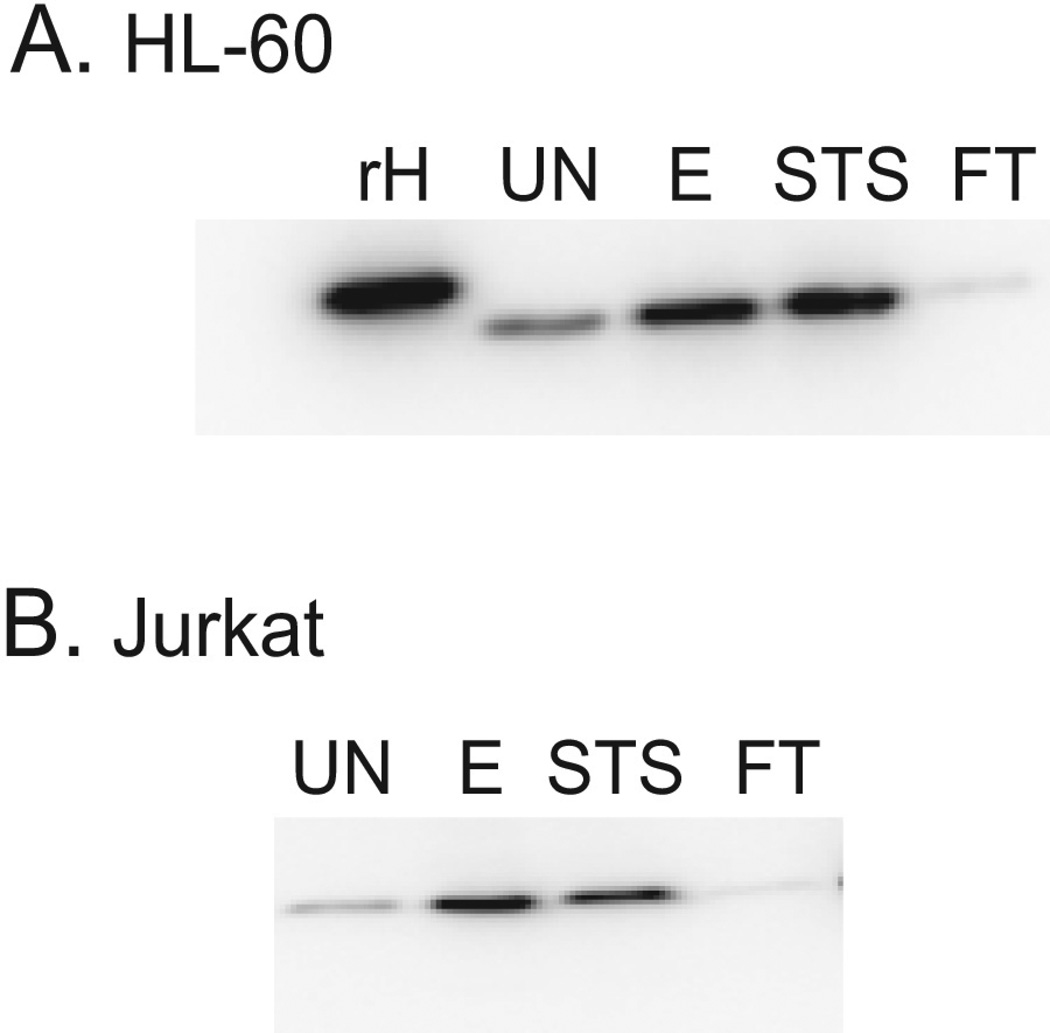
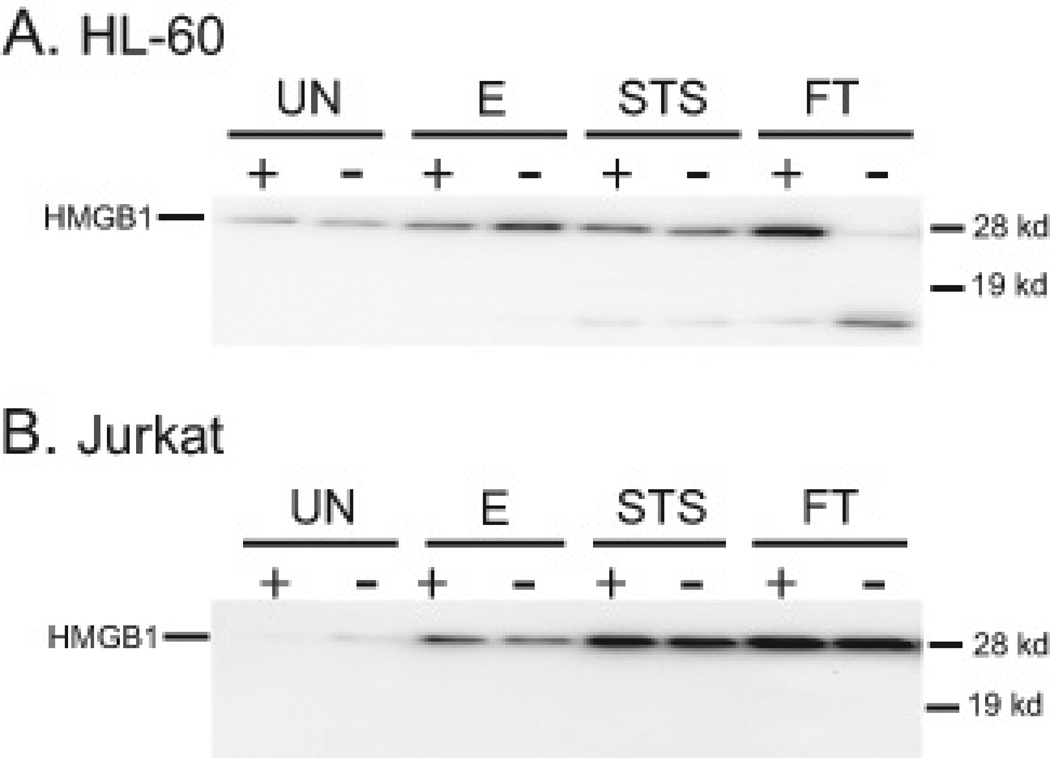
To determine the expression of HMGB1 on MPs, particles from the supernatants were prepared by differential centrifugation and then analyzed by immunoblotting. Figure 4 presents these results. As these data indicate, HMGB1 is present on particles from cells treated with etoposide or staurosporine as well as control cells, although the amounts of HMGB1 on MPs from apoptotic cells were greater than the amounts of HMGB1 on MPs from control cultures. These findings are consistent with HMGB1 translocation to particles during apoptosis. In interpreting these experiments, it is important to consider the nature of the control particles. MPs from control cultures may in fact derive from dying cells since, in untreated cultures, we consistently found cells undergoing apoptosis as indicated by cellular staining with annexin V and propidium iodide. These findings could suggest that the extent of HMGB1 translocation may be increased by agents such as etoposide or staurosporine. In contrast to the findings with MPs from apoptotic cells, levels of HMGB1 on MPs from cultures undergoing necrosis by freeze-thaw were less than the amounts of HMGB1 seen on MPs from control cultures.
Figure 4. Expression of HMGB1 on microparticles from HL-60 and Jurkat cells.
Microparticles from post-treatment supernatants [untreated (UN), etoposide (E), STS and freeze-thaw (FT)] of HL-60 (A) and Jurkat cells (B) were isolated by centrifugation and were then analyzed for HMGB1 content by western blot. A recombinant his-tagged human HMGB1 sample (rH) was analyzed as a positive control for the antibody and to indicate the expected location of human HMGB1 on the gel. Equal concentrations of sample protein were loaded in each lane. The data shown are representative of three separate experiments.
To assess the contribution of particles to the total amount of HMGB1 released from cells, we analyzed the supernatants of the apoptotic and necrotic cells, with or without centrifugation to remove particles. In the experiments performed, centrifugation removed greater than 91–99 % of particles (data not shown). As the results in Figure 5 indicate, the contribution of MPs to HMGB1 released during apoptosis appeared limited. Centrifugation to remove MPs did not reduce the amount of HMGB1 in the supernatants of either HL-60 or Jurkat cells treated with etoposide or staurosporine. The results with freeze-thaw treatment, however, differed for the two cell types. With Jurkat cells, removal of MPs did not change significantly the amount of HMGB1 in the supernatant, which is in agreement with results seen for cultures undergoing apoptosis. In contrast, supernatants from cultures of HL-60 cells subjected to freeze-thaw treatment, centrifugation significantly reduced the amount of HMGB1 detected. These studies also indicate a change in the apparent molecular weight of HMGB1 remaining in the supernatant; the detection of this band and its amount were variable, however. These findings could suggest protein degradation during necrosis of these cells as well as sample processing.
Figure 5. Assessment of the amount of HMGB1 in supernatants on microparticles.
Supernatants from untreated (UN) or treated [etoposide (E), STS or freeze-thaw (FT)] HL-60 (A) and Jurkat cells (B) were collected. The supernatants were divided into two equal aliquots. One aliquot was centrifuged to remove microparticles, thus generating a microparticle-free supernatant. The supernatants, with microparticles (+) and without microparticles (−), were concentrated using centrifugal filters. The samples were then analyzed by Western blot analysis for the presence of HMGB1. A protein with a molecular weight lower than that of the main protein band detected by anti-HMGB1 antibody was present in the blot of HL-60 supernatants (A). The presence of this band appears to be unique to HL-60 cells under these experimental conditions. The data shown are representative of three separate experiments.
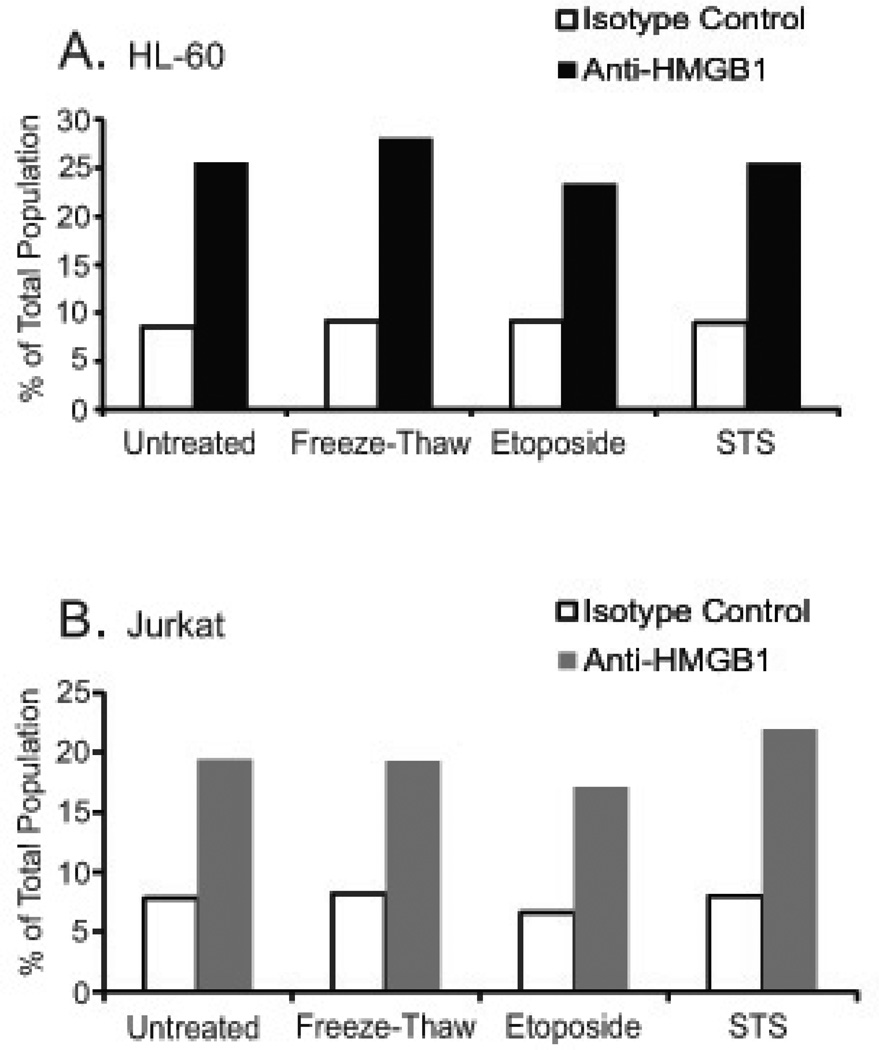
As another approach to assess the presence of HMGB1 on particles, MPs were stained with a murine monoclonal anti-HMGB1-PE conjugated antibody and analyzed by flow cytometry. These studies demonstrated low levels of staining although an increase in the percentage of particles positively staining with HMGB1 was consistently observed in repeat experiments (Figure 6). These findings support the presence of HMGB1 on particles and indicate its disposition in a locale accessible to protein binding.
Figure 6. Detection of microparticle HMGB1 by flow cytometry.
To assess microparticle HMGB1 by flow cytometry, the microparticles from untreated (UN) and treated [freeze-thaw (FT), etoposide (E) and STS] HL-60 (A) and Jurkats (B) cells were stained with anti-HMGB1-PE conjugated antibody (black bars) and analyzed by flow cytometry. Microparticles were also stained with an isotype control, also conjugated with PE, to determine background staining (white bars). The data shown are representative of three separate experiments.
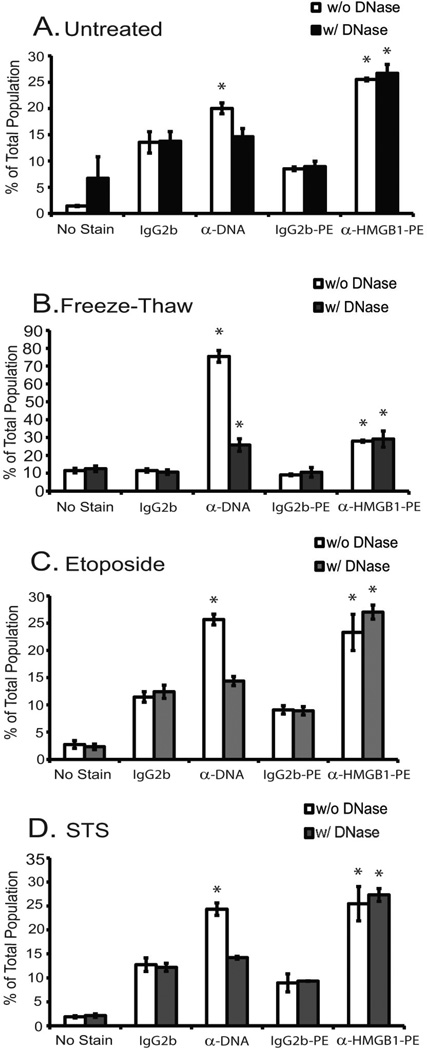
We then extended this system to assess the relationship between DNA and HMGB1 on the particles and to determine whether HMGB1 binding reflects association with or attachment to DNA (Figure 7). As shown previously, monoclonal anti-DNA antibodies can bind particles in a form susceptible to digestion by DNase [24,25]. For this experiment, purified MPs were incubated with or without DNase. We then compared the staining of these MPs with a monoclonal anti-DNA antibody or monoclonal anti-HMGB1-PE conjugated antibody; to detect the binding of the anti-DNA antibody, a PE-conjugated, anti-mouse IgG secondary reagent was used. For these experiments, we only used MPs produced by HL-60 cells. As results presented in Figure 7 indicate, while DNase digestion reduced staining by the anti-DNA antibody, it did not affect the staining by the anti-HMGB1 antibody. These results suggest that the presence of HMGB1 on MPs does not depend on the presence of DNA; alternatively, this HMGB1 may be attached to a form of DNA resistant to nuclease digestion.
Figure 7. The effect of rDNase on HMGB1 binding to microparticles.
Microparticles from untreated (A) and treated (freeze-thaw (B), etoposide (C) and STS (D)) HL-60 cells were incubated with and without rDNase I. The microparticles were then stained with anti-DNA and anti-HMGB1-PE conjugated antibodies, followed with analysis by flow cytometry. Microparticles that were unstained or labeled with isotype controls (IgG2b and IgG2b-PE conjugated) were also examined to show background staining levels. The experiment was repeated three times and data from all 3 experiments were used to calculate the standard error bars shown. Treatments that resulted in statistically significant antibody binding above that of its corresponding isotype control are denoted by an asterisk.
Discussion
In these studies, we demonstrated the presence of HMGB1 in microparticles released during in vitro death of Jurkat and HL-60 cells, indicating that extracellular HMGB1 may exist in both a particulate and non-particulate form. Thus, we showed using Western blotting that particles from cells undergoing apoptosis with staurosporine or etoposide contained HMGB1 in a form that is accessible to antibody binding and resistant to enzymatic removal of DNA, a molecule that HMGB1 binds in the nucleus. We also showed that, while particles contained HMGB1, most of the HMGB1 in the supernatants of apoptotic cells is present in a non-sedimentable or soluble form; similar findings were observed with cultures of Jurkat cells although the possible proteolysis with HL-60 cells limited interpretation of findings with that cell type. Together, these findings are consistent with other studies suggesting concomitant expression of HMGB1 and MPs during cell death (as well as activation) and establish further that particles can be a source of bioactive molecules such as HMGB1 to stimulate innate immunity.
As shown previously by immunological and biochemical techniques, MPs contain a wide variety of cellular components and, indeed, contain significant amounts of extracellular DNA and histones [26–28]. While these molecules are usually confined to the nucleus, they can undergo rearrangement and degradation during apoptosis as well as translocation into vesicle structures that most likely correspond to blebs [29,30]. Blebbing is a prominent feature of apoptosis, occurring at various stages in this process. The physiological role of blebbing is not clear although it may regulate the surface to volume relationship of cells during apoptotic shrinkage; blebbing may also facilitate clearance of the debris of dead and dying cells by packaging it in a more compact form. Previous studies have shown that MPs from the Jurkat and HL-60 cell lines contain DNA and histone [24,25]. The presence of HMGB1 is not unexpected especially in view of evidence that, during apoptosis, HMGB1 may become more adherent to chromatin in contrast to its usual free mobility in the nucleus of living cells. Studies by Schiller et al have provided evidence for this possibility [31].
We demonstrated the presence of HMGB1 in particles from apoptotic cells with both immunoblotting and flow cytometry although, for technical reasons, we used different antibodies for these experiments. In contrast, analysis of particle preparations from cells undergoing freeze-thaw indicated a different pattern of HMGB1 expression depending upon assay. Thus, Western blotting showed much lower levels of HMGB1 in particle preparations from freeze-thaw HL-60 cells than apoptotic cells and, with Jurkat and HL-60 cells, lower levels than particles from control cultures. With flow cytometry, however, particles from necrotic HL-60 cells showed staining levels similar to those of apoptotic cells. For these experiments, we used particles exposed to freeze-thaw as a model for necrosis since our previous studies indicated that this treatment, among other approaches to induce necrosis, produced the greatest amount of HMGB1 as well as number of particles in the supernatant [21]. We have previously discussed the relevance of freeze-thaw as a model for necrosis although, interestingly, freeze-thaw has been used to treat tumors, resulting in “cryo-shock” depending on the details of the procedure [32].
The difference in the presence of HMGB1 in particles or its detection by different reagents and analytic techniques could have a number of explanations. As shown in a variety of studies, the form of HMGB1 emanating from apoptotic and necrotic cells differs in redox state and other post-translational modifications that affect intra-nuclear mobility, interaction with chromatin and immunological activity [1,5–9]. It is thus possible that the overall content of HMGB1 as well as its composition in terms of modified forms in particles from apoptotic and necrotic sources may vary. Furthermore, depending on the specificity of the antibody used for detection as well as assay (i.e., immunoblotting vs. flow cytometry), the ability to measure different forms of HMGB1 (including degradation products) may vary.
For apoptotic cells, however, it appears that particles represent only a limited amount of the total HMGB1 released since centrifugation to remove particles did not significantly reduce the amount of HMGB1 in the supernatants. This situation also pertains to cultures of Jurkat cells treated with freeze-thaw; with HL-60 cultures, degradation of HMGB1 appears to be occurring although its extent varied among experiments. This degradation may reflect the action of enzymes released during killing since previous studies have shown that necrosis can lead to cleavage of nuclear proteins, with differences depending on inducing mechanisms (i.e., treatment with chemical agents vs. detergent lysis) [33]. Our findings would suggest that freeze-thaw may lead to more extensive proteolytic activity; we have previously shown that freeze-thaw preparations have high amounts of nuclease activity [21]. In this regard, HMGB1 cleavage occurs with other enzymes including caspase 1 and thrombin-thrombomodulin complexes. Such degradation may modify HMGB1’s immunological activity during different death processes [34–36]. This issue is now under study.
Studies using flow cytometry provide additional insight into the presence and disposition of HMGB1 in the particles. In recent studies, we investigated the expression of microparticles in the blood of healthy volunteers treated with LPS as part of studies to assess the effects of glucocorticoids and inhaled nitric oxide on mediators produced in response to immune activation [37]. Using plasma collected at various times from these volunteers, we measured the production of MPs by flow cytometry. As results of these studies showed, following LPS treatment, levels of MPs from a number of cell populations rose significantly in the blood, with the presence of HMGB1 demonstrated by flow cytometry on MPs from platelets and monocytes. Interestingly, levels of HMGB1 in the blood of these subjects did not show elevations by an ELISA, suggesting that particle HMGB1 may be a more sensitive measure of HMGB1 translocation than free levels in the blood. In these studies, the process leading to particle release is not clear and may result from pyroptosis as well as activation or apoptosis.
It is important to note that the application of flow cytometry to protein analysis of particles can be problematic since particles are very small and have as much as 104 times less material than the cell from which it derives [38,39]. Under these circumstances, measuring levels of proteins is complicated, with binding by isotype controls also limiting assessment. Despite these potential technical limitations, nevertheless, our findings strongly indicate that HMGB1 is present in these structures and is accessible to antibody binding either because of surface expression or its presence on the interior of particles with a porous membrane. Like apoptotic cells, particles can stain with propidium iodide, indicating that the membrane is permeable, likely from changes during apoptosis. In this situation, distinguishing surface expression vs. an interior locale is not possible.
Using flow cytometry, we showed further that DNase digestion did not affect the binding of the anti-HMGB1 antibody although it did reduce the binding of a monoclonal anti-DNA. These findings suggest that the presence of HMGB1 in particles may not be dependent on an interaction with DNA although alternative explanations are possible, however. Thus, in the context of a particle, DNase may not efficiently degrade DNA because of the nature of the microenvironment. Furthermore, HMGB1 (or other nuclear proteins) may protect from DNase digestion the DNA that anchors the HMGB1 to the particle. In this regard, we cannot exclude the possibility that HMGB1 on particles derives from a soluble form of HMGB1 that binds to the particle by interaction with another component. Such binding may also explain the presence of HMGB1 on particles in subjects administered with LPS [37].
While only a limited amount of HMGB1 released from cells may be in a particle form, it can nevertheless be biologically important, depending on its redox state. Thus, in the context of a particle, HMGB1 may be in close proximity with other bioactive molecules such as cytokines like IL-1β [40–41]. Previous studies have shown that particles (depending on their origin) contain IL-1 and that HMGB1 can interact with IL-1β, with simultaneous signaling enhancing activity afforded by display on particle structure and potential for interaction with multiple receptors. Since particles likely contain other DAMPs, these structures may represent a particular effective element in signaling in innate immunity as a reservoir of cytokines, DAMPs and potential PAMPs. Future studies will define more precisely the role of MPs in immune cell activation and the respective roles of soluble and particulate HMGB1.
Acknowledgments
This work was supported by a VA Merit Review Grant, a grant from Alliance for Lupus Research (ALR), and NIH 5U19-AI056363.
References
- 1.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8:195–202. doi: 10.1038/nrrheum.2011.222. [DOI] [PubMed] [Google Scholar]
- 3.Štros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochem Soc Trans. 2012;40:341–346. doi: 10.1042/BST20120014. [DOI] [PubMed] [Google Scholar]
- 5.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 6.Bonaldi T, Talamo F, Scaffidi P, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venereau E, Casalgrandi M, Schiraldi M, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209:1519–1528. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H, Antoine DJ, Andersson U, Tracey KJ. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol. 2013;93:1–9. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JS, Svetkauskaite D, He Q, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279:7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Hreggvidsdottir HS, Palmblad K, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A. 2010;107:11942–11947. doi: 10.1073/pnas.1003893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wähämaa H, Schierbeck H, Hreggvidsdottir HS, et al. High mobility group box protein 1 in complex with lipopolysaccharide or IL-1 promotes an increased inflammatory phenotype in synovial fibroblasts. Arthritis Res Ther. 2011;13:R136. doi: 10.1186/ar3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hreggvidsdottir HS, Lundbert AM, Aveberger AC, Klevenvall L, Andersoon U, Harris HE. High mobility group box protein 1 (HMGB1)-partner molecule complexes enhance cytokine production by signaling through the partner molecule receptor. Mol Med. 2012;18:224–230. doi: 10.2119/molmed.2011.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leclerc P, Wähämaa H, Idborg H, Jakobsson PJ, Harris HE, Korotkova M. IL-1β/HMGB1 complexes promote the PGE2 biosynthesis pathway in synovial fibroblasts. Scand J Immunol. 2013;77:350–360. doi: 10.1111/sji.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran TT, Groben P, Pisetsky DS. The release of DNA into the plasma of mice following hepatic cell death by apoptosis and necrosis. Biomarkers. 2008;13:184–200. doi: 10.1080/13547500701791719. [DOI] [PubMed] [Google Scholar]
- 16.Kutcher ME, Xu J, Vilardi RF, Ho C, Esmon CT, Cohen MJ. Extracellular histone release in response to traumatic injury: implications for a compensatory role of activated proteinC. J Trauma Acute Care Surg. 2012;73:1389–1394. doi: 10.1097/TA.0b013e318270d595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antoine DJ, Jenkins RE, Dear JW, et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol. 2012;56:1070–1079. doi: 10.1016/j.jhep.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Bell CW, Jiang W, Reich CF, III, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. Am J Physiol Cell Physiol. 2006;29:C1318–C1325. doi: 10.1152/ajpcell.00616.2005. [DOI] [PubMed] [Google Scholar]
- 19.Lamkanfi M, Sarkar A, Walle LV, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010;185:4385–4392. doi: 10.4049/jimmunol.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Romo GS, Caielli S, Vega B, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyer C, Stearns NA, Giessl A, Distler JH, Schett G, Pisetsky DS. The extracellular release of DNA and HMGB1 from Jurkat T cells during in vitro necrotic cell death. Innate Immun. 2012;18:727–737. doi: 10.1177/1753425912437981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pisetsky DS, Ullal AL, Gauley J, Ning TC. Microparticles as mediators and biomarkers of rheumatic disease. Rheumatology (Oxford) 2012;51:1737–1746. doi: 10.1093/rheumatology/kes028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reich CF, III, Pisetsky DS. The content of DNA and RNA in microparticles released by Jurkat and HL-60 cells undergoing in vitro apoptosis. Exp Cell Res. 2009;315:760–768. doi: 10.1016/j.yexcr.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Ullal AJ, Reich CF, III, Clowse M, et al. Microparticles as antigenic targets of antibodies to DNA and nucleosomes in systemic lupus erythematosus. J Immunol. 2011;36:173–180. doi: 10.1016/j.jaut.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Schiller M, Bekeredijian-Ding I, Heyder P, Blank N, Ho AD, Lorenz HM. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death Differ. 2008;15:183–191. doi: 10.1038/sj.cdd.4402239. [DOI] [PubMed] [Google Scholar]
- 27.Suber T, Rosen A. Apoptotic cell blebs: repositories of autoantigens and contributors to immune context. Arthritis Rheum. 2009;60:2216–2219. doi: 10.1002/art.24715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane JD, Allan VJ, Woodman PG. Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J Cell Sci. 2005;118:4059–4071. doi: 10.1242/jcs.02529. [DOI] [PubMed] [Google Scholar]
- 29.Charras GT. A short history of blebbing. J Microsc. 2008;231:466–478. doi: 10.1111/j.1365-2818.2008.02059.x. [DOI] [PubMed] [Google Scholar]
- 30.Charras GT, Coughlin M, Mitchison TJ, Mahadevan L. Life and times of a cellular bleb. Biophys J. 2008;94:1836–1853. doi: 10.1529/biophysj.107.113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiller M, Heyder P, Ziegler S, et al. During apoptosis HMGB1 is translocated into apoptotic cell-derived membraneous vesicles. Autoimmunity. 2013 Jan 17; doi: 10.3109/08916934.2012.750302. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Seifert JK, France MP, Zhao J, et al. Large volume hepatic freezing: association with significant release of the cytokines interleukin-6 and tumor necrosis factor a in a rat model. World J Surg. 2002;26:1333–1341. doi: 10.1007/s00268-002-6139-5. [DOI] [PubMed] [Google Scholar]
- 33.Casiano CA, Ochs RL, Tan EM. Distinct cleavage products of nuclear proteins in apoptosis and necrosis revealed by autoantibody probes. Cell Death Differ. 1998;5:183–190. doi: 10.1038/sj.cdd.4400336. [DOI] [PubMed] [Google Scholar]
- 34.Sparatore B, Patrone M, Passalacqua M, et al. Extracellular processing of amphoterin generates a peptide active on erythroleukaemia cell differentiation. Biochem J. 2001;357:569–574. doi: 10.1042/0264-6021:3570569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito T, Kawahara K, Okamoto K, et al. Proteolytic cleavage of high mobility group box 1protein by thrombin-thrombomodulin complexes. Artetrioscler Thromb Vasc Biol. 2008;28:1825–1830. doi: 10.1161/ATVBAHA.107.150631. [DOI] [PubMed] [Google Scholar]
- 36.Leblanc PM, Doggett TA, Choi J, et al. An immunogenic peptide in the A-box of HMGB1 protein reverses apoptosis-induced tolerance through RAGE receptor. J Biol Chem. 2014;289:7777–7786. doi: 10.1074/jbc.M113.541474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soop A, Hällström L, Frostell C, Wällén H, Mobarrez F, Pisetsky DS. Effect of lipopolysaccharide administration on the number, phenotype and content of nuclear molecules in blood microparticles of normal human subjects. Scand J Immunol. 2013;78:205–213. doi: 10.1111/sji.12076. [DOI] [PubMed] [Google Scholar]
- 38.Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracing analysis. Nano Med J. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ullal AJ, Pisetsky DS, Reich CF., III Use of SYTO 13, a fluorescent dye binding nucleic acids, for the detection of microparticles in in vitro systems. Cytometry A. 2010;77:294–301. doi: 10.1002/cyto.a.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacKenzie A, Wilson HL, Kiss-Toth E, Dower Sk, North RA, Surprenant A. Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 41.Berda-Haddad Y, Robert S, Salers P, et al. Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin 1α. Proc Natl Acad Sci U S A. 2011;108:20684–20689. doi: 10.1073/pnas.1116848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang JG, Williams JC, Davis BK, et al. Monocytic microparticles activate endothelial cells in an IL-1β-dependent manner. Blood. 2011;118:2366–2374. doi: 10.1182/blood-2011-01-330878. [DOI] [PMC free article] [PubMed] [Google Scholar]