Abstract
CLP1 is a RNA kinase involved in tRNA splicing. Recently, CLP1 kinase-dead mice were shown to display a neuromuscular disorder with loss of motor neurons and muscle paralysis. Human genome analyses now identified a CLP1 homozygous missense mutation (p.R140H) in five unrelated families, leading to a loss of CLP1 interaction with the tRNA splicing endonuclease (TSEN) complex, largely reduced pre-tRNA cleavage activity, and accumulation of linear tRNA introns. The affected individuals develop severe motor-sensory defects, cortical dysgenesis and microcephaly. Mice carrying kinase-dead CLP1 also displayed microcephaly and reduced cortical brain volume due to the enhanced cell death of neuronal progenitors that is associated with reduced numbers of cortical neurons. Our data elucidate a novel neurological syndrome defined by CLP1 mutations that impair tRNA splicing. Reduction of a founder mutation to homozygosity illustrates the importance of rare variations in disease and supports the clan genomics hypothesis.
INTRODUCTION
In order to translate genomic information, tRNAs undergo essential post-transcriptional modifications including chemical alterations, excision of introns followed by exon ligation, removal of 5’ leader and 3’ trailer sequences, and CCA addition (Phizicky and Hopper, 2010). Although tRNA synthesis and processing are essential to all cells, mutations in genes involved in tRNA transcription and maturation appear to preferentially affect the function of neurons. There is an emerging class of neurological disorders resulting from abnormal tRNA biogenesis. Mutations in genes encoding various aminoacyl tRNA synthetases result in Charcot-Marie-Tooth neuropathy (GARS, KARS, YARS, AARS and HARS) (Antonellis et al., 2003; Jordanova et al., 2006; Lee et al., 2006; McLaughlin et al., 2010; Vester et al., 2013), spastic ataxia with leukoencephalopathy (MARS2) (Bayat et al., 2012), distal spinal muscular atrophy (GARS) (Antonellis et al., 2003), or pontocerebellar hypoplasia type 6 (PCH6) (RARS2) (Edvardson et al., 2007). Another distinct group of disorders due to abnormal tRNA maturation are pontocerebellar hypoplasias associated with defects in genes encoding subunits of the tRNA splicing TSEN complex including mutations in TSEN2, TSEN34, and TSEN54 (Budde et al., 2008; Cassandrini et al., 2010) .
CLP1 was the first mammalian RNA kinase to be discovered (Weitzer and Martinez, 2007). We have recently shown that CLP1 acts in concert with the TSEN complex to remove introns present within the anticodon loop of numerous pre-tRNAs (Hanada et al., 2013). Clp1 kinase-dead (Clp1K/K) mice exhibit a progressive loss of spinal motor neurons associated with axonal degeneration in peripheral nerves, denervation of neuromuscular junctions, ultimately resulting in impaired motor functions, muscle weakness, paralysis and fatal respiratory failure (Hanada et al., 2013). Human CLP1 mutations have not been reported and it is not known whether such mutations would also affect the human nervous system. Here, we report the identification of a human CLP1 mutation that impairs tRNA splicing in vitro and causes a novel neurological syndrome involving both the central and peripheral nervous systems. All five families share a haplotype of common variants surrounding CLP1, supporting the notion that clan genomics contributed to the molecular pathology of disease in this lineage (Lupski et al., 2011).
RESULTS
A novel neurological syndrome defined by CLP1 mutations in humans
We studied patients with evidence of brain malformations and microcephaly, who shared similar facial characteristics, global growth and developmental delays, severe intellectual disabilities, and seizures refractory to treatment (Figures 1 and S1A). Magnetic resonance imaging revealed brain abnormalities of differing severities, including cortical dysgenesis marked by a simplified gyral pattern, particularly in the antero-temporal regions, mild or focal cerebellar vermian volume loss (BAB3520 and BAB4771), and thinning of the brain stem (BAB3520) (Figure 1). None of the eleven subjects studied could either ambulate or sit without support. They all have poor (10 to 15 seconds) head control, whereas the youngest patient (BAB5318; 6 months old) has not yet achieved head support. All patients had hypertonia and increased deep tendon reflexes. Electrophysiological studies revealed objective evidence for marked axonal sensorimotor neuropathies (Figure 2 and Table S1). In addition to these characteristic neurologic findings, careful clinical evaluation revealed that the patients exhibit facial features that appear distinctive including high arched eyebrows, prominent eyes, long palpebral fissures and eyelashes, broad nasal roots and hypoplasic alae nasi (Figure 1). Individual patient clinical details are provided in Supplementary notes.
Figure 1. Clinical features and brain MRI images of patients.
Pedigrees of five nuclear families and morphological features of patients showing similar dysmorphic facial features including high arched eyebrows and broad nasal roots. Mid-sagittal and axial views of cranial MRIs are also shown for each patient, revealing brain abnormalities of differing severities, including cortical dysgenesis marked by a simplified gyral pattern, particularly in the antero-temporal regions, shortening and thinning of the corpus callosum that is more prominent in the body segment, and vertical clivus in all patients. Also note focal volume loss of the cerebellar vermis in patient BAB4771 and mild cerebellar volume loss with thinning of the brain stem in patient BAB3520. See also Figures S1A and B.
Figure 2. Nerve conduction studies of patients with CLP1 mutations.
Left. Results of sensory nerve conduction studies of the right median, right ulnar and right sural nerves. Velocity and amplitude values are shown for individual patients as percentages of the minimum normal value (100% - dotted line). Abnormal values are highlighted in red hues. Values that were not recordable despite being tested are indicated as “NR”. Patients that did not have a given nerve tested are blank. Center. Schematic showing the locations of the nerves tested. Right. Results of motor nerve conduction studies of the right median, right common peroneal and right tibial nerves. Patient numbers are indicated. See also Table S1.
Targeted exome capture and whole exome sequence (WES) of four affected individuals (BAB3401, 3402, 3421, and 3422), two from each nuclear family (HOU1338 and HOU1333, respectively) (Figures 1 and S1B), identified a homozygous c.G419A (p.R140H) (chr11:g.57,427,367 G>A [hg19], NM_006831.2) non-synonymous substitution in the CLP1 gene located on chromosome 11q12.1 (Figure 3A), in which mutations have not been associated previously with human disease. The R140H mutation is novel and has not been reported in the 1000 Genomes Project (http://www.1000genomes.org) or other large-scale exome sequencing projects including the Exome variant server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (http://evs.gs.washington.edu/EVS/), and the ‘in-house’ generated exomes from over 2,500 individuals at the Baylor College of Medicine (BCM) Human Genome Sequencing Center and BCM Whole Genome Laboratory Database (MGL; http://www.bcm.edu/geneticlabs/; with over 1,000 individuals tested for diagnostic purposes) and the Atherosclerosis Risk in Communities Study (ARIC) Database (http://drupal.cscc.unc.edu/aric/).
Figure 3. Allele frequencies and modeling of the CLP1 mutation.
(A) B-allele frequency plots for chromosome 11 in the five families. The eleven affected individuals share a common 11.5 Mb region of absence of heterozygosity (AOH) in the proximal long arm of the chromosome, including the CLP1 gene. The location of the gene is marked with a red line. The shared region of AOH extends across the centromere to the proximal short arm of the chromosome. AOH figure for family HOU1981 was created from whole exome data. (B) Interactome of CLP1 with subunits of the TSEN complex (green), mRNA splicing factors and components of the mRNA 3' end cleavage and polyadenylation complex (red), or genes involved in cell cycle control and cell death such as p53, ATM1, BRCA1, MDM1, or VRK1 (blue). (C) Sequence alignment of human CLP1 with CLP1 in other species. R140 is a conserved residue across all vertebrates. (D) Crystal structure of the yeast Clp1 proteins (Noble et al., 2007) illustrating its domain architecture. The highlighted Lys149 is positioned at the interface of the N-terminal and middle domains and is predicted to define the relative orientation of these domains. The zoomed-in image shows two glutamate residues of the N-terminal domain interacting with Lys149 that protrudes from the middle domain.
Since all four subjects from the original two kindreds undergoing exome sequencing presented with the same clinical syndrome, we hypothesized that CLP1 mutations could cause a novel clinically distinguishable phenotype. We therefore re-analyzed our brain malformation cohort focusing on patients with similar central and peripheral nervous system abnormalities and dysmorphic features and found three additional families with consanguinity; these subjects were unrelated to the former two families and had no known relation to each other. Using this phenotype-directed approach, Sanger sequencing of CLP1 in the latter three families revealed the same p.R140H mutation in the homozygous state in all seven additional affected individuals, heterozygous carrier states in the parents and one unaffected male sibling, and homozygous wild type status in one unaffected female sibling (HOU1380, HOU1926, and HOU1981) (Figures 1 and S1B); consistent with Mendelian recessive expectations.
None of the families in our study were previously aware of a relationship. All five families belong to the same ethnic group and reside in the same geographic area of Eastern Turkey. We therefore hypothesized that each family inherited the same mutation from a common ancestor in which this founder mutation occurred. To investigate this, we performed genome-wide SNP microarray analysis on nineteen individuals. Pairwise identity-by-descent (IBD) estimation in PLINK (Purcell et al., 2007) matched reported familial relationships exactly, but did not suggest recent common ancestry among these seemingly unrelated individuals (pi-hat 0 – 0.04). Notably all families share an 11.5 Mb haplotype in the CLP1 region, which spans the centromere and is homozygous in the affected children (Figure 3A). For the last family (HOU1981; samples BAB4980 and BAB4981) the regions with absence of heterozygosity were experimentally determined from WES data that revealed homozygosity of the CLP1 region in these two affected individuals (Figure 3A). Thus, we have identified a defined clinical syndrome with progressive central and peripheral nervous system defects in 11 affected children from five families, all of whom carry a homozygous CLP1 R140H mutation.
Molecular modeling
Analysis of the human protein-protein interaction network revealed interactions between CLP1 and of both mRNA 3' end processing and the tRNA splicing TSEN complex (Figure 3B). In silico analyses suggest CLP1 is also connected to a number of human disease associated genes involved in DNA repair and cell cycle control such as p53 and VRK1, as well as tRNA metabolism (Figure 3B). Analysis on the molecular evolution of the CLP1 mutation showed that the R140H amino acid change affects an arginine residue that is highly conserved from human to zebrafish (Figure 3C).
The CLP1 R140H mutation is predicted to be “damaging or disease causing” by different bioinformatics algorithms (PolyPhen-2, SNAP, SIFT and Mutation Taster). Inspection of the yeast Clp1 crystal structure provides a possible explanation for this effect. In the yeast protein, the corresponding lysine residue (position 149) protrudes from the middle domain towards the N-terminal domain, determining the relative orientation of the two domains to each other (Figure 3D). Whereas an arginine, as seen in higher eukaryotes, may exert a similar organizing effect, a histidine side-chain is too short to mediate this interaction. We thus speculate that the R140H mutation results in an altered domain arrangement that is incompatible with binding potential partner proteins. We further predict that the R140H mutation should not abolish the RNA kinase activity of the enzyme, which is associated with the middle domain containing the catalytic site.
Perturbed CLP1-TSEN complex integrity impairs pre-tRNA cleavage
Based on structure-prediction analysis, we investigated whether the R140H mutation affects CLP1 function. We first assayed RNA kinase activity of recombinant GST-tagged wild type CLP1, kinase-dead CLP1 (K127A mutation), and CLP1 with the R140H mutation expressed in E. coli (Figures 4Aand 4B) and FLAG-CLP1 complexes containing these CLP1 versions ectopically expressed and affinity-purified from HEK293 cells (Figure 4C). Whereas kinase activity was abolished in the K127A mutants, as previously reported (Weitzer and Martinez, 2007), the R140H mutant protein retained kinase activity, albeit at a reduced level when compared to the wild type CLP1 protein (Figures 4B and 4C). We next tested for interactions between mutant CLP1 and members of the TSEN complex. Intriguingly, the interaction of R140H CLP1 with TSEN2, TSEN54 or TSEN34 was markedly reduced (Figure 4D). In line with loss of CLP1-TSEN complex integrity, pre-tRNA cleavage activity of purified R140H CLP1 was almost abolished (Figure 4E). Of note, the kinase-dead CLP1 K127A mutation also showed impaired interaction with TSEN proteins (Figure 4D), correlating with decreased pre-tRNA cleavage activity (Figure 4E) (Hanada et al., 2013).
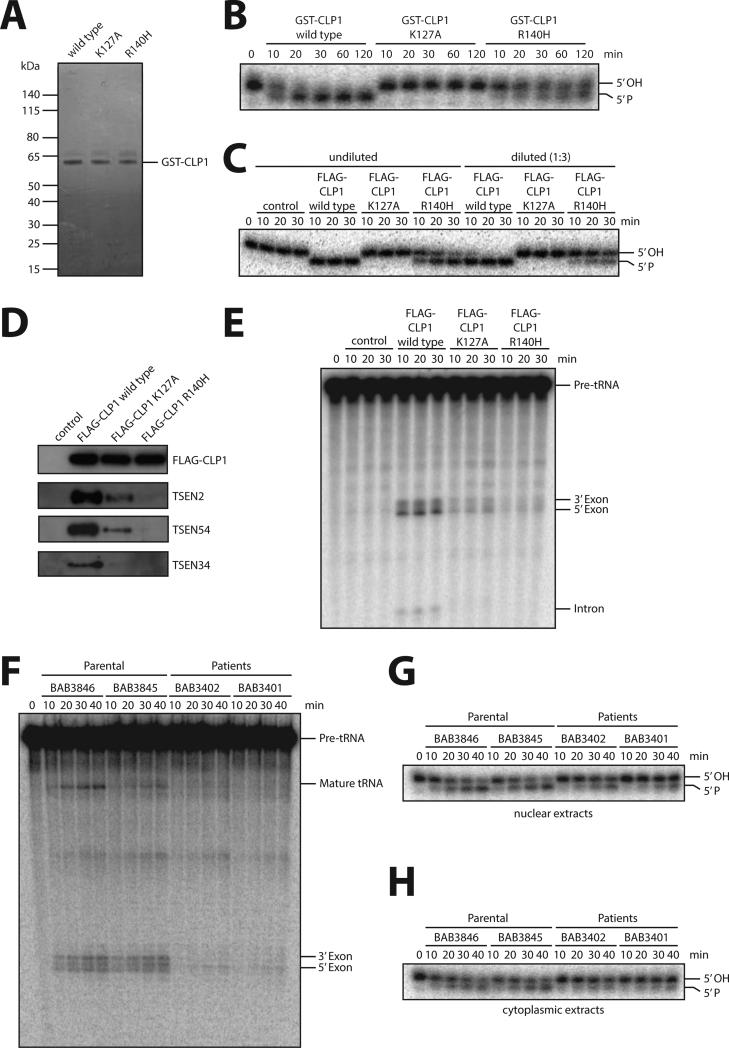
Figure 4. Biochemical studies on purified CLP1 R140H and patient fibroblasts.
(A) Coomassie-blue staining of purified recombinant GST-tagged wild type CLP1, kinase-dead K127A CLP1, and the CLP1 R140H mutant protein. (B) RNA kinase assay using the indicated recombinant CLP1 versions showing that CLP1 R140H is still able to phosphorylate RNA. Recombinant proteins were incubated with an RNA duplex bearing a 5’-OH group and [32P]Cp 3’end-label at one strand for the indicated time points. RNA phosphorylation results in a migration shift after running the reaction products in a denaturing acryamide gel. Note that RNA phosphorylation is completely abolished by the CLP1 K127A mutation. The panel is a representation of two technical replicates. C) RNA kinase assay using protein complexes containing FLAG-CLP1 wild type, FLAG-CLP1 K127A and FLAG-CLP1 R140H affinity-purified from stably expressing HEK293 cells. HEK293 cells without expression of any tagged proteins served as a control. Assays were carried out with undiluted and 1:3-diluted eluates as indicated. (D) Western blotting for TSEN components interacting with affinity-purified FLAG-CLP1 wild type, FLAG-CLP1 K127A and FLAG-CLP1 R140H. (E) Pre-tRNA cleavage assay of affinity-purified FLAG-CLP1 wild type, FLAG-CLP1 K127A and FLAG-CLP1 R140H complexes incubated with an internally labeled intron-containing yeast pre-tRNAPhe. Pre-tRNA processing was monitored by denaturing gel electrophoresis. Panels C-E are representative examples of two replicates. (F) tRNA splicing assay of nuclear extracts of parental (BAB3845 and 3846) and patient (BAB3401 and 3402) fibroblasts incubated for the indicated time points with an internally labeled intron-containing yeast pre-tRNAPhe. Pre-tRNA processing was monitored by denaturing gel electrophoresis. (G, H) RNA kinase activity assay of nuclear (G) or cytoplasmic (H) extracts derived from parental and patient fibroblasts. Extracts were incubated with a 3’ end-labeled 5’ OH group containing RNA duplex for the indicated time points. RNA phosphorylation was monitored by denaturing gel electrophoresis. Panels F-H are representative examples of triplicate experiments. See also Figures S2A to C.
We next isolated and established fibroblast cultures using skin biopsies from patients (BAB3401 and BAB3402) and their parents (BAB3845 and BAB3846), and prepared extracts equalizing for protein concentrations. In nuclear extracts from patient fibroblasts there was only a minor detectable pre-tRNA cleavage activity (Figure 4F) and RNA kinase activity was reduced (Figure 4G). Since the CLP1-TSEN complex can be purified from cytoplasmic extracts of HeLa cells (Weitzer and Martinez, 2007), we also tested RNA kinase activity in the cytoplasm of patient fibroblasts. Here, the activity was reduced in comparison to parental cells (Figures 4H). Another tRNA splicing related biochemical activity, RNA ligation activity as assessed by an interstrand ligation assay in nuclear extracts (Popow et al., 2011), did not correlate with the occurrence of the CLP1 R140H mutation in fibroblasts (Figure S2A). Western blot analysis confirmed the presence of CLP1, TSEN proteins and the tRNA ligase HSPC117 in nuclear extracts of patient cells (Figure S2B). While levels of CLP1 appeared reduced in nuclear extracts, CLP1 was present at similar levels in the cytoplasm of parental and patient cells (Figure S2C). Thus, our in vitro assays for analyzing purified protein complexes showed that the CLP1 R140H mutation does not abrogate kinase function but severely affects the association of CLP1 with components of the TSEN complex, resulting in impaired pre-tRNA cleavage. This is consistent with impaired tRNA splicing observed in nuclear extracts from patient fibroblasts.
CLP1 R140H mutant fibroblasts accumulate introns derived from different tRNA genes
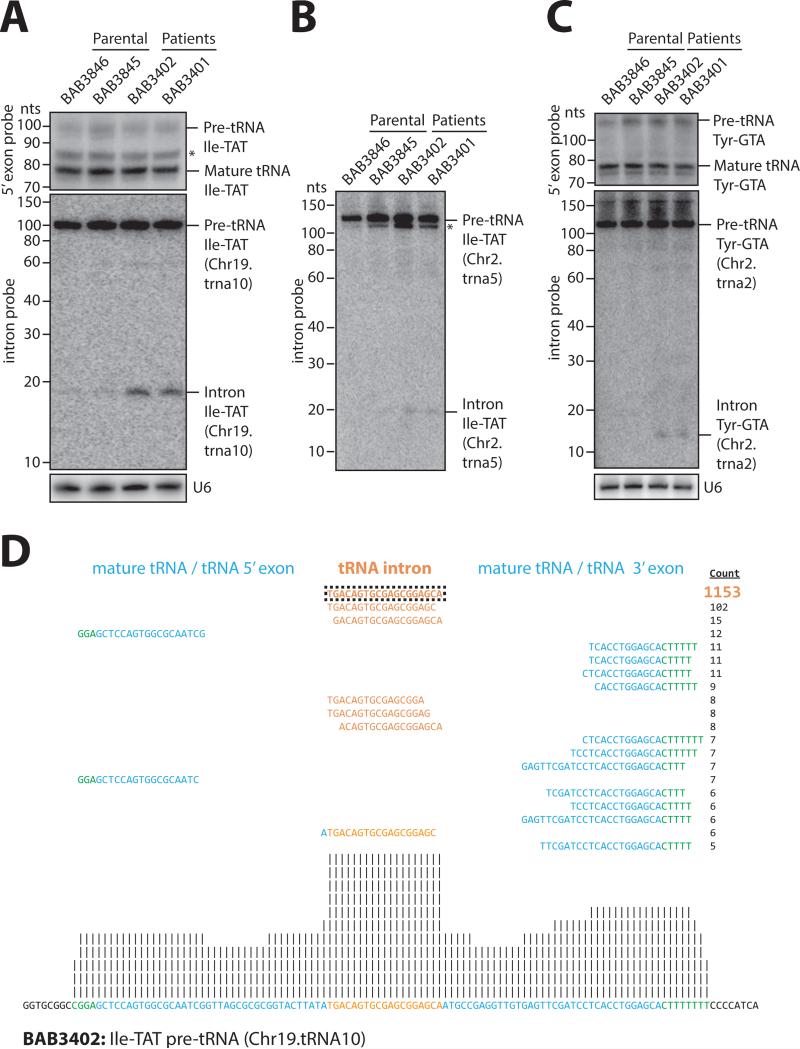
We next isolated total RNA from parental and patient fibroblasts and performed Northern blot analysis using probes detecting tRNAs genomically encoded as intron-less versions (i.e. Met-CAT) or intron-containing versions (Ile-TAT, Tyr-GTA, Leu-CAA, Arg-TCT). There was no significant difference in mature tRNA steady-state levels as exemplified by using probes against the exon junction and the 5' exon detecting multiple tRNAs of the same isotype, so called pan-probes, in parental and patient fibroblasts (Figure 5A-C, top panels; Figures S2D and S2F). In addition, we performed RNA deep sequencing and subjected reads to a quantitative bioinformatics pipeline that failed to reveal consistent differences in global mature tRNA levels between both parental and patient cell lines (Supplementary Table 2). We also performed Northern blot analysis to assess pre-tRNA levels using 5’ exon pan-probes and intron probes detecting specific tRNA genes. We were not able to detect consistent global differences neither in steady-state levels of pre-tRNAs between both parental and patient fibroblast cell lines nor by comparing results obtained from either approaches (Figures 5A and 5C, middle panels; Figures S2D-E and S2G, and data not shown). Interestingly, both by Northern blotting and deep sequencing analysis we reproducibly detected an accumulation of tRNA introns derived from three particular tRNA genes (Ile-TAT Chr19.trna10; Ile-TAT Chr2.tRNA5 and Tyr-GTA Chr2.tRNA2; Figures 5A and C, middle panels; Figure 5B; Figures S3A to C and Table S3) in patient fibroblasts. Biochemical analysis using RNase R treatment that digests linear but not circular RNA revealed that Ile-TAT (Chr19.trna10) introns are linear (Figure S3D). Since these introns can be ligated to a 3’ hydroxy-terminating RNA linker only upon prior 5’ phosphorylation, we conclude that introns display a 5’ hydroxy group (Figure S3E). Thus, our Northern analysis and deep sequencing data indicate that the CLP1 R140H mutation in fibroblasts influences processing of pre-tRNAs resulting in the accumulation of linear tRNA introns, whereas pre- and mature tRNA levels remain largely unaffected.
Figure 5. tRNA analysis of patient fibroblasts.
(A-C) Northern blot analyses of RNA from parental and patient fibroblasts. A probe complementary to the 5’ exon of isoleucine-TAT and tyrosine-GTA tRNAs was used to detect mature and pre-tRNA species (top panels in [A] and [C]). Probes specifically directed against intron sequences were used to detect pre-tRNAs and tRNA introns of isoleucine-TAT Chr.19.tRNA10 ([A], middle panel) and Chr2.tRNA5 (B), and tyrosine-GTA Chr2.tRNA2 ([C], middle panel; human February 2009 [hg19] genome assembly). U6 snRNA served as loading control (bottom panels in [A] and [C]). Asterisks denote truncated pre-tRNA species. See also Figure S2D-G and Figure S3A-B. (D) Example for an alignment of RNA-seq reads of patient (BAB3402) fibroblasts against precursor tRNA isoleucine-TAT (Chr19.tRNA10). Total RNA was subjected to partial alkaline hydrolysis prior to cloning and sequencing. Reads were aligned against an in-house curated list of mature and pre-tRNAs. The mature tRNA (blue), the tRNA intron (orange), and the 5’ leader and 3’ trailer sequences (green) are shown. The frequency of each read is presented (count). All reads mapped uniquely to the identified positions. Upstream and downstream nucleotides with no sequencing evidence are shown in black. Vertical lines represent the relative frequency of binned, normalized read counts in log2 increments. See also Figure S3C to compare the accumulation of intron reads relative to parental (BAB3846) fibroblasts.
Microcephaly in Clp1 defective mice
We recently reported progressive loss of motor neurons, axonal motor neuropathy, and muscle paralysis leading to death in kinase-defective K127A CLP1 (Clp1K/K) mutant mice (Hanada et al., 2013), a phenotype consistent with the observed impairment of motor functions in our patients, including features of sensorimotor axonal neuropathy. Of note, on a B6 background all Clp1K/K newborn mice die due to impaired innervations of the diaphragm, whereas, on a CBA/J background, Clp1K/K mutant mice grow to adulthood and progressively lose spinal motor neurons and motor functions (Hanada et al., 2013). Given the significant microcephaly noted in patients carrying the CLP1 R140H mutation (Figures 1and S1A), we wondered whether we had missed a brain phenotype in our mutant Clp1K/K mice, also considering fundamental differences between human and rodent brain development (Lancaster et al. 2013).
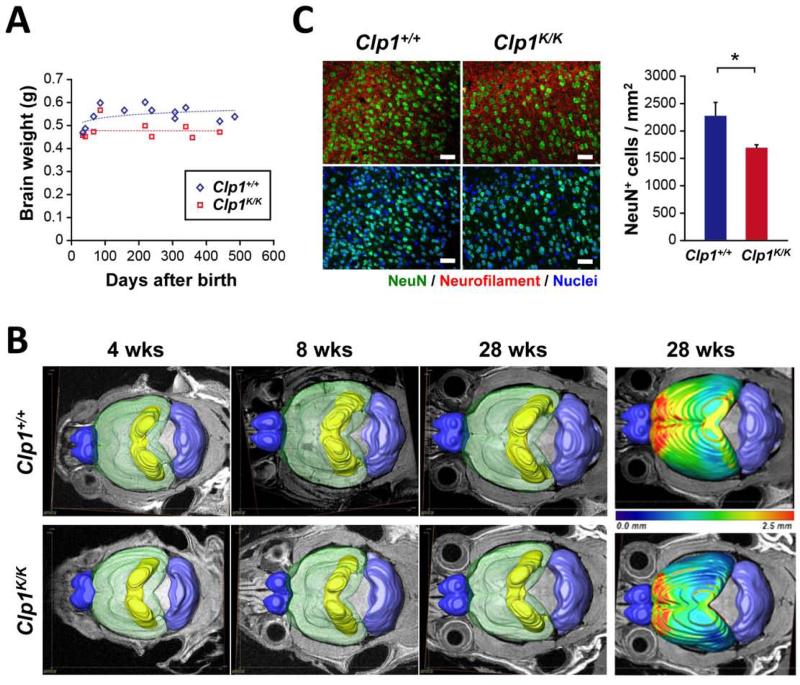
To analyze potential microcephaly in Clp1K/K mice we first determined the brains sizes of control and viable adult Clp1K/K mice on the CBA/J background. In all littermate pairs analyzed, we observed reduced brain weights in mice carrying a kinase-dead version of Clp1 (Figures 6A and S4A). To confirm reduced brain weights, we used 15.2 Tesla MRI imaging. MRI-3D brain reconstructions showed significantly reduced brain volumes at 8, 12, and 28 weeks after birth, but not at 4 weeks after birth (Figures 6B and S4B). Clp1K/K mice also did not show a continuous increase in the brain aspect ratio (Figure S4C), which describes the elongation of the brain during normal development. Moreover, using geometric structural analysis, we observed markedly reduced cortical thickness throughout the entire cortex (Figures 6B and S4D). This difference is particularly prominent in the frontal and somatosensory-motor areas of the cortex (see red and yellow areas in Figures 6B and S4D). The reduction of cortical thickness was confirmed histologically (Figures S4E and S4F).
Figure 6. Microcephaly in adult kinase-defective Clp1 mice.
(A) Scatter plots showing brain weights of Clp1+/+ and K127A mutant Clp1K/K mice on the viable CBA/J background. The ages of mice at the time of analysis are indicated; matching ages correspond to littermate pairs. (B) Surface renderings of bulbus olfactorius (dark blue), cortex (transparent green), hippocampus (yellow), and cerebellum (light blue) from MRI datasets of adult mice at 4, 8 weeks and 28 weeks old Clp1+/+ and Clp1K/K littermate mice. The right panels additionally show for 28 week old animals a pseudo-colour coded mapping of cortical thickness ranging from blue (0.0 mm) to increased thickness shown in red (2.5 mm). (C) Immunohistochemical analysis with antibodies against NeuN to detect neurons in the cortex of 12 weeks old littermate Clp1+/+ and Clp1K/K mice. Left panels show representative images of NeuN+ neurons (green). Sections are also stained for neurofilament and counterstained with Hoechst 33342 to visualize nuclei (blue). Scale bars: 50 μm. Right panel shows quantification (mean values +/− SEM) of NeuN+ neuron numbers in the neocortex. n = 6 mice per genotype. * P < 0.05. See also Figures S4 and S5.
Immunohistochemical analysis with antibodies against neuronal nuclear antigen (NeuN) to detect neurons showed normal cortical layering (Figure S5A). Importantly, in line with reduced brain size and reduced cortical thickness, we found reduced numbers of NeuN+ neurons in the neocortex of adult Clp1K/K mice as compared to their wild type littermate controls (Figure 6C). By contrast, microglial cells, as detected by immunostaining for ionized calcium binding adaptor molecule 1 (Iba-1), appeared to be increased in the neocortex of Clp1K/K mice, albeit not to significant levels (Figures S5B and S5C). Numbers of glial fibrillary-acidic protein (GFAP)-positive astrocytes were apparently not affected in the hippocampus (Figures S5D and S5E), nor in the cortex (data not shown) of Clp1K/K mutant mice. Of note, neuronal, microglial, and astrocyte numbers and distributions were not affected in the cerebellum of Clp1K/K mice (Figures S5F to J, and not shown); the cerebellar volume was also not affected in Clp1K/K mutant mice as determined by MRI (Figure 6B, and not shown). Thus, similar to humans with CLP1 R140H mutations, these data show that mice carrying kinase-dead Clp1 mutation exhibit microcephaly, in particular due to reduced numbers of cortical neurons.
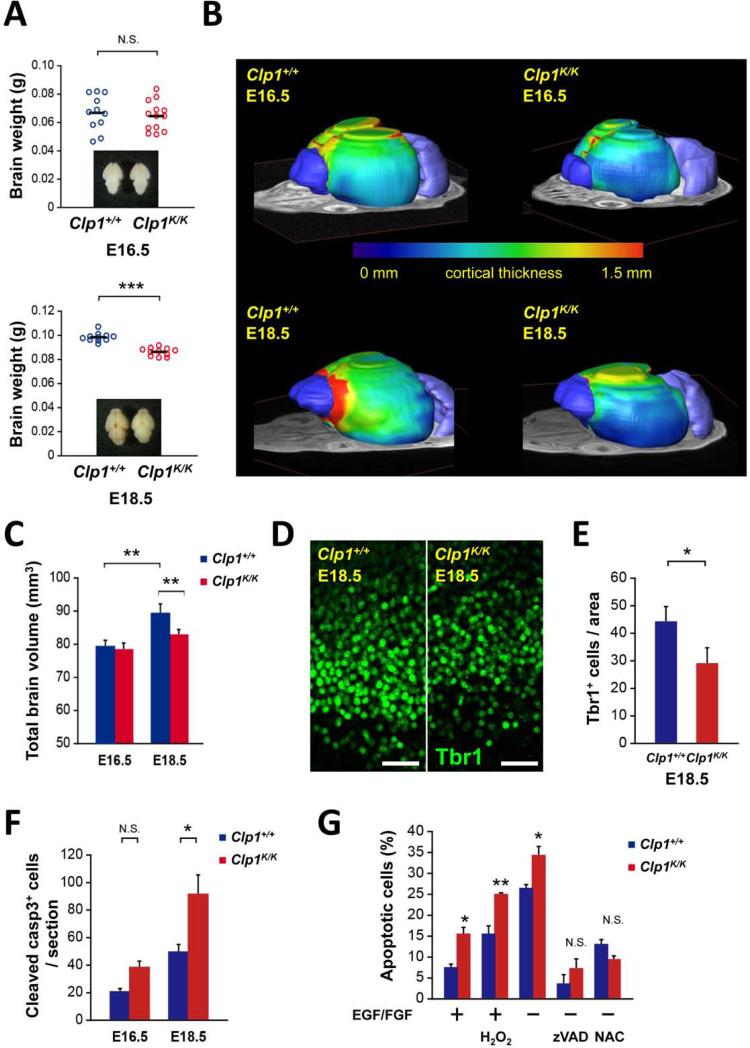
When we analyzed Clp1K/K embryos on the lethal B6 mouse background, we did not observe significant total brain weight differences between wild type and Clp1K/K mice at embryonic day 16.5 (E16.5); however, Clp1K/K mice showed significantly decreased brain weights at E18.5 and reduced brain sizes as determined by histology (Figures 7A and S6A to S6C). The results were confirmed using MRI showing comparable brain volumes at E16.5 (64.4mm2 for Clp1+/+ vs 62.3mm2 for Clp1K/K embryos) and a 13.7% reduction in mutant embryos at E18.5 (95.9mm2 for Clp1+/+ vs 84.3mm2 for Clp1K/K embryos) (Figure S6D). Moreover, in 3D whole brain reconstructions using 15 Tesla MRI imaging, we also observed markedly reduced brain volumes at E18.5, but comparable brain volumes at E16.5 (Figures 7B and 7C). At E18.5, the main reduction in brain volume was observed in the cortex of Clp1K/K mice, with additional significant reductions in the volume of the bulbus olfactorius, whereas the volumes of the cerebella were comparable between control and Clp1K/K littermates (Figures 7B and S6E). Cortical thickness was comparable between E16.5 Clp1+/+ and Clp1K/K embryos, but it was markedly reduced in E18.5 Clp1K/K embryos as compared to their wild type littermate controls (Figure 7B). Shape analysis of the brain geometry by principal component analysis (PCA) further revealed that in the control all three spatial axes increased significantly from E16.5 to E18.5, but in the Clp1K/K embryos the third PCA (dorso-ventral direction) had even decreased, whereas the first PCA (anterior-posterior direction) and the second PCA (caudal-rostral direction) were apparently normal in the Clp1K/K mice (Figure S6F), indicating impaired dorso-ventral expansion of the cortex. The latter is also reflected by the fact that only wild type control brains exhibit a significant decrease in cortical roundness due to changes in brain geometry (Figure S6G). These data show that mice carrying a homozygous CLP1 K127A kinase-dead mutation exhibit impaired expansion of the brain cortex resulting in microcephaly.
Figure 7. Microcephaly in kinase-defective Clp1 embryos.
(A) Brain weights and representative dorsal whole brain views (insets) of Clp1+/+ control and Clp1K/K E16.5 and E18.5 mouse embryos on the neonatal lethal C57BL/6 (B6) background. (B,C) 3D brain evaluations via MRI. (B) Representative visualizations of individual E16.5 and E18.5 Clp1+/+ and Clp1K/K mouse embryos on the B6 background. On top of an MRI slice iso-surface, 3D renderings are shown for the bulbus olfactorius (dark blue), cerebellum (light blue), and the cortex. The cortex is rainbow color-coded to illustrate the cortical thickness from 0 mm (blue) to 1.5 mm (red, more thick). (C) Quantification of brain volumes (mean values +/− SEM) determined via MRI. n = 8. (D) Representative images from immunohistochemical analysis with antibodies against Tbr1 to detect neurons in the cortical plate of Clp1+/+ control and Clp1K/K E18.5 embryos on the B6 background. Scale bars: 50 μm. (E) Quantification (mean values +/−SEM) of Tbr1+ neuron numbers in the neocortex of Clp1+/+ control and Clp1K/K E18.5 embryos on the B6 background. Of note, the area of Tbr1+ cells encompasses a region from the lateral ventricle to the brain surface (width of 300 pixels, ~ 100 μm). n = 8 mice per genotype. (F) Quantification (mean values +/− SEM) of cleaved Caspase 3+ cell numbers, indicative of apoptosis, in coronal sections of Clp1+/+ control and Clp1K/K E16.5 and E18.5 embryos on the B6 background. n = 3 mice per genotype. (G) Cell death of neuronal progenitors isolated from E14.5 Clp1+/+ and Clp1K/K embryos. Cells were cultured with (+) or without (−) EGF/FGF (each 20 ng/ml) and challenged with H2O2 (100 μM). Death was determined by assaying for cleaved Caspase 3 and blocked using zVAD (100 μM) or NAC (10 mM). Data are shown as mean values +/− SEM of triplicate cultures. * P < 0.05. ** P < 0.01. *** P < 0.001. N.S., not significant. See also Figures S6 and S7.
Similar to adult Clp1K/K mice, mutant E18.5 embryos had reduced numbers of Tbr1+ neurons as compared to control littermates; at E16.5, the numbers of Tbr1+ neurons were comparable among control and Clp1K/K embryos (Figures 7D, 7E; and S6H). Numbers and distributions of Iba-1+ microglial cells were apparently not altered in E16.5 and E18.5 Clp1K/K embryos (Figures S7A and S7B), nor did we observe any obvious alterations in GFAP+ astrocytes (Figure S7C). We also observed normal numbers of Pax6+ neuronal progenitor cells in E16.5 and E18.5 Clp1K/K mice as compared to their littermate controls (Figures S7D to S7F). Numbers of proliferating cells as determined by Ki67 staining were also normal, albeit slightly reduced in E16.5 as well as E18.5 Clp1K/K embryos (Figure S7G). Importantly, we detected a higher number of apoptotic cells throughout the brain of E16.5 and, in particular, E18.5 Clp1K/K embryos (Figure 7F). To test whether neuronal progenitors are more susceptible to death in Clp1 mutant embryos, we prepared neural precursor cells (NPCs) from E14.5 control and Clp1K/K embryos. Importantly, Clp1K/K neural precursor cells exhibited enhanced cell death in basal growth conditions (using EGF and FGF), under oxidative stress, as well as following growth factor withdrawal; this enhanced cell death could be blocked using the pan-caspase inhibitor zVAD as well as the antioxidant N-acetyl-L-cysteine (NAC) (Figure 7G). These data indicate that Clp1K/K embryos have normal numbers and proliferation of neuronal progenitors, but neuronal progenitor cells undergo enhanced cell death resulting in reduced numbers of cortical neurons.
DISCUSSION
In this study, we provide genomic, genetic, molecular modeling, biochemical, and animal model evidence for the involvement of CLP1 in a complex neurological phenotype that includes both the central and peripheral nervous systems. We identified the same homozygous rare variant, R140H, of the CLP1 gene in eleven individuals affected with this novel neurological syndrome from five families using WES and candidate gene sequencing. Further genetic analysis revealed that these families share the same haplotype block encompassing the CLP1 region. These data support the concept of clan genomics (Lupski et al., 2011) in that a rare variant in the population is concentrated in a lineage which introduced the variant, either by de novo mutation or immigration and subsequent founder effect. In the patients identified here the rare R140H mutation was reduced to homozygosity. Our genomic and genetic data indicate that a rare variant allele arose in a distant common ancestor, segregated through the generations, and was reduced to homozygosity by apparent consanguinity. Genomic approaches to rare variant detection in Mendelizing disease traits may eventually provide new insights into both the genetic and molecular basis of disease as well as reveal unexpected consequences of dysfunction of basic biological processes such as RNA metabolism.
Our findings reveal cortical dysgenesis with predominant forebrain involvement by MRI and no overt evidence for midbrain or prominent cerebellar abnormalities. Our mouse studies support predominant cortical cellular apoptosis likely responsible for the uniform microcephaly observed during development in mice and postnatally in all human subjects with homozygous R140H alleles. A parallel study (Schaffer et al. this issue) reported CLP1 mutation in association with a clinical phenotype of pontocerebellar hypoplasia; they also report a R140H founder mutation. In a zebrafish model organism study of Clp1 mutation they show TUNEL staining revealing dramatic increase specific to forebrain and hindbrain. Forebrain staining supports both our mouse and human findings in which predominant cortical involvement is observed.
Our data provide insights into the biology of disease including cortical dysgenesis. Mutations in proteins involved in RNA modifications have been associated previously with preferentially or exclusively either central or peripheral nervous system pathology. For instance, mutations in, EXOSC3, TSEN54, TSEN2 and TSEN34 are known to cause neurological phenotypes that manifest in the brain stem and cerebellum causing Pontocerebellar hypoplasia (PCH) PCH1B, PCH2A, PCH4, PCH2B and PCH2C , but little is known regarding potential peripheral nervous system involvement as the results of nerve conduction studies have not been reported (Budde et al., 2008; Renbaum et al., 2009; Wan et al., 2012). Conversely, mutations in tRNA synthetase genes, such as GARS, KARS, YARS, AARS and HARS are predominantly associated with Charcot Marie Tooth neuropathy, distal spinal muscular atrophy, and other peripheral nervous system disorders, without significant central nervous system involvement (Antonellis et al., 2003; Jordanova et al., 2006; Lee et al., 2006; McLaughlin et al., 2010; Vester et al., 2013).
Our biochemical experiments provide evidence that the CLP1 R140H mutation impairs tRNA exon generation, most likely due to decreased interaction between mutant CLP1 and the TSEN complex. Together with our previous study showing that a kinase-dead Clp1 K127A mutation results in reduced pre-tRNA cleavage by affecting CLP1-TSEN complex integrity (Hanada et al., 2013), we propose that an intact CLP1-TSEN association is essential for efficient tRNA splicing. The fact that the CLP1 R140H mutant shows RNA kinase activity but is unable to associate with TSEN components will support the need for future studies on modeling potential CLP1-TSEN interaction platforms that are, in contrast to the kinase-dead CLP1 K127A mutation, uncoupled from its ATP binding and/or hydrolysis activity.
We further show that the global steady-state levels of mature tRNAs in fibroblasts are not influenced by the CLP1 R140H mutation, suggesting the existence of backup mechanisms that ensure sufficient levels of mature tRNAs despite the undetectable pre-tRNA cleavage activity observed in nuclear extracts from patient cells. No obvious increase in general pre-tRNA levels was observed in patient fibroblasts and therefore it is unlikely that enhanced Pol III transcription of intron-containing tRNA genes could compensate for inefficient pre-tRNA cleavage. Analogous to the accumulation of 5’ leader-exon tRNA fragments in cells and tissues of Clp1K/K mice (Hanada et al., 2013), we detected increased levels of isoleucine and tyrosine tRNA introns in R140H patient fibroblasts. These introns are linear and harbor 5’ OH groups, properties that are expected for a reaction product after pre-tRNA cleavage and prior to a potential 5’ phosphorylation by CLP1 (Weitzer and Martinez, 2007) or circularization by the tRNA ligase (Popow et al., 2011). Future work will have to address whether accumulation of linear introns is a consequence of a defective release from a TSEN complex in CLP1 R140H patient cells, thus being not exposed to cellular nuclease activities. Further studies may also provide insights into whether generation of tRNA processing intermediates is a consequence of mutations in the CLP1 gene, and potential functions of these tRNA splicing by-products.
We demonstrated that mutations in CLP1 impair functioning of neurons and lead to abnormalities of both the central and peripheral nervous systems. Our patients present with peripheral nerve dysfunction as evidenced by abnormal electrophysiological studies showing axonal neuropathy affecting both motor and sensory nerves. These observations were universal in the eight affected individuals with CLP1 mutations for whom NCVs were assessed. Moreover, all patients exhibit global cortical dysgenesis and microcephaly which is suggestive of global CNS dysfunction. Based on these human phenotypes, we speculated that our mutant mice carrying a kinase-inactivating CLP1 mutation might also exhibit abnormalities of both the central and peripheral nervous systems. Indeed, using histological analyses and 3D reconstruction from MRI images, mice carrying a homozygous CLP1 K127A kinase-dead mutation exhibit marked microcephaly, in particular due to reduced numbers of cortical neurons. Mechanistically, this phenotype appears to be progressive and can be attributed to enhanced apoptosis of neuronal progenitor cells at basal conditions and in response to oxidative stress and growth factor withdrawal. Whether additional mechanisms contribute to the observed microcephaly in mice and whether these uncovered mechanisms also extend to humans will need to be evaluated in future experiments. It is striking that the clinical presentation of patients with CLP1 mutations is nearly identical to one we observed in patients with VRK1 mutations (Gonzaga-Jauregui et al., 2013) especially in the neurological presentation (progressive weakness), brain imaging studies (cortical dysgenesis, microcephaly) and NCV (axonal neuropathy). From our network analysis, CLP1 and VRK1 are molecular interactors and it has been reported that VRK1 can phosphorylate p53 (Valbuena et al., 2011; Valbuena et al., 2006). We recently reported that kinase-dead CLP1 sensitizes motor neurons and fibroblasts to death following oxidative stress via a p53-mediated cell death pathway; genetic inactivation of p53 completely rescued the neonatal lethal phenotype of Clp1K/K mice on a B6 background (Hanada et al., 2013). Thus, based on our published genetic data in mice (ClpK/K) now complemented by genomic approaches in human rare disease studies (CLP1, VRK1), it appears that the neurotoxic CLP1-p53 pathway we have identified in mouse, is also conserved and operates in humans and is critical to neuron function.
EXPERIMENTAL PROCEDURES
Patients
This study was approved by the Institutional Review Board at Baylor College of Medicine and informed consent was obtained from all subjects prior to enrollment in the project. All subjects were evaluated by one or more pediatric neurologists and clinical geneticists experienced with brain malformation phenotypes. Genomic DNA was extracted from blood based on the manufacturer's protocol (QIAGEN Sciences, Maryland, USA).
Whole exome sequencing
Four initial affected individuals (BAB3401, BAB3402, BAB3421, and BAB3422) underwent targeted whole exome capture using the BCM HGSC Core design followed by Illumina HiSeq massively parallel sequencing through the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) initiative. An average of 10Gb of raw sequence data were produced and subsequently mapped and aligned to the reference human genome sequence GRCh37/Hg19 using the BWA algorithm with an average depth of coverage of 120× (median coverage = 91×); ~92% of the bases were covered at 20×. Variants were called and annotated using an in-house developed bioinformatics pipeline. Analysis of variants was initially performed in search for shared variants between the pairs of affected siblings under a recessive model of inheritance and later filtered for high-frequency and commonly observed variants.
Sanger PCR confirmation and segregation studies
To confirm the mutation detected by exome sequencing and to perform segregation analysis, standard PCR was carried out as previously described (Pehlivan et al., 2012) by using CLPF1: 5`-AGAGCTGACCCGAAACAAGA-3` and CLPR1: 5`-CCAGCTGAGAAAATGCAGTG-3` primers. Amplification products were electrophoresed on 0.8% agarose gels. PCR products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA) and analyzed by standard Sanger dideoxy nucleotide sequencing (DNA Sequencing Core Facility at Baylor College of Medicine, Houston, TX).
Genome-wide genotyping
To further investigate family relationships and the haplotype structure of variant alleles, we performed genome-wide SNP genotyping on Illumina HumanOmniExpress (families HOU1338 and HOU1333) and HumanOmni2.5 (families HOU1926 and HOU1380) microarrays (Illumina Inc.). Whole genome amplification, fragmentation, hybridization, enzymatic single base extension, slides staining and washing were performed according to the manufacturer's instructions (Illumina, Inc.). Microarrays were scanned on an Illumina iScan System. GenomeStudio software v2011.1 (Illumina Inc.) was used for SNP clustering, genotype calling, data intensity analysis, and the generation of the B-allele frequency plots in Figure 3A. CNV- partition was used for copy number and absence of heterozygosity analyses. For the last family (HOU1981; samples BAB4980 and BAB4981) B-allele frequencies were determined from the whole exome data, i.e. by computing the variant/total reads ratio for each SNP. The regions with absence of heterozygosity were identified using in-house scripts in R language (www.r-project.org) and Circular Binary Segmentation algorithm implemented in DNAcopy R package (Olshen et al., 2004).
Identity-by-descent estimation
PED and MAP files were exported from GenomeStudio and analyzed in PLINK. Because two different microarrays were used in the study, merged PED and MAP files were generated which included only the 668,800 SNPs that were common to both arrays. Genotyping rate for this subset of SNPs, after merging the data for the four families, was 0.99984. Heterozygous haploid (sex-chromosome) SNPs were excluded. Linkage disequilibrium-based pruning was then performed, excluding SNP pairs with r2 values > 0.2. After LD-based pruning, the remaining 28,581 SNPs were used to estimate pairwise identity-by-descent metrics (171 pairwise comparisons for 19 individuals).
Bioinformatics
The functional impact of the p.R140H substitution was assessed by considering conservation scores from PhyloP (Siepel, 2006), GERP (Cooper et al., 1998) and LRT (Chun and Fay, 2009), and additionally functional prediction scores from PolyPhen-2 (Adzhubei et al., 2010), SNAP (Bromberg et al., 2008), SIFT (Kumar et al., 2009) and Mutation Taster (Schwarz et al., 2010), which are computational algorithms that estimate the detrimental effect of non-synonymous substitutions based on statistical and machine learning models. Graphical representations of molecular models were prepared with PyMol (The PyMOL Molecular Graphics System, Schrödinger, LLC).
OFC calculations
WHO head circumference data is only available from birth to age 5 years. Therefore, we used head circumference data from the literature for boys and girls starting from birth to 18 years (Roche et al., 1987). For time points that were not available in the primary data, we interpolated the values for both mean and standard deviation using the R statistical programing language (R Core Development Team).
Histology and immunohistochemistry
Clp1K/K mutant mice were generated as described (Hanada et al., 2013) and maintained according to institutional guidelines. Whole brains were fixed in 4% paraformaldehyde, washed in PBS, dehydrated, and paraffin embedded according to standard protocols. For all procedures, 2-5 μm thick coronal sections of formalin-fixed paraffin-embedded mouse brains were used. For H&E staining, routine protocols were used. For immunohistochemical analysis, blocking of endogenous peroxidase activity was performed with 0.3% H2O2 for 15 min on deparaffinated sections at room temperature, followed by treatment in a microwave oven (750 W) for 15 min in 10 mM citrate buffer (pH 6.0) for antigen retrieval before applying primary antibodies. Then, the first antibody in blocking buffer (5% goat serum / 45% Tris buffered saline pH 7.6 [TBS] / 0.1% Triton X-100 in antibody diluent solution [Zytomed]) was applied overnight at 4°C. The following antibodies were used: Rabbit polyclonal anti-Iba-1 (Iba-1, Wako Chemicals), mouse monoclonal anti-GFAP (Dako), mouse monoclonal antibody against neuronal nuclear antigen (NeuN, Chemicon), mouse monoclonal anti-Calbindin D-28K antibody (Sigma-Aldrich), mouse monoclonal anti-Neurofilament 200 antibody (Sigma-Aldrich), rabbit polyclonal anti-Pax6 (Covance), rabbit polyclonal anti-Tbr1 (Abcam), rabbit polyclonal anti-cleaved caspase 3 (Cell Signaling Technology), and anti-Ki67 (Novocastra). Histofine universal immuno-peroxidase polymer and Histofine Mouse Stain Kit (Nichirei Biosciences) were applied after washing with TBS. The peroxidase reaction was detected using diaminobenzidine (Sigma-Aldrich) as chromogen. Counterstaining was performed using alum-haematoxylin. For immunofluorescence staining, Alexa Fluor 488 or 555 (Molecular Probes) was used as a secondary antibody with Hoechst 33342 for nuclear staining. Some quantifications were done using the public NIH Image J 1.46 software on comparable sections taken at the same, defined anterior-posterior level.
Magnetic resonance imaging
Clp1K/K mutant mice and wild-type mice were analyzed using magnetic resonance imaging (MRI). To our knowledge these are the first data to be published using a new ultra high field 15.2 Tesla horizontal Biospec scanner (152/11, Bruker, Ettlingen, Germany). The magnet has a free bore of 110 mm and is equipped with actively shielded gradients (1000 mT/m) and third order shims. All scans were performed using a whole body transmitter/receiver quadrature coil with an inner diameter 35 mm. The SNR improvement of the high field allowed us to obtain from E16.5 and E18.5 mouse embryos 3D diffusion weighted spin echo sequences (TR/TE = 500/16.5 ms, 16 averages , b factor = 650 s/mm2) with FOV 12x12x8 mm with an imaging matrix 240×240×40 resulting in a spatial resolution of 50×50×200 μm. The same approach was taken to scan adult mouse brains, with the modification that the FOV (16×16×8 mm) and consequently the imaging matrix (320×320×40) had to be adjusted to keep the same resolution and total measurement time (NEX 9). For the adult brains a b factor of 300 s/mm2 resulted in better contrast of the different brain structures. For images of whole embryos, we used a 7T whole body scanner. These images were acquired with a standard turbo spin echo sequence (TE 48ms, TR 380ms) with a voxel resolution of 78×78×500 μm3 (Friske, 2012).
MRI image analysis
For each 3D image dataset the bulbus olfactorius, the cortex, the cerebellum and, for the adult brains, the hippocampus were manually segmented using the AMIRA software (V 5.5, VSG). Based on these segmentations, surface renderings of the individual segmented brain structures were displayed onto a representative horizontal MR slice at the ventral eye position using AMIRA visualization modules. The thickness of the cortex was determined at each vertex by computing the distance along the vertex normal to the normal's intersection with the closest triangle of the cortex. The resulting scalar field was surface mapped onto the surface rendering of the cortex. Geometric properties (volume, principal axes, form factors) of each segmented brain structure were calculated using MagnAn (BioCom GbR).
Preparation of neural precursor cells (NPCs)
Preparation of embryonic neuroepithelial cells containing NPCs was performed as previously described (Shiraishi et al., 2010) Cerebral cortices from embryonic day 14.5 pups were dissected in Hank's balanced salt solution and mechanically dissociated using a micropipette. After centrifugation, cells were plated in a 60 mm culture dish coated with poly-L-ornithine (Sigma) and recombinant human fibronectin (R&D). Cells were cultured in a neural progenitor maintenance medium (NPMM) bullet kit (Lonza). The medium was changed every other day. After 4 days of expansion of NPCs, cells were re-plated and cultured with 100 μM H2O2 or with and without EGF and FGF for induction of cell death. 100 μM zVAD-fmk (BD bioscience) or 10 mM N-acetylcysteine (Sigma) were used as pan-caspase inhibitor or antioxidant, respectively. After 24 hours, cell survival was evaluated with immunostaining for cleaved caspase 3.
Statistics
Comparisons between groups were made by 2-sided Student's t test. A P value less than 0.05 was considered significant.
Supplementary Material
Research Highlights.
A novel neurological syndrome affecting both CNS and PNS defined by CLP1 mutations.
R140H mutation impairs tRNA exon generation, most likely decreasing CLP1- TSEN complex interaction.
K127A kinase-dead mice exhibit marked microcephaly due to reduced numbers of cortical neurons.
ACKNOWLEDGEMENTS
We thank the patients and families who participated in this study. We thank the CSF/PIF MRI imaging facility at the campus Vienna BioCenter. This work was supported in part by US National Institute of Neurological Disorders and Stroke (NINDS) grant R01NS058529 and US National Human Genome Research Institute (NHGRI) grant U54HG006542 to JRL. HS is supported in part by the fellowship of Astellas Foundation for Research on Metabolic Disorders and The Mochida Memorial Foundation for Medical and Pharmaceutical Research. WW is supported by a K23NS078056 grant from the NINDS. MG is supported by grants from the DFG (FG885 and GRK 1459). JM and SW are supported by IMBA. JMP is supported by an Advanced ERC grant and IMBA. LMF is supported by a Career Development Award (1K23AI087821-01) from the NIAID. JRL is a paid consultant for Athena Diagnostics, has stock ownership in 23andMe and Ion Torrent Systems, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the clinical exome sequencing offered in the Medical Genetics Laboratory (MGL; http://www.bcm.edu/geneticlabs/).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information is available in the online version of the paper.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat V, Thiffault I, Jaiswal M, Tetreault M, Donti T, Sasarman F, Bernard G, Demers-Lamarche J, Dicaire MJ, Mathieu J, et al. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- Cassandrini D, Biancheri R, Tessa A, Di Rocco M, Di Capua M, Bruno C, Denora PS, Sartori S, Rossi A, Nozza P, et al. Pontocerebellar hypoplasia: clinical, pathologic, and genetic studies. Neurology. 2010;75:1459–1464. doi: 10.1212/WNL.0b013e3181f88173. [DOI] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Ball EV, Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardson S, Shaag A, Kolesnikova O, Gomori JM, Tarassov I, Einbinder T, Saada A, Elpeleg O. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friske J, Berg A, Welsch GH, Pachowsky M, Gelse K, Trattnig S. High-field high-resolution and microscopic T2-maps of a cartilage repair sheep model on a 7 human scanner: first results. European Society for Magnetic Resonance in Medicine and Biology Meeting. 2012 [Google Scholar]
- Gonzaga-Jauregui C, Lotze T, Jamal L, Penney S, Campbell IM, Pehlivan D, Hunter JV, Woodbury SL, Raymond G, Adesina AM, et al. Mutations in VRK1 Associated With Complex Motor and Sensory Axonal Neuropathy Plus Microcephaly. JAMA neurology. 2013 doi: 10.1001/jamaneurol.2013.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Liu C, Igarashi T, Pehlivan D, Chu K, Iyer R, Cruz P, Cherukuri PF, Hansen NF, et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Beuth B, Taylor IA. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 2007;35:87–99. doi: 10.1093/nar/gkl1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Pehlivan D, Hullings M, Carvalho CM, Gonzaga-Jauregui CG, Loy E, Jackson LG, Krantz ID, Deardorff MA, Lupski JR. NIPBL rearrangements in Cornelia de Lange syndrome: evidence for replicative mechanism and genotype-phenotype correlation. Genet Med. 2012;14:313–322. doi: 10.1038/gim.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes & Development. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J., Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P, Kellerman E, Jaron R, Geiger D, Segel R, Lee M, King MC, Levy-Lahad E. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am J Hum Genet. 2009;85:281–289. doi: 10.1016/j.ajhg.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche AF, Mukherjee D, Guo SM, Moore WM. Head circumference reference data: birth to 18 years. Pediatrics. 1987;79:706–712. [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Shiraishi H, Okamoto H, Hara H, Yoshida H. Alternative cell death of Apaf1-deficient neural progenitor cells induced by withdrawal of EGF or insulin. Biochimica et biophysica acta. 2010;1800:405–415. doi: 10.1016/j.bbagen.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Siepel A, Pollard KS, Haussler D. New methods for detecting lineage-specific selection Proceedings of the 10th International Conference on Research in Computational Molecular Biology (RECOMB) 2006:190–205. [Google Scholar]
- Valbuena A, Sanz-Garcia M, Lopez-Sanchez I, Vega FM, Lazo PA. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell Signal. 2011;23:1267–1272. doi: 10.1016/j.cellsig.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Valbuena A, Vega FM, Blanco S, Lazo PA. p53 downregulates its activating vaccinia-related kinase 1, forming a new autoregulatory loop. Mol Cell Biol. 2006;26:4782–4793. doi: 10.1128/MCB.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester A, Velez-Ruiz G, McLaughlin HM, Program NCS, Lupski JR, Talbot K, Vance JM, Zuchner S, Roda RH, Fischbeck KH, et al. A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Human mutation. 2013;34:191–199. doi: 10.1002/humu.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Yourshaw M, Mamsa H, Rudnik-Schoneborn S, Menezes MP, Hong JE, Leong DW, Senderek J, Salman MS, Chitayat D, et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat Genet. 2012;44:704–708. doi: 10.1038/ng.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3' transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.