Abstract
Papaya (Carica papaya L.) is a climacteric fleshy fruit that undergoes dramatic changes during ripening, most noticeably a severe pulp softening. However, little is known regarding the genetics of the cell wall metabolism in papayas. The present work describes the identification and characterization of genes related to pulp softening. We used gene expression profiling to analyze the correlations and co-expression networks of cell wall-related genes, and the results suggest that papaya pulp softening is accomplished by the interactions of multiple glycoside hydrolases. The polygalacturonase cpPG1 appeared to play a central role in the network and was further studied. The transient expression of cpPG1 in papaya results in pulp softening and leaf necrosis in the absence of ethylene action and confirms its role in papaya fruit ripening.
Introduction
Papaya (Carica papaya L.) is a fleshy fruit that undergoes severe pulp softening during ripening. Although softening increases the sensory and nutritional properties of the fruit, it also contributes to post-harvest deterioration and product losses because the fruit becomes susceptible to physical injury and mold growth [1].
The softening of fleshy fruits during ripening is characterized by the degradation of the plant's cell wall, which is mainly formed by polysaccharides organized in the pectic (amorphous) and the hemicellulosic/cellulosic (crystalline) fractions and the loss of the rigidity of cell wall structure is attributed to the solubilization of both fractions [2]. This solubilization is achieved by the interactive action of several enzymes on determined polysaccharides, such as polygalacturonases (PGs), pectinesterases (PMEs) and pectate lyases (PLs) on homogalacturonans, arabinofuranosidases (ARFs) and galactanases (GALs) on heterogalacturonans and xyloglucan endotransglucosylases/hydrolases (XTHs), endoxylanases (EXYs) and cellulases (CELLs) on hemicelluloses [3], [4].
In the case of papaya fruit, the investigation of polysaccharide changes during ripening has revealed that the disassembly of the cell wall is predominately caused by the degradation and solubilization of pectin [5]. In this regard, polygalacturonase (cpPG1) [6] and β-galactosidase (cp_b-GAL) [7] play a central role in pectin solubilization during papaya ripening. However, other studies have provided evidence that the solubilization of the hemicellulosic fraction by endoxylanases (EXY) is a major contributor to the softening of papaya fruit [8]. This apparent contradiction between the experimental evidence provided by different studies suggests that the disassembly of the cell wall in papaya fruit is a more complex process that involves several enzymes and various cell wall components. In fact, the investigation of several genes or proteins simultaneously may provide better insight on the physiological process, as revealed by data from an XSpecies microarray of papaya ripening that indicated that several cell wall-related genes exhibit similarities to the gene expression profiles observed during the transition of 5- to 11-day-old hypocotyls in Arabidopsis thaliana [9], [10]. Moreover, evaluating the correlations and co-expression of a set of genes may provide clues to the interactive role of those genes.
Therefore, investigating the network of cell wall-related genes, specifically those encoding depolymerization or degradation enzymes, may further elucidate the mechanism of cell wall disassembly during papaya ripening. Thus, this study describes the qPCR analysis of the mRNA levels of some genes of enzymes that act on homogalacturonans (polygalacturonases, pectinesterases and pectate lyases), heterogalacturonans (arabinofuranosidases and galactanases) and hemicellulose (xyloglucan endotransglucosylases/hydrolases and xylanases), which covers the diversity of polysaccharides from the amorphous and crystalline fractions of cell wall, and the correlations and co-expression network of 25 cell wall-related genes during papaya ripening. In addition, the protein levels and enzymatic activity of endopolygalacturonase and the overexpression of the cpPG1 gene were measured in agroinfiltrated papaya fruit pulp and leaves.
Results
Cloning cell wall-related genes in papaya fruit
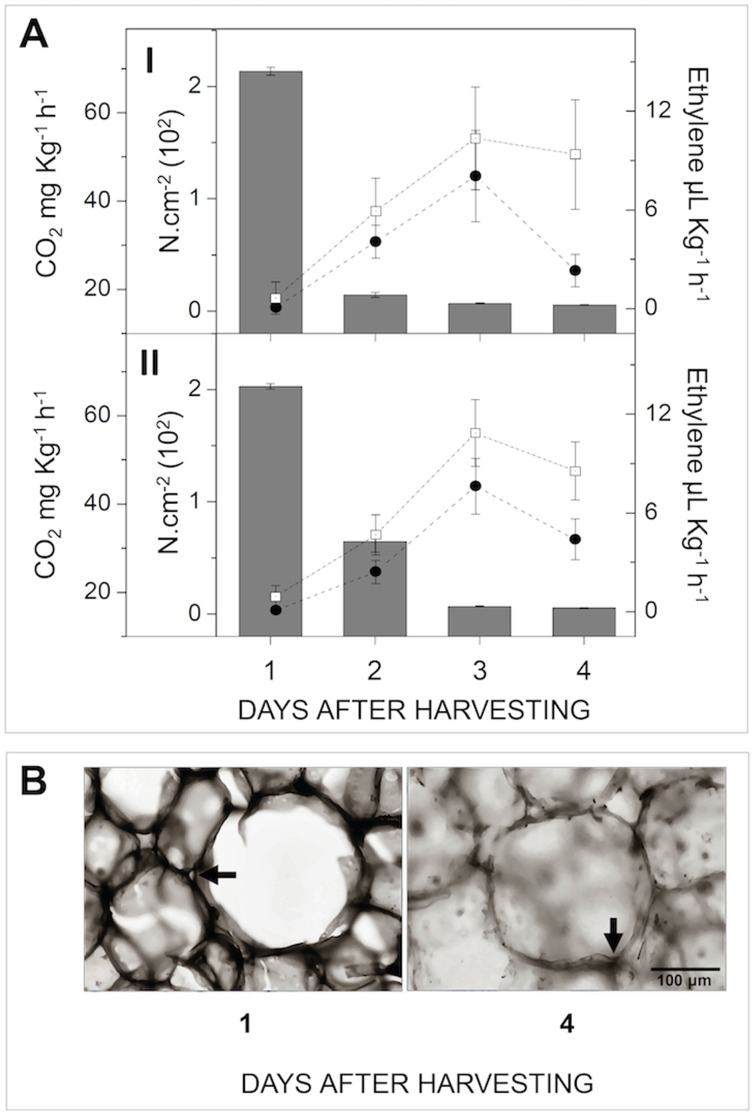
The fruit used for the cloning and expression analysis presented typical softening resulting from the extensive solubilization of cell wall components, as indicated by the climacteric decrease in pulp firmness [1] and cell wall thinning ( Figure 1 ).
Figure 1. Ripening of papaya fruit.
A) The amount of CO2 produced by respiration (Open squares – mg Kg−1 h−1), the production of endogenous ethylene (Black circles - µL Kg−1 h−1), and the pulp firmness (Bars – N.cm−2) were monitored throughout ripening. Error bars indicate SDs of the mean (n = 12) for each sampling (I and II). B) Sections of frozen papaya pulp (12–15 µm) in two different stages (unripe and ripe – 1st and 4th day after harvesting, respectively) were stained with 0.05% of toluidine blue and documented using light microscope. Different colorations indicate cell wall thinning. Arrows indicate the junction zones of cell walls. Bar indicates the scale. The image is a representation of, at least, a triplicate of experiment.
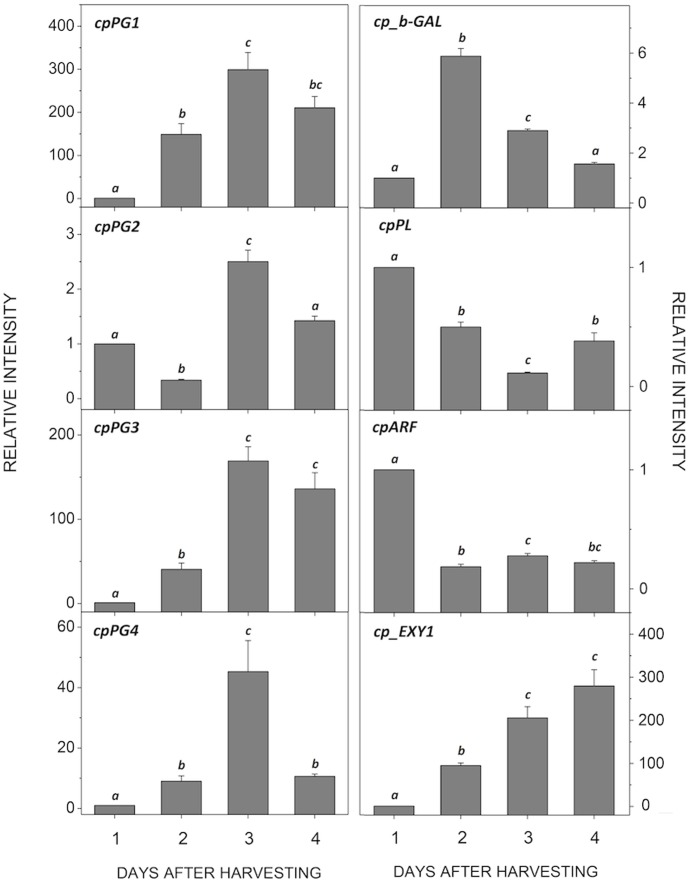
A BLAST search of the papaya genome allowed the cloning of cpPG1 (FJ007644 – Fabi et al., 2009), cpPG2 (GQ479794), cpPG3 (GQ479795), cpPG4 (GQ479796), cpARF (GQ479793), the previously reported cp_b-GAL (AF064786 – [7]) and cpPL (DQ660903). As summarized in Table 1 , the properties of the encoded proteins are similar to those from other plants. The four PGs were differentially expressed and peaked in the climacteric papaya (3DAH) ( Figure 2 ). CpEXY1 was also differentially expressed during ripening, but in a linear increasing manner. Papaya cp_b-GAL was also up-regulated, although it was brought to the initial levels after second day after harvesting (2DAH), whereas both cpPL and cpARF were down-regulated. When the amount of the transcripts of the up-regulated genes was compared, cpPG1 and cp_b-GAL were determined to be the most abundant (Figure S1 in File S1), and the amount of cpPG1 exceeded that of the less abundant cpPG4 by six orders of magnitude.
Table 1. Cell wall-related proteins identified in papaya pulp and their probable biological properties.
| Gene name | Length † | Accession Number | Chromosomes | TAIR | Cleavage Site | Molecular Weight (KDa)† | Isoelectric Point †† | Blast Best Match |
| cpPG1 | 397 | ACH82233 | Un contig 23855 (ABIM01023821) | AT3G59850 | Ala29 | 38.98 | 9.14 | Polygalacturonase [P. communis] CAH18935 |
| cpPG2 | 494 | ACV85695 | LG4 contig 16510 (ABIM01016487) | AT3G57510 | Ser33 | 51.03 | 5.21 | Polygalacturonase A [A. chinensis] AAF71160 |
| cpPG3 | 444 | ACV85696 | LG4 contig 19176 (ABIM01019150) | AT3G07970 | Gly31 | 45.78 | 9.22 | Polygalacturonase QRT2 [A. thaliana] NP_187454 |
| cpPG4 | 389 | ACV85697 | LG1 contig 4651 (ABIM01004644) | AT2G43870 | Ala31 | 38.67 | 8.54 | Polygalacturonase 3 [P. communis] BAF42034 |
| cpARF | 710 | ACV85694 | LG4 contig 9013 (ABIM01009003) | AT3G10740 | Ala43 | 74.16 | 4.94 | α-L-arabinofuranosidase [M. domestica] AAP97437 |
| cp-b-GAL | 721 | AAC77377 | LG7 contig 16621 (ABIM01016598) | AT3G13750 | Ala21 | 78.49 | 8.26 | β-galactosidase 1 [P. persica] ABV32546 |
| cpPL | 369 | ABG66730 | LG6 contig 2998 (ABIM01002995) | AT3G07010 | Ser19 | 40.52 | 9.14 | Pectate lyase [P. persica] BAF43573 |
Amino acids numbers;
The Molecular Weight and isoelectric points were estimated using the mature proteins.
Figure 2. Expression of cell wall-related genes during papaya ripening.
Real-time PCR (qPCR) was used to analyze the mRNA levels of various genes during ripening. The column heights indicate the relative mRNA abundance; the expression values for unripened fruit one day after harvest were set to 1. The error bars on each column indicate the SD of four technical replicates from samplings I and II. The different letters represent samples that were significantly different from those collected on other days post-harvest (within the same gene), as determined by one-way ANOVA and Tukey's test (α<0.05, n = 4).
Gene expression and correlation analysis
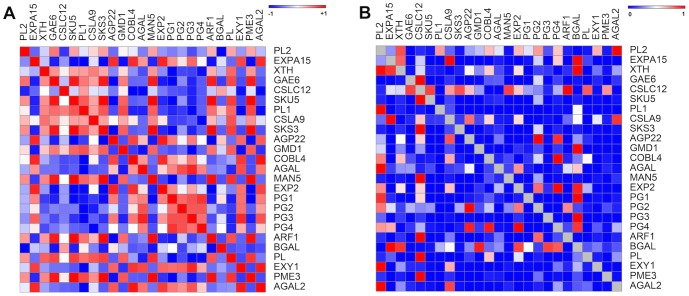
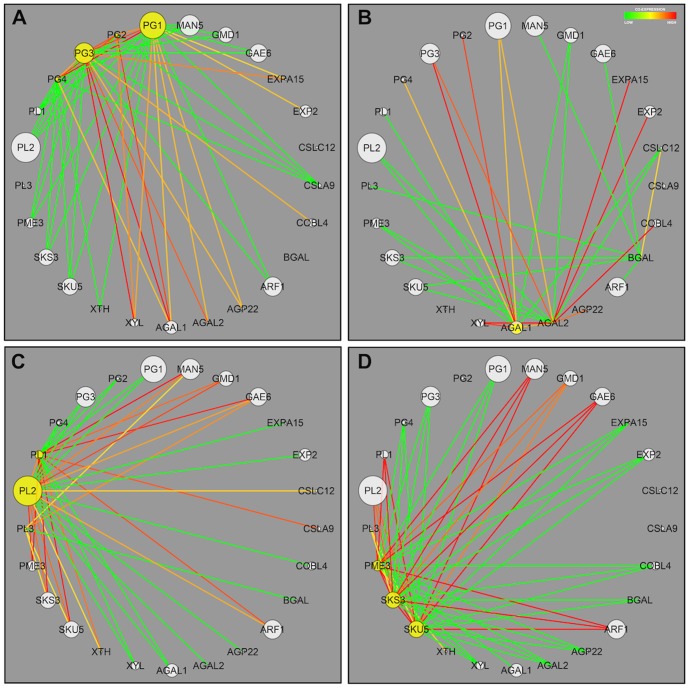
The expression pattern of a collection of 25 cell wall-related genes that were previously described as ripening-related ones [11], [12], [10] was analyzed for co-expression during ripening. Among the 600 possible correlations, 300 had p-values less than 5% and were further analyzed ( Figure 3 ), which revealed that PGs, galactanases, PMEs and PLs significantly impact the network assembly ( Figure 4 ). The PGs were positively correlated to galactanases, xylanases and, to a lesser degree, expansins and negatively correlated to a second group of genes encompassing PMEs, PLs and ARF. Because the expression of papaya cell wall disassembling genes was similar to Arabidopsis from 5 to 11-day-old hypocotyls growth [9], [10], a new comparison of papaya data was done with newly Arabidopsis data [13]. The comparison (Table S1 in File S1) showed changes in the expression of 11 out of 16 genes, which were similar between the ripening fruit and the growing plant. The genes that were most significantly affected in both developmental processes were cpPG1 and its homologues.
Figure 3. Pearson correlations and associated p-values of papaya cell wall-related genes.
The R package for weighted correlation network analysis (weighted gene co-expression network analysis - WGCNA) was used to calculate the Pearson correlation (Figure A) and the corresponding p-values for the 25 papaya cell wall-related genes that were identified by our group (Figure B). From the 600 possible gene expression correlations, 300 had p-values less than or equal to 0.05 and were further analyzed. Figure 1 A: The heat map is described as positive values set to red color and negative values set to blue color. Figure B: The heat map is described as values near to one set to red color and values near to zero set to blue color. The twenty-five genes are described in Table S1 in File S1.
Figure 4. Co-expression network of papaya cell wall-related genes given by the weighted gene co-expression network analysis (WGCNA).
The R package of the weighted gene co-expression network analysis (WGCNA) program was used to produce a gene association network for 25 papaya cell wall-related genes. The size of the circle indicates the weight of the gene on the network. In Figures A, B, C and D, the genes considered for the network assembly are highlighted with yellow backgrounds. Figure A describes the network for the papaya PG genes. Figure B describes the network for the papaya GAL genes. Figure C describes the network for the papaya PME genes. Figure D describes the network for the papaya PL genes. The color lines are described as positive co-expression values set to red color and negative co-expression values set to green color. The twenty-five genes are described in Table S1 in File S1.
Transient expression of papaya cpPG1
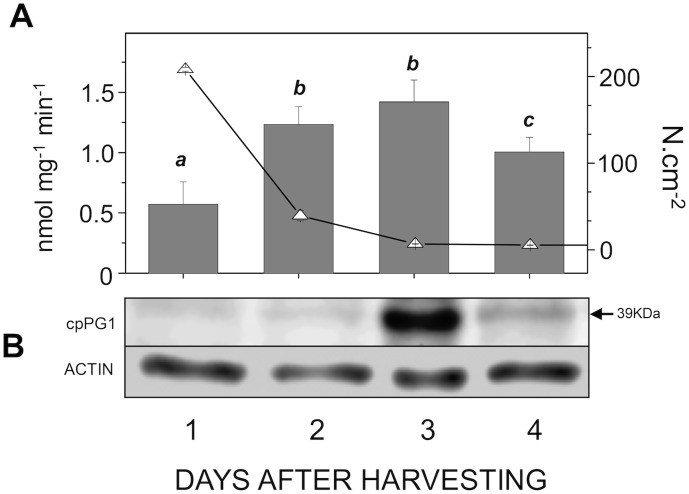
We identified four papaya PG genes that have AT-rich intronic regions with two specific patterns, one for cpPG1 and cpPG4 and another for cpPG2 and cpPG3 (Figure S2 in File S1). Notably, the proteins encoded by cpPG1/cpPG4 were separated in a distinct clade from cpPG2/cpPG3 when the PGs from diverse plants were aligned (Figure S3 in File S1). The expression of cpPG1 in prokaryotic cells resulted in a recombinant product with the expected size and identity of the predicted cpPG1 protein, as revealed by MS sequencing (Figure S4 in File S1). This protein was used to produce polyclonal antibodies utilized in western blotting (Figure S5 in File S1), which showed that the amount of PG protein was correlated to that of the cpPG1 transcript and its enzymatic activity and with pulp softening ( Figure 5 ).
Figure 5. Correlation between pulp softening, enzymatic activity and cpPG1 protein expression during papaya ripening.
A) The pulp firmness (open triangles) was monitored during the ripening process using a texturometer (Fabi et al., 2007), and the measurements are given by N.cm−2. PG enzymatic activities was done according to Fabi et al. (2009). The error bars indicate the SDs of the mean. The different letters represent samples that were significantly different from those collected on other days post-harvest, as determined by one-way ANOVA and Tukey's test (α<0.05, n = 4). B) Western blotting experiment was done using the monoclonal anti-actin for plants as a control experiment (ACTIN), and the images are representative of at least a triplicate experiment. The arrow indicates the molecular weight of the mature protein cpPG1 with the corresponding molecular weight.
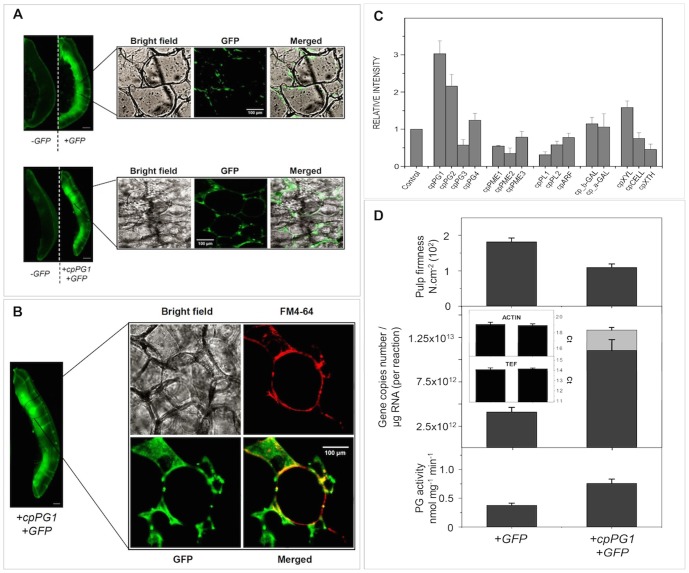
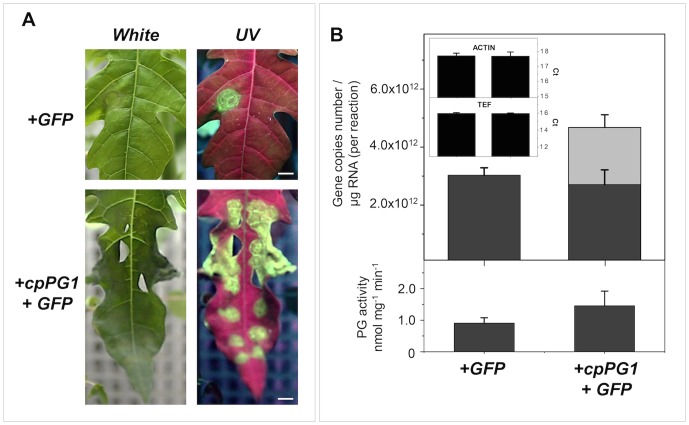
To verify the role played by this PG gene in pulp softening, 1-MCP-treated fruits were transiently transformed to overexpress cpPG1. After three days of agroinfiltration, GFP signals were observed in both the control and cpPG1-GFP-transformed fruits, which showed transient cpPG1-GFP protein delivered to the extracellular space ( Figure 6A and B ). The transcript analysis of 15 cell wall-related genes showed that cpPG1 was up-regulated after treatment, and cpPG2 also appeared to be affected ( Figure 6C ). The absolute quantitation of the transcript revealed a 20% excess of cpPG1 mRNA ( Figure 6D ), which was correlated with a two-fold increase in PG activity and pulp softening. Papaya leaves that were agroinfiltrated with the same construct to overexpress cpPG1 in 1-MCP-treated fruit exhibited a 45% increase in cpPG1 expression, demonstrated increased activity and survived for only two days, whereas the control leaves survived for six days ( Figure 7 ).
Figure 6. Transient expression of the cpPG1 gene in the pulp of papaya fruits treated with 1-MCP.
Constructs carrying GFP or cpPG1-GFP genes were agroinfiltrated into papaya fruits treated with 1-MCP, as described in the Materials and Methods. GFP and cpPG1-GFP fusion proteins were transiently expressed and observed with the Versa Doc Gel Imaging System and a confocal laser scanning microscope (Figures A and B). Contrast with the vital dye FM4-64 shows that the transiently expressed cpPG1-GFP proteins were targeted to the extracellular compartment (Figure B). The scale bars in the images of the fruits are 1 cm, and the scale bars in the confocal images indicate the scale. The images are representative of triplicate experiments performed on each agroinfiltrated fruit (n = 3). Figure C shows the relative mRNA abundance of 15 cell wall-related genes and indicates that cpPG1 and cpPG2 may be the main contributors to papaya pulp softening in agroinfiltrated fruits. The expression values for control fruit expressing only GFP (closed vector) were set to 1 for all genes. The absolute quantification of the cpPG1 mRNA levels in Figure D demonstrates that cpPG1 (gray bar) is transiently expressed. The light gray bar indicates data from transient cpPG1 expression, the gray bars indicate data from endogenous cpPG1 expression, and the black bars indicate the threshold cycle values (Ct) for the two genes used as internal controls (cpACT and cp_EF1).
Figure 7. Transient expression of the cpPG1 gene in papaya leaves.
Constructs carrying GFP or cpPG1-GFP genes were agroinfiltrated into papaya leaves, and the GFP expression in 20-day-old leaves was observed under UV light excitation (Figure A). The images are representative of triplicate experiments that were performed on each agroinfiltrated leaf (n = 3). The absolute quantification of the cpPG1 mRNA levels in Figure B demonstrate that cpPG1 (gray bar) is transiently expressed. The light gray bar indicates data from the transient expression of cpPG1, the gray bars indicate data from endogenous cpPG1 expression, and the black bars indicate the threshold cycle values (Ct) for the two genes used as internal controls (cpACT and cp_EF1). The images are representative of triplicate experiments that were performed on each agroinfiltrated leaf (n = 3). The figure bars are scaled to 1 cm.
Discussion
Pulp softening is a fast and marked change that occurs in ripening papayas and is denoted by the severe loss of firmness after just a few days. The thinning of the cell wall, which results from the substantial action of enzymes on the polysaccharide structure, causes a loss in pulp tissue rigidity. Considering its complexity, which is derived from various polysaccharide chains and the arrangement of pectic and hemicellulosic fractions, several enzymes are expected to act as a result of the interactions of co-expressed cell wall-related genes.
According to the evaluation of the gene expression profiles of various cell wall enzymes, the depolymerizing capacity is predominately attributed to the increased expression of PGs, β-gal and endoxylanase. This result suggests that the massive solubilization of pectin may be the predominant event, although a minor contribution of the hemicellulose component should not be disregarded, as indicated by the increase in endoxylanase. Notably, the gene encoding PL, an enzyme responsible for pectin depolymerization through a β-elimination reaction, was down-regulated. In fact, WGCNA analysis revealed both positive and negative correlations among the expression patterns of the genes of pectin-acting enzymes, in contrast from what was observed for strawberries, in which a concomitant action of PL and PG might be essential for pulp softening [14], [15]. In addition to PL, the co-expression of PGs was also negatively correlated to that of PMEs. Therefore, the action of papaya PGs does not require the simultaneous removal of pectin methyl groups, which would be consistent with the stable amount of methoxylated pectin during ripening [1], [12], [10], [5].
Another interesting result that emerged from the WGCNA analysis was the co-expression of galactanases, which may act on the heterogalactans of pectins. Alpha-galactosidase genes only marginally contribute to pulp softening [16] and were strongly correlated to PG expression. In contrast, an up-regulated β-galactosidase that is partially responsible for papaya softening [17] demonstrated increased co-expression with genes related to pectin and hemicellulose modifications, such as GAE6, ARF1, PL2, PL3 and XTH [18], [19], [20], [21], [22]. This finding might indicate that cp_b-GAL plays a ubiquitous role in papaya plant cell wall mobilization that is not limited to pectin depolymerization during fruit ripening.
The data from the gene expression and co-expression network analysis suggest a cooperative network of cell wall-related genes during papaya pulp softening that is mainly composed of endoPGs, which is consistent with the massive pectin solubilization [5] and the release of uronic acid oligosaccharides [23]. Softening dependence on massive PG action has already been seen in apple fruit, as denoted by the inhibition of softening in transgenic lines with PG suppression [24]. The up-regulation of cp_EXY1 corroborates previous data reported by Manenoi and Paull [8], and the encoded enzyme accounts for the hydrolysis of β-1,4-D-xylosyl that is bounded to polysaccharides in the cell wall matrix, which are mainly hemicelluloses. However, because cellulase expression decreases during ripening [10], cp_EXY1 would contribute to the minor disassembly of hemicellulose fractions by the release of heteroglucan chains but not the depolymerization of the cellulose backbone, which is mostly found in a crystalline phase. Therefore, the action of pectinases on less compacted pectin in the amorphous phase would have a more significant effect on cell wall disassembly during ripening.
In addition to a previous observation of the similarities between ripening papayas and Arabidopsis hypocotyl gene expression [10], new data from Arabidopsis development (young and mature plants – [13] – Table S1 in File S1) indicate similar co-expression patterns for several cell wall-related papaya genes. In this regard, the cpPG1 orthologue from Arabidopsis (AT3G59850) appears to play a central role in cell wall disassembly because the transgenic expression of this gene and the co-expression of expansin (EXP2 - AT2G39700 - [25]) markedly changed the pulp firmness and increased the tomato susceptibility to Botrytis cinerea [26].
The occurrence of different endoPGs in various fleshy fruit organs has previously been reported [27], [28], [29] and suggests a degree of redundancy or specialization in pectin depolymerization. In this regard, cpPG1 appears to play a predominant role in papaya softening as it occurs for apple softening [30], [24]. Thus, the more abundant transcript would account for most of the enzymatic activity observed in the fruit pulp, although it was not so well adjusted to the protein peak abundance at the third day after harvest, probably as a result of the interference of other PG proteins, such as cpPG3 and cpPG4, in the assays. Furthermore, since the performed activity assay measures reducing end formation, we cannot exclude a minor contribution of exopolygalacturonases. In relation to cpPG2, that gene was separated from cpPG1 in a distinct clade, and the encoded protein had a Lys346 catalytic residue instead of a Glu, which allowed speculation that this product may act differently on pectin depolymerization (Table S2 and Figure S6 in File S1).
The transient transformation of papaya demonstrated the effect of cpPG1 on pulp softening. Treatment with 1-MCP precluded ethylene synthesis and the triggering of PG genes. Fruit agroinfiltrated with pGBPGWG_cpPG1 transiently expressed cpPG1-GFP proteins that were delivered to the extracellular compartment, and the pulp of the fruit softened compared with those that received the empty vector. Notably, the up-regulation of cpPG2 may reveal some level of responsiveness or dependence on cpPG1 and insensitivity to ethylene because the gene was expressed during late ripening in non-transformed fruit ( Figure 2 ). At the same time, it is possible that other ethylene-independent cell wall-related genes had contributed to partial softening of the pulp. The results of the transient transformation of papaya leaves with pGBPGWG_cpPG1 were similar to those of tobacco leaves that overexpress polygalacturonases [31], which cause cell death and leaf necrosis ( Figure 7A ). Moreover, it is likely that the overexpression of the cpPG1 gene reduced cell wall adhesion causing disruption of leaf organization as was observed in apple tree leaves constitutively expressing a fruit-specific PG [30]. The above-mentioned results demonstrate that the abundantly expressed cpPG1 gene may exert profound changes in the papaya cell wall structure and play a central role in pectin disassembly during fruit ripening. The softening of fleshy fruit, particularly pectin-rich fruit, may now be studied through a broader approach by considering a key group of cell wall-related genes and their interactions with each other and other cell wall-related genes, as was demonstrated with papayas.
Materials and Methods
Plant material and ripening parameters
Papayas (Carica papaya L. cv. Golden) were obtained from a producer in Linhares City, Espírito Santo, Brazil. The fruits were harvested with up to 25% yellow peel (150 days post-anthesis). The fruits were stored in 240 L chambers with controlled temperature and humidity (22°C±0.1°C and 95%, respectively) for seven days. Daily analysis was performed on six fruits (two distinguishable biological replicates from two different seasons). The CO2, ethylene and pulp firmness were measured according to Fabi et al. [1], and the remaining pulp was N2-frozen, pooled and stored at −80°C. Sections of frozen tissue (12–15 µm) were cut using a refrigerated microtome, stained with toluidine blue (0.05% in 0.1 M phosphate buffer, pH 6.8) and visualized using a light microscope (Zeiss, Germany).
Bioinformatics and sequence cloning and analysis
The papaya genomic sequences were identified by comparing a partial papaya genome [32] with polygalacturonases from diverse fleshy fruits and Arabidopsis thaliana using Blast tools and primers that were designed to flank the coding regions (Table S3 in File S1). Papaya pectate lyase (cpPL1) and β-galactosidase (cp_b-GAL) genes were downloaded from the GenBank database (DQ660903 and AF064786, respectively). For papaya α-L-arabinofuranosidase, a pair of degenerated primers was used to identify part of the coding region (Table S3 in File S1). The total RNA was extracted according to Fabi et al. [6], and first-strand cDNAs were synthesized from 1 µg of total RNA using an Improm-II Reverse Transcription System kit (Promega, Madison, WI, USA) and oligo-dT primers. The coding regions were PCR-amplified using KOD Hot Start DNA Polymerase (Merck KGaA, Darmstadt, Germany), and sequences with a high GC content (cpPG2 and cpPG3) were PCR-amplified using KAPA HiFi DNA Polymerase (Kappa Biosystems, Boston, MA, USA). The PCR fragments were digested with XhoI and ligated on a pET-45b(+) (Merck KGaA, Darmstadt, Germany) plasmid. After cloning and sequencing, the sequences were analyzed with the TargetP 1.1 tool [33] and the SignalP tool [34]. Finally, the DNA was extracted according to Fabi et al. [6], PCR-amplified as described above, cloned and genomically sequenced.
Quantitative analysis of gene expression by real time-PCR (qPCR)
Gene expression analyses were performed according to [10] and following the ‘Minimum Information for Publication of Quantitative Real-Time PCR Experiments – MIQE’ [35]. The primer sequences are shown in Table S4 in File S1. The gene expression of a previously identified papaya endoxylanase (cpEXY1 - AY138968; [8]) was also analyzed. The actin gene (ACT) located on chromosome LG9 contig 1059 (GenBank accession no. ABIM01001059) and the elongation factor 1-alpha gene (EF1) located on chromosome LG9 (GenBank accession no. ABIM0101268) were used as internal controls. To evaluate the agroinfiltration experiments using the pGBPGWG vector, a pair of primers enclosing the GFP gene was synthesized (Table S4 in File S1). For primer testing and identity confirmation, all of the fragments from the PCR reactions were cloned and sequenced. Real-time PCR was performed using a four-channel Rotor-Gene 3000 multiplexing system (Corbett Research, Sydney, Australia). The non-template controls (NTCs) and melting curve analyses of the amplicons were monitored for all experiments. The threshold cycle (Ct) values (four technical replicates and two biological replicates) were averaged using the Rotor-Gene 3000 software, and the quantification was performed using the relative standard curve method [36]. The results of the standard curves calculations are shown in Table S5 in File S1. To assess the gene copy numbers, the corresponding cloned coding regions were used to construct calibration curves (Table S6 in File S1). The results are expressed as gene copy numbers/microgram of RNA for each PCR reaction. All of the data were analyzed by one-way ANOVA, and the means were compared using Tukey's test. The statistical analyses were performed using OriginPro version 8 software (OriginLab).
Comparative biology and co-expression patterns
ClustalW phylogenetic trees [37] were calculated from the alignment of the corresponding proteins with proteins from fleshy fruits and A. thaliana. To search for co-expression patterns between genes related to papaya cell wall metabolism, the log2 values of the gene expression data from 25 genes were analyzed using the weighted gene co-expression network analysis (WGCNA) in the R package [38]. In summary, pair-wise gene (Pearson) correlations were calculated using gene expression data from genes identified in this study and three previous studies [10], [11], [12]. The correlations and associated p-values were calculated using the WGCNA R package and the “corAndPvalue” module. The GENE-E software was loaded with values, and corresponding heat map figures were generated (http://www.broadinstitute.org/cancer/software/GENE-E). A weighted adjacency matrix was then constructed by the WGCNA package using default parameters from the “adjacency” module and a soft-threshold parameter equal to 3, which forces the program to emphasize stronger correlations over weaker ones [39]. The obtained co-expression networks were visualized using Cytoscape [40].
Heterologous Protein Expression, Purification and Sequencing
Protein expression was performed by transforming Rosetta 2 (DE3) pLysS (Novagen) E. coli with a pET-45b(+) construct containing cpPG. Bacteria were grown in LB medium (Cm+/Amp+), and inductions were performed with isopropyl β-D-1-thiogalactopyranoside (IPTG) when the OD600 nm reached 0.6. A control experiment was performed using a closed pET-45b(+) plasmid. Images of polyacrylamide gels were captured using a Versa Doc Gel Imaging System (Bio-Rad, CA, USA). The molecular masses of corresponding bands were estimated by comparing them with standard bands of molecular weight proteins using the Quantity One software (version 4.6.7) (Bio-Rad, CA, USA). The pelleted bacterial suspensions were disrupted using the commercial BugBuster Protein Extraction Reagent (Merck KGaA, Darmstadt, Germany) supplemented with the protease inhibitor cocktail ProteoBlock (Fermentas, Thermo Fisher Scientific Inc., Vilnius, Lithuania). The recombinant proteins were extracted from inclusion bodies using 8 M urea with 24 hours of agitation and purified in a nickel column (Protino NI-TED 1000 - Macherey-Nagel GmbH & Co KG, Düren, Germany) according to the manufacturer's instructions. The samples were analyzed by SDS-PAGE on a 12.5% acrylamide gel [41]. The bands of interest were excised from the gels, and the digestion and protein sequencing were performed according to [42].
Production of Polyclonal Antibodies
After confirming the identities of the recombinant proteins, 1 mg of the purified protein was separated using SDS-PAGE. The band was excised and the rabbits were immunized according to [43].
Western Blotting and Enzymatic assays
The total protein from the pulp of papayas was extracted according to Carpentier et al. [44] by adding ProteoBlock (Fermentas) protease inhibitor. The protein concentrations were determined using a 2-D Quant Kit (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions. Twenty micrograms of total protein were separated by SDS-PAGE and transferred to nitrocellulose membranes (Hybond-ECL, GE Healthcare, Piscataway, NJ, USA). The membranes were incubated with antiserums and secondary anti-rabbit ECL Plex Goat-the-Rabbit Cy5 antibody (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions. Monoclonal anti-actin for plants (Sigma, #A0480, St. Louis, MO, USA - 1∶5000 dilution) with a secondary anti-mouse ECL Plex Goat-a-Mouse Cy3 antibody (GE Healthcare, Piscataway, NJ, USA) was used as a control experiment. Images of the membranes were captured with a Versa Doc Gel Imaging System (Bio-Rad) using the corresponding channels for Cy5 and Cy3 detection (multiplexing). Images were taken after 120 seconds of exposure for Cy5 and 30 seconds of exposure for Cy3, and the negative image files were obtained with the Quantity One (Bio-Rad) software. The bands observed were compared with the standard molecular weight ECL Plex Fluorescent Rainbow Marker (GE Healthcare). The proteins for enzymatic assays were extracted as previously described [6]. The polygalacturonase activity was assayed according to Fabi et al. [6].
Transient gene expression of cpPG1 in papaya fruits and leaves
The cpPG1 gene was transiently expressed in papaya fruits and leaves by agroinfiltration. The binary vector used in the experiments was pGBPGWG expressing green fluorescent protein (GFP) under the regulation of the 35S promoter (GenBank accession number AM884372), and the cpPG1 ORF was cloned into the vector by recombination, as previously described by Zhong et al. [45]. The agroinfiltration of the papaya fruits was performed according to Spolaori et al. [46]. Agrobacterium tumefaciens GV3101 containing a pSoup plasmid (Tet+/Gent+) was transformed with a binary vector that enclosed cpPG1 ORF (pGBPGWG_cpPG1 – Cm+/Kan+) and used in both experiments. Moreover, bacterial suspensions were co-infiltrated (1∶1) with A. tumefaciens GV2260 that was transformed with the 35S: p19 binary vector (Rif+/Kan+) to decrease the post-transcriptional gene silencing of exogenous cpPG1 [47].
One day after harvesting, the papaya fruits were treated with 100 ppb of 1-methylcyclopropene (1-MCP) for 12 hours to reduce cpPG1 endogenous gene expression, as previously described [1]. After treatment, three fruits (n = 3) were agroinfiltrated using sterile syringes and a short needle (1 mL bacteria suspension per 4 cm2). The fruits were allowed to ripen at 22°C for a 16-h photoperiod. A previous test using a mixture of Agrobacterium and methylene blue was conducted to calculate the extent of agroinfiltration. Three days post-infiltration, the fruits were sliced and photographed using a Versa Doc Gel Imaging System (Bio-Rad) with parameters for GFP detection. The pulp firmness was measured according to Fabi et al. [1]. The remaining slices were N2-frozen for future gene expression quantification. A control experiment (n = 3) was similarly conducted using the pGBPGWG-closed vector. Sections of pulp were manually cut, stained with FM4-64 vital dye and analyzed using a confocal laser-scanning microscope (Zeiss, Jena, Thuringia, Germany, LSM 510). The images were processed using the LSM image browser software.
The agroinfiltration of young papaya leaves (20 days and 50 days after germination) was performed according to Boudart et al. [31]. The abaxial spaces of the leaves (n = 6) were agroinfiltrated using sterile syringes (without the needle). The plants were maintained at 22°C for a 16 h photoperiod, and the leaves were collected 2 days post-infiltration (20-day-old leaves). A control experiment (six leaves) was similarly conducted using the pGBPGWG closed vector.
Supporting Information
These are the legends for Supporting Tables / Figures presented in File S1. Table S1. Cell wall-related genes from ripe papaya and mature A. thaliana plant. Table S2. Similarity percentage of amino acid from papaya and other plants PGs. Table S3. Nucleotide sequences used in PCR reactions. Table S4. Nucleotide sequences used in qPCR. Table S5. Calibration curves for relative gene expression. Table S6. Calibration curves for absolute gene expression. Figure S1. Up-regulation of cell wall-related genes during papaya ripening. Real-time PCR (qPCR) was used to determine the absolute quantitation of the mRNA levels of various genes during papaya ripening. The quantification is represented by the column height. The error bars on each column indicate the SD from four technical replicates from samplings I and II. The different letters represent samples that were significantly different from those collected on other days post-harvest (within the same gene) as determined by one-way ANOVA and Tukey's test (α<0.05, n = 4). Figure B shows the threshold cycle values (Ct) for the two genes used as internal controls (actin gene – cpACT and elongation factor 1-alpha gene - cp_EF1). Figure S2. Genomic and mRNA organization of different PGs from papaya fruit. Grey boxes represent coding regions (exons), while black lines represent non-coding regions (introns). White boxes represent the mRNA sequences concatenated from the above compared exons. Figure S3. Unrooted phylogram encompassing PGs from papaya, Arabidopsis and several other plant organisms. A phylogenetic tree was calculated using the neighbor-joining method based on the ClustalW alignment of the deduced amino acid sequences. The putative signal peptide from all of the proteins was removed from the sequence. The following proteins and their corresponding GenBank IDs were used: A. thaliana 1, 2 and 3 (NP_191544, NP_191310, NP_187454), P. persica 1 and 2 (AAC64184, CAA54448), P. communis 1 and 2 (CAH18935, BAC22688), D. carota (BAC87792), S. lycopersicum 1 and 2 (ABW38780; AC28947), C. melo 1 and 2 (AAC26510; AAC26512), V. vinifera 1 and 2 (XP_002263164, XP_002282759), B. napus (CAA65072), G. Max 1 and 2 (ABC70314, AAL30418), D. kaki (ACJ06506), R. communis 1, 2 and 3 (XP_002529090, XP_002529616, XP_002517823), A. deliciosa (P35336), O. europaea (ACA49228), P. trichocarpa 1, 2 and 3 (XP_002331621, XP_002322711, XP_002313308) and C. papaya 1, 2, 3 and 4 (FJ007644, GQ479791, GQ479794 e GQ479795). The values for the branch lengths are based on the scale bar; there are 0.1 residue substitutions per site. Letters A and B show the two distinct clades resulting from the phylogenetic analyses. Figure S4. Recombinant proteins sequencing. Figures show the query sequences of cpPG1 (A) protein, and the correspondent peptides that were visualized in protein sequencing. Residues highlighted by gray boxes and black letters are from expression vector; residues highlighted by black boxes and white letters are those obtained in protein sequencing. The histidine tags are underlined and asterisks indicate the end of protein. Figure S5. Expression of recombinant proteins from papaya pulp. The figures show representative gels from heterologous expression of cpPG1 (A) gene. Figure A. Proteins profiles from bacteria carrying pET45-cpPG1 before IPTG induction (PG), after 16 hs growth non-induced (PG-NI) and after 16 hs growth IPTG-induced (PG-I); Ni-purified recombinant proteins can be seen after properly elution (E 1 and 2). Proteins markers are highlighted with weight numbers. Figure B. The figure displays the hybridization of antiserum for cpPG1 recombinant protein. The arrow indicates the molecular weight for commercially available protein markers. The quantities of proteins are also indicated. Figure S6. Protein alignment of four distinct papaya PGs. The corresponding proteins from cpPG1 (PG1 - FJ007644), cpPG2 (PG2 - GQ479791), cpPG3 (PG3 - GQ479794) and cpPG4 (PG4 - GQ479795) genes were aligned using ClustalW and viewed with BOXSHADE program and then manually edited. Gaps were introduced to optimize alignment. Identic amino acids are highlighted by black boxes and white letters, similar amino acids are highlighted by grey boxes and white letters and different amino acids are highlighted by white boxes and black letters. The conserved domain of glycosil hydrolases from family 28 (PS00502) is indicated by a dotted-line box. The probable catalytic residues are indicated by asterisks, being possible to observe the cpPG2 protein has a Glu346 instead of a Lys346 as the other three polygalacturonases have.
(ZIP)
Acknowledgments
We would like to thank Prof. David Baulcombe, SL and The Gatsby Charitable Foundation for donating the p19 construction.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was financially supported by grants #2002/12452-9 and #2008/54476-8, scholarships to JPF (#2003/00932-9 and #2007/56515-8), SGB (#2009/53329-4) and SLGLS (#2008/58653-1), São Paulo Research Foundation (FAPESP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fabi JP, Cordenunsi BR, Barreto GPM, Mercadante AZ, Lajolo FM, et al. (2007) Papaya fruit ripening: response to ethylene and 1-methylcyclopropene (1-MCP). J Agric Food Chem 55: 6118–6123. [DOI] [PubMed] [Google Scholar]
- 2. Gapper NE, McQuinn RP, Giovannoni JJ (2013) Molecular and genetic regulation of fruit ripening. Plant Mol Biol 82(6): 575–91. [DOI] [PubMed] [Google Scholar]
- 3. Brummell DA (2006) Cell wall disassembly in ripening fruit. Funct Plant Biol 33: 103–119. [DOI] [PubMed] [Google Scholar]
- 4. Hyodo H, Terao A, Furukawa J, Sakamoto N, Yurimoto H, et al. (2013) Tissue Specific Localization of Pectin-Ca(2+) Cross-Linkages and Pectin Methyl-Esterification during Fruit Ripening in Tomato (Solanum lycopersicum). PLoS One 13 8(11): e78949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shiga TM, Fabi JP, Nascimento JRO, Petkowicz CLO, Vriesmann LC, et al. (2009) Changes in cell wall composition associated to the softening of ripening papaya: evidence of extensive solubilisation of large molecular mass galactouronides. J Agric Food Chem 57: 7064–7071. [DOI] [PubMed] [Google Scholar]
- 6. Fabi JP, Cordenunsi BR, Seymour GB, Lajolo FM, Nascimento JRO (2009) Molecular cloning and characterization of a ripening-induced polygalacturonase related to papaya fruit softening. Plant Physiol Biochem 47: 1075–1081. [DOI] [PubMed] [Google Scholar]
- 7. Othman R, Choo TS, Ali ZM, Zainal Z, Lazan H (1998) A full-length beta-galactosidase cDNA sequence from ripening papaya. Plant Physiol 118: 1102–1102. [Google Scholar]
- 8. Manenoi A, Paull RE (2007) Papaya fruit softening, endoxylanase gene expression, protein and activity. Physiol Plant 131: 470–480. [DOI] [PubMed] [Google Scholar]
- 9. Jamet E, Roujol D, San-Clement H, Irshad M, Soubigou-Taconnat L, et al. (2009) Cell wall biogenesis of Arabidopsis thaliana elongating cells: transcriptomics complements proteomics. BMC Gen 10: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabi JP, Seymour GB, Graham NS, Broadley MR, May ST, et al. (2012) Analysis of Ripening-related Gene Expression in Papaya using an Arabidopsis-based Microarray. BMC Plant Biol 12: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fabi JP, Lajolo FM, Nascimento JRO (2009) Cloning and characterization of transcripts differentially expressed in the pulp of ripening papaya. Sci Hort 121: 159–165. [Google Scholar]
- 12. Fabi JP, Mendes LRBC, Lajolo FM, Nascimento JRO (2010) Transcript profiling of papaya fruit reveals differentially expressed genes associated with fruit ripening. Plant Sci 179: 225–233. [Google Scholar]
- 13. Chen CC, Fu SF, Lee YI, Lin CY, Lin WC, et al. (2012) Transcriptome analysis of age-related gain of callus-forming capacity in Arabidopsis hypocotyls. Plant Cell Physiol 53(8): 1457–69. [DOI] [PubMed] [Google Scholar]
- 14. Santiago-Doménech N, Jiménez-Bermúdez S, Matas AJ, Rose JKC, Muñoz-Blanco J, et al. (2008) Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening. J Exp Bot 59: 2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quesada MA, Blanco-Portales R, Posé S, García-Gago JA, Jiménez-Bermúdez S, et al. (2009) Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiol 150: 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soh CP, Ali ZM, Lazan H (2006) Characterisation of an alpha-galactosidase with potential relevance to ripening related texture changes. Phytochem 67: 242–254. [DOI] [PubMed] [Google Scholar]
- 17. Lazan H, Ng SY, Goh LY, Ali ZM (2004) Papaya beta-galactosidase/galactanase isoforms in differential cell wall hydrolysis and fruit softening during ripening. Plant Physiol Biochem 42: 847–853. [DOI] [PubMed] [Google Scholar]
- 18. Mandaokar A, Thines B, Shin B, Lange BM, Choi G, et al. (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46(6): 984–1008. [DOI] [PubMed] [Google Scholar]
- 19. Bassel GW, Fung P, Chow TF, Foong JA, Provart NJ, et al. (2008) Elucidating the germination transcriptional program using small molecules. Plant Physiol 147(1): 143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yadav RK, Girke T, Pasala S, Xie M, Reddy GV (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci U S A 106(12): 4941–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dalchau N, Hubbard KE, Robertson FC, Hotta CT, Briggs HM, et al. (2009) Correct biological timing in Arabidopsis requires multiple light-signaling pathways. Proc Natl Acad Sci U S A 107(29): 13171–13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ng S, Giraud E, Duncan O, Law SR, Wang Y, et al. (2013) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288(5): 3449–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sañudo-Barajas JA, Labavitch J, Greve C, Osuna-Enciso T, Muy-Rangel D, et al. (2009) Cell wall disassembly during papaya softening: Role of ethylene in changes in composition, pectin derived oligomers (PDOs) production and wall hydrolases. Postharvest Biol Technol 51: 158–167. [Google Scholar]
- 24. Atkinson RG, Sutherland P, Johnston SL, Gunaseelan K, Hallett IC, et al. (2012) Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuber H, Davidian JC, Aubert G, Aimé D, Belghazi M, et al. (2010) The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol 154(2): 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, et al. (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea . Proc Natl Acad Sci U S A 105(3): 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hiwasa K, Kinugasa Y, Amano S, Hashimoto A, Nakano R, et al. (2003) Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. J Exp Bot 54: 771–779. [DOI] [PubMed] [Google Scholar]
- 28. Morgutti S, Negrini N, Nocito FF, Ghiani A, Bassi D, et al. (2006) Changes in endopolygalacturonase levels and characterization of a putative endo-PG gene during fruit softening in peach genotypes with nonmelting and melting flesh fruit phenotypes. New Phyt 171: 315–328. [DOI] [PubMed] [Google Scholar]
- 29. Sekine D, Munemura I, Gao M, Mitsuhashi W, Toyomasu T, et al. (2006) Cloning of cDNAs encoding cell-wall hydrolases from pear (Pyrus communis) fruit and their involvement in fruit softening and development of melting texture. Physiol Plant 126: 163–174. [Google Scholar]
- 30. Atkinson RG, Schroder R, Hallett IC, Cohen D, MacRae EA (2002) Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol 129: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boudart G, Charpentier M, Lafitte C, Martinez Y, Jauneau A, et al. (2003) Elicitor activity of a fungal endopolygalacturonase in tobacco requires a functional catalytic site and cell wall localization. Plant Physiol 131(1): 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 452: 991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 34. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Bio 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 35. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4): 611–622. [DOI] [PubMed] [Google Scholar]
- 36. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 (9): 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thompson JD, Higgis DG, Gibson TJ (1994) CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langfelder P, Horvath S (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bio 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: 17. [DOI] [PubMed] [Google Scholar]
- 40. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11): 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hames BD, Rickwood D (1990) Gel electrophoresis of proteins – A practical approach. 2nd ed. IRL Press, Oxford University Press. 372p.
- 42. Nogueira SB, Labate CA, Gozzo FC, Pilau EJ, Lajolo FM, et al. (2012) Proteomic analysis of papaya fruit ripening using 2DE-DIGE. J Proteomics 75(4): 1428–1439. [DOI] [PubMed] [Google Scholar]
- 43. Nascimento JRO, Cordenunsi BR, Lajolo FM (1997) Purification and partial characterization of sucrose-phosphate synthase from pre-climacteric and climacteric bananas (Musa acuminata). J Agri Food Chem 45: 1103–1107. [Google Scholar]
- 44. Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, et al. (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5: 2497–2507. [DOI] [PubMed] [Google Scholar]
- 45. Zhong S, Lin Z, Fray RG, Grierson D (2008) Improved plant transformation vectors for fluorescent protein tagging. Transgenic Res 17(5): 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spolaore S, Trainotti L, Casadoro G (2001) A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J Exp Bot 52(357): 845–850. [DOI] [PubMed] [Google Scholar]
- 47. Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33(5): 949–956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
These are the legends for Supporting Tables / Figures presented in File S1. Table S1. Cell wall-related genes from ripe papaya and mature A. thaliana plant. Table S2. Similarity percentage of amino acid from papaya and other plants PGs. Table S3. Nucleotide sequences used in PCR reactions. Table S4. Nucleotide sequences used in qPCR. Table S5. Calibration curves for relative gene expression. Table S6. Calibration curves for absolute gene expression. Figure S1. Up-regulation of cell wall-related genes during papaya ripening. Real-time PCR (qPCR) was used to determine the absolute quantitation of the mRNA levels of various genes during papaya ripening. The quantification is represented by the column height. The error bars on each column indicate the SD from four technical replicates from samplings I and II. The different letters represent samples that were significantly different from those collected on other days post-harvest (within the same gene) as determined by one-way ANOVA and Tukey's test (α<0.05, n = 4). Figure B shows the threshold cycle values (Ct) for the two genes used as internal controls (actin gene – cpACT and elongation factor 1-alpha gene - cp_EF1). Figure S2. Genomic and mRNA organization of different PGs from papaya fruit. Grey boxes represent coding regions (exons), while black lines represent non-coding regions (introns). White boxes represent the mRNA sequences concatenated from the above compared exons. Figure S3. Unrooted phylogram encompassing PGs from papaya, Arabidopsis and several other plant organisms. A phylogenetic tree was calculated using the neighbor-joining method based on the ClustalW alignment of the deduced amino acid sequences. The putative signal peptide from all of the proteins was removed from the sequence. The following proteins and their corresponding GenBank IDs were used: A. thaliana 1, 2 and 3 (NP_191544, NP_191310, NP_187454), P. persica 1 and 2 (AAC64184, CAA54448), P. communis 1 and 2 (CAH18935, BAC22688), D. carota (BAC87792), S. lycopersicum 1 and 2 (ABW38780; AC28947), C. melo 1 and 2 (AAC26510; AAC26512), V. vinifera 1 and 2 (XP_002263164, XP_002282759), B. napus (CAA65072), G. Max 1 and 2 (ABC70314, AAL30418), D. kaki (ACJ06506), R. communis 1, 2 and 3 (XP_002529090, XP_002529616, XP_002517823), A. deliciosa (P35336), O. europaea (ACA49228), P. trichocarpa 1, 2 and 3 (XP_002331621, XP_002322711, XP_002313308) and C. papaya 1, 2, 3 and 4 (FJ007644, GQ479791, GQ479794 e GQ479795). The values for the branch lengths are based on the scale bar; there are 0.1 residue substitutions per site. Letters A and B show the two distinct clades resulting from the phylogenetic analyses. Figure S4. Recombinant proteins sequencing. Figures show the query sequences of cpPG1 (A) protein, and the correspondent peptides that were visualized in protein sequencing. Residues highlighted by gray boxes and black letters are from expression vector; residues highlighted by black boxes and white letters are those obtained in protein sequencing. The histidine tags are underlined and asterisks indicate the end of protein. Figure S5. Expression of recombinant proteins from papaya pulp. The figures show representative gels from heterologous expression of cpPG1 (A) gene. Figure A. Proteins profiles from bacteria carrying pET45-cpPG1 before IPTG induction (PG), after 16 hs growth non-induced (PG-NI) and after 16 hs growth IPTG-induced (PG-I); Ni-purified recombinant proteins can be seen after properly elution (E 1 and 2). Proteins markers are highlighted with weight numbers. Figure B. The figure displays the hybridization of antiserum for cpPG1 recombinant protein. The arrow indicates the molecular weight for commercially available protein markers. The quantities of proteins are also indicated. Figure S6. Protein alignment of four distinct papaya PGs. The corresponding proteins from cpPG1 (PG1 - FJ007644), cpPG2 (PG2 - GQ479791), cpPG3 (PG3 - GQ479794) and cpPG4 (PG4 - GQ479795) genes were aligned using ClustalW and viewed with BOXSHADE program and then manually edited. Gaps were introduced to optimize alignment. Identic amino acids are highlighted by black boxes and white letters, similar amino acids are highlighted by grey boxes and white letters and different amino acids are highlighted by white boxes and black letters. The conserved domain of glycosil hydrolases from family 28 (PS00502) is indicated by a dotted-line box. The probable catalytic residues are indicated by asterisks, being possible to observe the cpPG2 protein has a Glu346 instead of a Lys346 as the other three polygalacturonases have.
(ZIP)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.