Abstract
Heparan sulfate (HS) is a polysaccharide modified with sulfation, acetylation, and epimerization that enable its binding with protein ligands and regulation of important biological processes. Tandem mass spectrometry has been employed to sequence linear biomolecules e.g., proteins and peptides. However, its application in structural characterization of HS is limited due to the neutral loss of sulfate (SO3) during collisional induced dissociation (CID). In this report, we studied the dissociation patterns of HS disaccharides and demonstrate that the N-sulfate (N–S) bond is especially facile during CID. We identified factors that influence the propensities of such losses from precursor ions and proposed a Free Proton Index (FPI) to help select ions that are able to produce meaningful backbone dissociations. We then investigated the thermodynamics and kinetics of SO3 loss from sulfates that are protonated, deprotonated, and metal-adducted using density functional theory computations. The calculations showed that sulfate loss from a protonated site was much more facile than that from a deprotonated or metal-adducted site. Further, the loss of SO3 from N-sulfate was energetically favored by 3–8 kcal/mol in transition states relative to O-sulfates, making it more prone to this process by a substantial factor. In order to reduce the FPI, representing the number of labile sulfates in HS native chains and oligosaccharides, we developed a series of chemical modifications to selectively replace the N-sulfates of the glucosamine with deuterated acetyl group. These modifications effectively reduced the sulfate density on the HS oligosaccharides and generated considerably more backbone dissociation using online LC/tandem MS.
Keywords: Glycan, Glycosaminoglycan, Heparan sulfate, Sulfate, Electrospray, Tandem MS, Glycomics
Introduction
Heparan sulfate (HS) is a long linear polysaccharide attached to the core proteins of proteoglycans. Together with chondroitin/dermatan sulfate (CS/DS), another member of glycosaminoglycan family, HS is highly expressed in proteoglycans on the cell surface and in the extra-cellular matrices. Proteoglycans, often associated with hyaluronan, constitute a heavily hydrated, gel-like medium in extracellular matrices, and help resist compressive forces in animal tissues [1, 2]. However, the more intriguing function of proteoglycans is their ability to bind a variety of protein ligands, including growth factors, growth factor receptors, morphogens, cytokines, chemokines, and others [3–5]. Through these binding interactions, for which HS is largely responsible, proteoglycans play important roles in cell proliferation, development, differentiation, and migration [4, 6–8]. Consequently, these functions of HS have great implications in cancer biology, and a number of studies have aimed to investigate HS and its roles in tumorigenesis [9–12].
The binding of polyanionic HS chains may cause conformational changes to proteins [13], to recruit multiple protein partners to one site [14], or to sequester one protein from another [15]. All these mechanisms depend on the binding affinities and expression levels of specific HS structure(s) and/or overall chain properties [16]. HS consists of repeating units of −4-GlcA-β/IdoA-α-1,4-GlcNAc(NS)-α-and its length can extend from 15 to 100 of these disaccharide blocks. The structural variety of HS comes from (1) the incomplete N-deacetylation and N-sulfation of the glucosamine, which result in N-sulfation, N-acetylation, and in rare cases, free -NH2 at this saccharide; (2) the sulfations at 2-OH of the glucuronic acid or iduronic acid and at 6-OH (and less frequently, 3-OH) of the glucosamine, and (3) the epimerization at C5 of GlcA to IdoA. These modifications create over 20 possible structural variants of the disaccharide, making the entire chain enormously polydisperse. Although more than a dozen enzymes have been identified to be involved, there is no apparent template in the biosynthetic pathway of HS. Therefore, the structural heterogeneity of HS from tissue to tissue, cell type to cell type, and possibly between same proteoglycans, stands as the major obstacle to correlate HS structure with biological function and to develop HS-based therapeutics.
There have been a number of studies and hypotheses that suggest HS chains are organized in domains characterized by the substitution groups on the amino of GlcN unit and the overall sulfation degrees [17–19]. There are NS domains, in which GlcN units are all N-sulfated, NA domains in which GlcN units are all N-acetylated, and hybrid NA/NA domains, with alternate N-sulfation and N-acetylation. Among them, NS domains are most heavily sulfated, and have high abundance of IdoA units. It is generally considered that the NS domains are the most likely binding sites on the HS chains due to their abundance of sulfate anions that can interact with basic residues on their binding proteins [20–23]. Therefore, with the inability to study the structure of the entire HS chains using current available technologies, the structural characterization of NS domains, typically generated by HS polysaccharide lyase digestion, produces the most relevant knowledge on HS–protein interactions.
Mass spectrometry (MS), especially tandem MS with different dissociation methods, has achieved great successes in proteomics [24, 25], glycoproteomics [26], and glycomics [27] during the past 15 years. Proteins can be sequenced with the aid of genomic databases. Phosphorylation sites, glycosylation sites, and other post-translational modifications can also be identified with tandem MS [28–30]. O- Linked and N-linked glycans have also been structurally identified using multi-stage mass spectrometry [31, 32] and emerging dissociation modes in recent years [33–35]. It was realized, however, from very early MS experiments of HS oligosaccharides and disaccharides, that the neutral sulfate loss is extremely prevalent during ionization and collision induced dissociation (CID) [36–39]. The loss of sulfates, even to a moderate extent, will diminish the structural information gathered from the fragments and prevent the deduction of the structure of the original precursor ions.
Previously, Huang et al. chemically modified the -OH and -SO3H in chondroitin sulfate (CS) oligosaccharides with permethylation, desulfation, and peracetylation [40]. In so doing, they eliminated sulfates while retained all the positional information of sulfates in the original oligosaccharide. After the modification, CS chains were converted to neutral sugars and were analyzed with reverse phase chromatography. Similar efforts were also carried out for HS disaccharides [41]. While they provide valuable alternate routes to circumvent the sulfate loss of native GAG anions, the complexity of the chemistry and presence of side reactions drive the need for complementary chemistry.
In this work, we aimed to understand the mechanisms by which neutral losses of sulfates occur, and explored other chemical modifications and mass spectrometric techniques to generate high quality tandem mass spectra of HS negative ions. Via tandem mass spectrometric experiments in CID mode and computational studies, we compared the energetics of sulfate loss in protonated, deprotonated, and metal-adducted sites. The results showed that the N-sulfate is more fragile than O-sulfate groups and allowed us to gain insight into the energetic barrier that the sulfate loss process must overcome in the gas phase. We propose a Free Proton Index (FPI), to account for the fact that the degree of backbone cleavage in CID of HS oligosaccharide ions is largely determined by the density of free and mobile protons. We followed by adopting chemical modifications to selectively substitute N-sulfate with N-acetate-d3. When coupled with charge state manipulation during electrospray ionization, this modification effectively reduces the number of sulfates, one important source of free protons and contributor to the FPI value. These combined strategies generate considerably more abundant ions from backbone dissociation, including glycosidic bond and cross-ring cleavages, for the majority of the HS oligosaccharides we studied.
Experimental
Materials
Heparan sulfate from porcine intestinal mucosa was purchased from Sigma-Aldrich (St. Louis, MO, USA). HS hexasaccharide (in degree of polymerization, dp6) and octasaccharide (dp8) were prepared as previously reported [23]. Pyridine, Amberlite 120H+ ion exchange resin, deuterated acetic anhydride, propionic anhydride, and DMSO were from Sigma-Aldrich.
HS Disaccharides and Oligosaccharides Nomenclature
HS disaccharide nomenclature follows the convention proposed by Lawrence and Esko et al. [42]. For example, D2S6 corresponds to ΔHexA2S-GlcNS6S. HS oligosaccharide composition follows the coding system that we created previously with five digits in a bracket [v,w,x,y, z], with each digit referring to the number of ΔHexA, HexA, GlcN, Ac, and SO3 in the molecule, respectively. Arixtra was purchased from Sanofi-Synthelabo (West Orange, NJ, USA).
Chemical Modification HS Oligosaccharides
Heparan sulfate oligosaccharides (2 to 50 μg) were dissolved in 0.5 mL saturated sodium bicarbonate (with some suspension of the solid) and 100 μL methanol at 0 °C, followed by addition of 100 μL of propionic anhydride. The mixture was stirred vigorously on ice. The caps of the reaction tubes were punctured with a needle to release the CO2 generated in the reaction. The pH of the reaction mixture was checked periodically, and a 150 μL volume of saturated NaHCO3 slurry was added every 45 min to maintain the pH at approximately 8. After 2.5 h, the solution was added to water to bring the total volume to 2.5 mL and passed through PD-10 columns (GE Healthcare Life Sciences, Piscataway, NJ, USA), and the products were collected in 3.5 mL water. A two-μL volume of each eluate solution was dried and profiled using HILIC LC/MS (described below). The rest of the solutions were loaded onto a H+ exchange columns which were prepared with about 7 mL of Amberlite 120H+ ion exchange resin packed tightly into a BioRad 10 mL empty column and washed excessively with double distilled water. After loading, 5 mL water was used to elute the acid forms of HS oligosaccharides. All the eluents, including flow-through, were collected immediately, and 70 μL of a 1:100 water:pyridine solution was added. The solutions were lyophilized to dryness. The pyridinium salts of heparin lyase generated HS oligosaccharides were dissolved in 1 mL DMSO/MeOH (9:1) and heated at 55 °C for 2 h. The synthetic Arixtra pentasaccharide was heated at 45 °C, at which temperature excessive sulfate loss was minimized. The de-N-sulfated oligosaccharides were then passed through a PD-10 column or G-10 column and eluted with water in order to remove DMSO. A small fraction of the solution was taken for HILIC LC-MS profiling. The rest aqueous solutions were dried in vacuum. The de-N-sulfated HS oligosaccharides were dissolved in 0.5 mL saturated sodium bicarbonate and re-acetylated using acetic anhdyrde-d6 using the identical procedure as the propionylation in the first step. After the reaction was complete, the mixture was diluted with water to a total volume of 2.5 mL and passed through a PD-10 or G-10 column. The re-N-acetylated HS oligosaccharides in aqueous solutions were dried in vacuum.
Mass Spectrometric Analyses
The tandem mass spectrometric experiments of HS disaccharides were performed using an Applied Biosystems/ SCIEX QSTAR Pulsar quadrupole time-of-flight mass spectrometer (Framingham, MA) in enhanced mode. The samples (50 pmol/μL in 1:1 water:acetonitrile) were directly infused through a Turbo-IonSpray interface at 15 μL/min with the nebulizer gas was set at 40, turbo gas at zero, curtain gas at 25 and ionization voltage at −3500 V. Different charge states and sodium adducts were selected at Q1 and the CID was carried out in Q2 with incrementally increasing collision energy (2 or 4 V).
For MS3 experiments, HS disaccharides standards (10 pmol/μL, 1:1 water:acetonitrile) were directly infused to a Bruker amaZon ion trap mass spectrometer (Bremen, Germany) at a flow rate of 200 μL/h. The capillary voltage was set at 5000 V, end plate offset at −500 V, temperature at 150 °C, nebulizer gas at 2 psi, and drying gas at 2 L/min.
HILIC-MS and -MS/MS experiments were conducted using an Agilent 6520 quadrupole time-of-flight mass spectrometer (Santa Clara, CA, USA), equipped with a chip cube interface system and make-up-flow setup that coupled the Agilent 1200 HPLC system with the Q-TOF. The dimensions of amide-80 chip, including the pulsed makeup-flow chip, LC conditions, and pulsing conditions were as previously reported [43]. The collision energies were empirically determined according to the charge state and size of the precursor ion. Typically, 26 V was applied to 2– ions, 17 for 3– ions, 13 for to 4– ions, and 8 or 10 V for 5– ions.
Computational Methods
The structural optimizations and thermodynamic properties were computed at the Scientific Computation Facility at Boston University using Gaussian 03 software suite [44]. The geometric optimizations were performed using a B3LYP/6-31g* basis set. The frequency analysis was also carried out at the same level of theory and basis set to ensure energy minima. The global minimums were surveyed by considering possible intermolecular hydrogen bonding between –OH, –COOH, and –SO3H and nearby hydroxyl groups or ring oxygen atoms. The single point energy was calculated at B3LYP/6-311+g** basis set. The enthalpies were scaled with a factor of 0.9804 for thermal corrections at B3LYP/6-31g* basis set [45]. Transition states were calculated by QST2 function in Gaussian and confirmed by frequency analysis. Thermal corrections at different temperatures (from 298 K up to 1000 K) were conducted for some reactions. The reaction rate constants of the unimolecular sulfate loss were calculated according the Erying-Polanyi equation under a series of different temperatures. Computational details, including coordinates, zero-point energies, thermal corrections, and frequencies are given in the Supplemental Information section.
Results and Discussion
Charge States and Cation Adduction Affect CID of HS Negative Ions
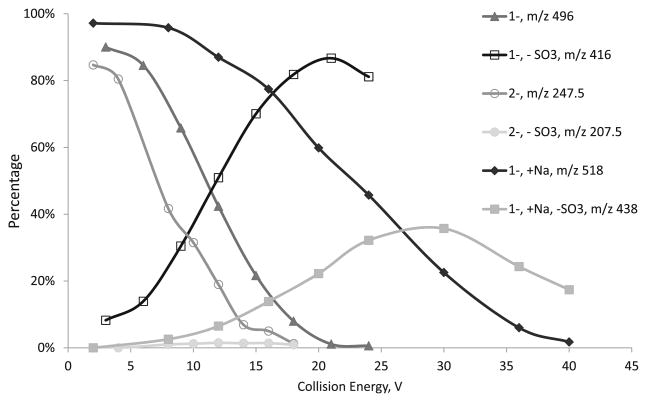
HS disaccharide D0S6 has one 6-O-sulfate at GlcNS and one N-sulfate and electrospray ionization of D0S6 produces four ionic species, including m/z 4961–([M – H]−),247.52–([M – 2H]2–), 4161– (loss of a neutral sulfate in or post source), and 5181– (sodium adduct). Precursor ions of m/z 4961–, 247.52–, and 5181– were selected to undergo CID in Q2. The relative abundances of the precursor ions and major product ions in percentages were plotted with regard to collision energy voltage. From Figure 1, it is apparent that the loss of sulfate from [M – H] − ion is extremely facile, as the diminishment of the product ion 4961– almost mirrors the increase of the product ion 4161–. Collision energy of 11 V was required in order for half of the precursor ions to lose one sulfate. For the [M – 2H]2– ion, the loss of sulfate was never significant even though overall m/z 247.52– is more fragile, as it takes 7.5 V to break 50 % of the precursor ion. The sodium adduct [M – 2H + Na]- ion, 5181–, appears to be more stable than both 4961– and 247.52–. It needs about 23 V to dissociate 50 % of the sodium adduct ions, while the relative abundance of the sulfate loss product, 4381–, did not peak until about 30 V. Meanwhile, the majority of the dissociations from 5181– are backbone cleavages. These behaviors appear to be similar for the isomer of D0S6, D2S0 (Figure S1).
Figure 1.
A plot of the relative abundances the precursor ions of D0S6, in 1–, 2–, and sodiated forms, and their product ions from sulfate loss versus the collision energy
These results suggest that the charge state and cation-adduction determine the CID behavior of HS negative ions, specifically, the propensity of sulfate loss, the abundances of backbone cleavages, and the energy needed to dissociate the precursor ions. Sulfate loss is a dominant process for the 1–charge state of doubly sulfated disaccharides, while such losses are insignificant for the 2– ions. The sodium adduct behaves somewhere in between, where sulfate loss is one of the major dissociation pathways along with backbone cleavage. In previous studies, we demonstrated the tandem mass spectrometry of synthetic heparin saccharides were influenced largely by the charge states of the precursor ions and the adduction of calcium or sodium cations, which stabilize the sulfate groups and produce more abundant backbone cleavage [46, 47]. Similar effects were also observed by Wolff et al. in electron detachment dissociations (EDD) experiments [48].
N-Sulfate is Especially Fragile During CID
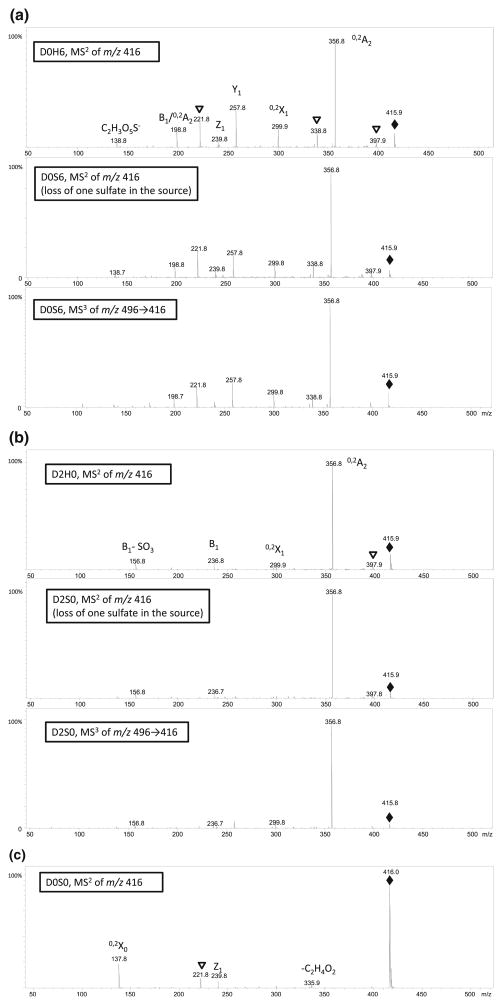
In order to reveal which sulfate was lost during CID in the experiments above, we performed multistage tandem mass spectrometric analysis using an ion trap instrument. We first compared tandem mass spectra of D0H6 with MS3 of D0S6→[M-SO3] produced by in-source dissociation and MS3 of D0S6→[M-SO3] produced by CID in the ion trap (Figure 2a). An analogous experiment for MS2 of D2H0, MS3 of D2S0→[M-SO3] generated in-source, and MS3 of D2S0→[M-SO3] generated by CID in the ion trap (Figure 2b) were then recorded. The characteristic product ions of these three species and their relationship with D2S0 and D0S6 are summarized in Table 1. Between isomers D2H0 and D0H6, under the same collision energy (amplitude=0.39), both their [M – H] − ions produce 0,2A2 (m/z 357) as the most abundant product ion. D2H0 also generates B1 (m/z 237) and B1-SO3 (m/z 139) as its characteristic ions, while D0H6 produces Y1 (m/z 258), Z1 (m/z 240), B1/ 0,2A2 (m/z 199), and C2H3O5S− (m/z 139, a 0,4-cleavage at GlcNH2). Under the amplitude, however, D0S0 appears to fragment much less (Figure 2c), as its [M – H] − remains the most abundant product ion, together with less abundant but distinct 0,2X ion (m/z 138), which contains the N-sulfate at the reducing end, and [M – H] −-C2H4O2 (m/z 336, a 0,4-cleavage at GlcNS).
Figure 2.
(a) Comparison of MS2 spectra of D0H6 and MS2 and MS3 spectra of D0S6. (b) Comparison of MS2 spectra of D0H6 and MS2, and MS3 spectra of D2S0. (c) MS2 spectra of D0S0. Diamonds denote precursor ions, triangles denote the loss of water ions from the immediate adjacent ions
Table 1.
Comparison of MS2 of Isomeric Disaccharides D2H0 and D0H6, and MS3 of Isomeric Disaccharides D2S0 and D2S6
| Precursor ion | N-S loss | O-S loss | MS3 Ions observed | ||
|---|---|---|---|---|---|
|
| |||||
| Product | Characteristic fragmentation | Product | Characteristic fragmentation | ||
| D2S0 | D2H0 | B1 (m/z 237) | D0S0 | 0,2X0 (m/z 138) | B1 (m/z 237) |
| B1-SO3(m/z 157) | M-C2H4O2 (m/z 336) | B1-SO3(m/z 157) | |||
| D0S6 | D0H6 | Y1 (m/z 258) | D0S0 | 0,2X0 (m/z 138) | Y1 (m/z 258) |
| Z1 (m/z 240) | M-C2H4O2 (m/z 336) | Z1 (m/z 240) | |||
| B1/ 0,2A2 (m/z199) | B1/ 0,2A2 (m/z199) | ||||
| C2H3O5S− (m/z139) | C2H3O5S− (m/z 139) | ||||
The MS3 profile of D0S6 m/z 496→416 (Figure 2a middle panel), together with MS2 spectrum of m/z 416 (from in- or post-source fragmentation, Figure 2a bottom panel) produces very similar spectra as MS2 of D0H6 (Figure 2a top panel). Meanwhile, both MS3 spectrum of D2S0 m/z 496→416 (Figure 2b middle panel) and MS2 spectrum of m/z 416 (from in or post-source fragmentation, Figure 2b bottom panel) very much resemble the MS2 spectrum of D2H0 (Figure 2b top panel). It is worth to note that the characteristic ion of N-sulfated D0S0, 0,2X0 (m/z 138), was not observed in multi-stage mass spectra of either D0S6 or D2S0. Therefore, these results indicate that the vast majority of sulfate loss in these experiments was preferentially to N-sulfate over 6-O or 2-O sulfate.
Computational Studies Reveal that Loss of N-Sulfate is Energetically More Accessible
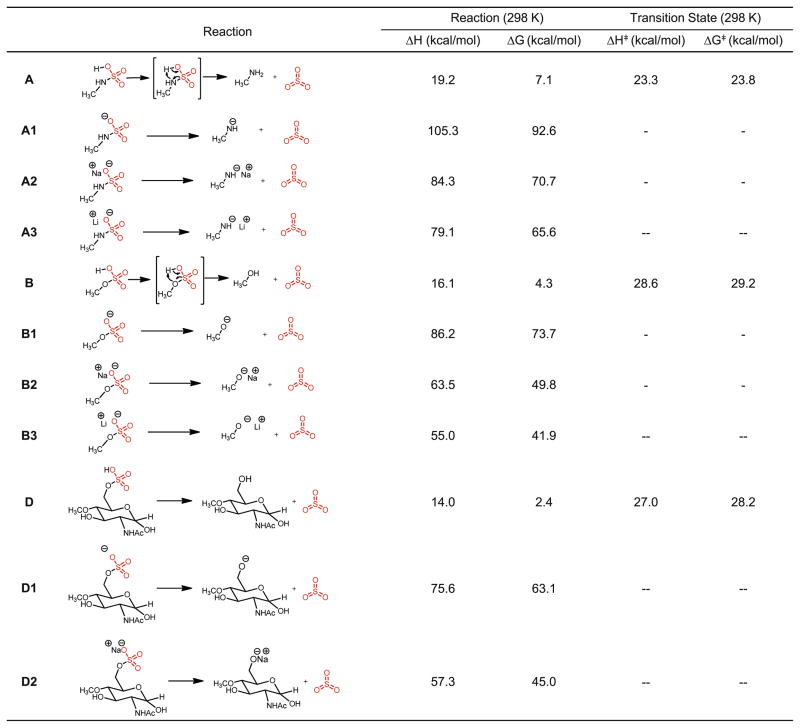
A series of HS model compounds and in silico reactions of sulfate loss in the gas phase were designed. Their reaction enthalpies, free energy changes, and transition state barriers were computed by density functional theory using Gaussian 03 software package. Because of the large sizes of the disaccharides and oligosaccharides and the existence of a heavy atom, sulfur, both of which demand substantial computational resources, monosaccharides were used instead, with the 1– or 4– position replaced with a methoxy group as the surrogates of other parts of the sugar chain.
In order to rationalize the observations in Figure 2, the simplest model of sulfate-containing compound CH3OSO3H and CH3NHSO3H and a monosaccharide GlcNAc with a 6-O sulfate and 4-O methoxy group were studied with regard to sulfate loss as neutral molecules, negative ions, and metal adducts (sodium and lithium). The reactions are depicted in Figure 3, with the calculated reaction enthalpy, free energy, and transition state energy barrier in enthalpy and free energy are also listed. From Figure 3, it can be seen that at 298K, both N-sulfate and O-sulfate loss in their protonated forms are endothermic by 16–19 kcal/mol. The transition states of these processes proceed through a 4-member ring, with the proton shifting from the sulfate oxygen to the hydroxyl oxygen and the elongation of the S–O bond. The transition state barriers, both ΔH⧧ and ΔG⧧, vary between 23 and 29 kcal/mol, a range quite accessible in typical low energy CID experiments. In contrast, both the N-sulfate loss and O-sulfate loss from the sulfate anion, sodium, or lithium adducts are much more endothermic, ranging from 55 to 105 kcal/mol. Furthermore, no single transition state can be located by QST2 in Gaussian 03 from deprotonated or metal-adducted sulfate to lose a neutral sulfate. A modredundant calculation was performed for reaction A1 in Figure 3 with the increasing S–O bond length, and it exhibits a monotonic energy increase when the S–O bond was elongated at fixed values until infinity (Figure S2). These results demonstrate not only the sulfate losses from deprotonated or metal-adducted ions are energetically disfavored by a wide margin compared with those of protonated sulfates, but also suggest that these reactions proceed, if they do occur, through other intermediates or transition states that are probably not easily accessible.
Figure 3.
SO3 loss from protonated, deprotonated sulfate sites, sodium adducts and lithium adducts of HS model compounds; “−” denotes transition state was not located
We continued to investigate the energetic differences between N-sulfate and O-sulfate loss. The generic reactions of sulfate loss of CH3OSO3H (ΔH=19.2 kcal/mol at 298 K) and CH3NHSO3H (ΔH=16.1 kcal/mol at 298 K) is moderately endothermic, with O-sulfate loss slightly less favored. The free energies of the transition state of sulfate loss, ΔG⧧, which determine the kinetic rate constants of sulfate loss processes, was calculated to be 23.8 kcal/mol for CH3NHSO3H and 29.2 kcal/mol for CH3OSO3H. From the Erying-Polanyi equation, the unimolecular reaction constants of sulfate loss can be calculated from ΔG⧧ at a specific temperature,
where kB is the Boltzmann constant, h is the Plank constant, R is the gas constant, and ΔG⧧ is the free energy barrier of the transition state. There is one variable in the equation, the temperature T at which these dissociations occur, that is unknown. We adopted Cooks’ “effective temperature” concept [49] and calculated a series of rate constants under different temperatures from 298 K to 1000 K, supposing somewhere in between represents a typical CID reaction in a mass spectrometer [50, 51]. A list of the ratios of N-sulfate loss and O-sulfate loss rate constants between reaction A and B is shown in Table 2. The ratio of the two rate constants decreases from 9447 at 298 K to 5.7 at 1000 K, with N-sulfate loss being the more facile and faster reaction.
Table 2.
The Calculated Rate Constants and Ratios for N- and O-sulfate Loss in Reaction A, B, C, and D at Different Temperatures
| Calculated rate constant
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 298 K | 350 K | 400 K | 500 K | 600 K | 700 K | 800 K | 900 K | 1000 K | |
| kA (NS) | 2.17×10−5 | 6.97×10−3 | 4.43×10−1 | 1.53×102 | 7.79×103 | 1.32×105 | 1.12×106 | 6.00×106 | 2.32×107 |
| kB (OS) | 2.30×10−9 | 3.91×10−6 | 8.17×10−4 | 1.45×100 | 2.21×102 | 7.23×103 | 9.97×104 | 7.44×105 | 4.04×106 |
| kA/kB | 9447 | 1785 | 542 | 105 | 35 | 18 | 11.2 | 8.06 | 5.73 |
| KE (NS) | 2.88×10−6 | 1.59×10−3 | 1.47×10−1 | 8.49×101 | 5.91×103 | 1.23×105 | 1.19×106 | 6.96×106 | 2.85×107 |
| KF (OS) | 1.46×10−8 | 1.54×10−5 | 2.26×10−3 | 2.49×101 | 2.70×1012 | 7.74×103 | 9.66×104 | 6.92×105 | 3.36×106 |
| KE/kF | 197 | 103 | 65 | 34.1 | 21.9 | 15.8 | 12.3 | 10.1 | 8.5 |
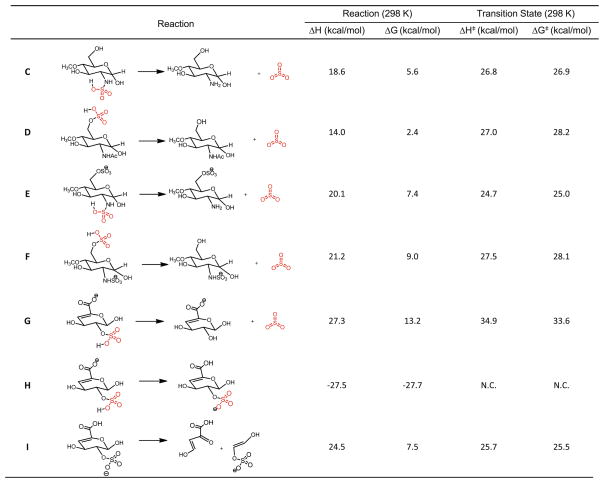
As a more relevant example, we compared reactions E and F (Figure 4), which constitute a pair of isomers with two reaction pathways, leading to N-sulfate loss and 6-O-sulfate loss, respectively. These two pathways, like other reactions A and B, and C and D, also feature more facile and faster N-sulfate loss, with the reaction rate constant ratio (N-S/O-S) equal to 197 at 298 K to 8.5 at 1000 K (Table 2). The lifetimes of these species that undergo sulfate loss as a first order reaction are on the order of milliseconds when the temperature is elevated to about 600 K for most of processes, while at room temperature, the reactions do not occur at meaningful rates. This is consistent with the millisecond time scale of typical CID experiments in mass spectrometers.
Figure 4.
N-sulfate loss versus O-sulfate loss for HS model compounds. “N.C.” denotes not calculated because of the proton scrambling
It should be noted that in reaction E and F the anion at N-sulfate, which will produce O-sulfate loss, is slightly more stable than the anion at 6-O-sulfate, which leads to the N-sulfate loss. These energetic differences between the starting structures, although minor, could change the landscape of the overall kinetics. Since in both ion traps and collision cells, ions subjected to CID undergo multiple collision events and proton scrambling is considered very facile and efficient, we could assume there is a fast equilibrium between two forms of ions as starting structures in reaction E and F. With the energies we have obtained, a reaction system can be constructed, as depicted in Figure S4. Using chemical kinetics simulation software KinFitSim, the final product ratios of N-sulfate and O-sulfate loss are obtained at different temperatures. The ratio of the N- and O-sulfate is simulated to be 1.14 at 600 K and 2.30 at 1000 K. Therefore, with the complication of the isomerization, N-sulfate loss is favored even though for this particular model compound in which the starting structure leading to N-sulfate loss is energetically less favored.
Another question to answer is the relative gas phase acidity of the carboxylic acid and sulfates in HS anions. This will determine which site of the oligosaccharide ions will be deprotonated. For proton scrambling reaction H in Figure 4, we calculated that the anion site at the 2-O sulfate is 27.5 kcal/mol lower than that at the carboxylic acid. This energy difference will likely make the anion exclusively a sulfate ion prior to CID. This energy is also significant enough that the scrambling of proton from carboxylic acid followed by sulfate loss does not occur in favor of other dissociation modes such as backbone cleavages. This is also demonstrated by tandem mass spectrometric experiments of D0A6 and D0S0, two singly sulfated HS dp2s, as the sulfate loss was never an important dissociation channel even under high collision energies (shown in Figure S3).
In order to gain some insight of the energetic barrier for sulfate loss compared with the glycan backbone dissociations (the glycosidic bond cleavages and cross-ring cleavages), one generic reaction was examined. The precursor ion in Reaction I of Figure 4, which possesses a 4–5 unsaturated bond, is present in the non-reducing end of all disaccharides and oligosaccharides generated by heparin lyase digestion. Ions of the type 0,2X are frequently observed to occur from 4–5-unsaturated HexA residues, and the mechanism has been described as a retro Diels-Alder reaction. Reaction I is calculated to have a reaction enthalpy ΔH=24.5 kcal/mol, while the ΔG⧧ equal to 25.5 kcal/ mol at 298 K. This energy barrier appears to be slightly lower than all transition state barriers for those for O-sulfate loss, and higher than two of the three of N-sulfate losses in Figure 4. The computational results suggest that retro-Diels-Alder reaction is in indeed very facile and is likely to be the mechanism for the 0,2X cross-ring cleavage, at 4–5-unsaturated HexA.
Overall, the results from thermodynamics calculations of model compounds indicate that the sulfate loss for both N-sulfate and O-sulfate have transition state barriers between 23 and 35 kcal/mol. In all cases, O-sulfate loss, compared with N-sulfate loss, has a higher transition state barrier and, hence, lower reaction rate constant. This explains why almost exclusively N-sulfate loss was observed in the previous experiments. Moreover, the retro-Diels-Alder reaction has a transition state that is slightly lower than that of a typical O-sulfate loss. Therefore, this type of cross-ring cleavage competes with sulfate loss as a reaction pathway for HS oligosaccharide ions during CID. For larger oligosaccharides ions, which may carry multiple charges, the rupture of the backbone bonds, including glycosidic bonds, will be assisted by electrostatic strains between the charges, making backbone cleavages even more favorable.
It has become clear that from both experiments and computational studies that protonated sulfate is most prone to the neutral loss. Both deprotonation and metal cation adduction displace the proton and prevent or decrease the tendency of sulfate loss. It is likely that acidic proton density, as a function of the number of sulfates, charge state, and metal adduction, dictates the degree of sulfate loss, and on the other hand, the relative abundances of backbone cleavage of HS negative ions. Since protons can scramble from the carboxylic groups to the deprotonated sulfates, it is also useful to include the contribution of carboxylic groups. We therefore propose the FPI, for the precursor ions for HS oligosaccharides, as well as other GAG precursor anions, to characterize and predict their tandem mass spectrometric behavior. The FPI is calculated by
In this definition, the sum of all acid protons from both sulfates (O- and N-) and carboxylic acids was subtracted by the charge state and the adducted metal ions, where valence is also considered. The net number of protons was then normalized by the total number of monosaccharide residues. The value of decreasing FPI is expected to be associated with increasing abundances of backbone cleavages and quality of CID tandem mass spectra of HS negative ions. We have previously evaluated the value of metal cationization and found it to be limited for online LC-MS/MS analyses [43]. Therefore, in order to reduce FPI and produce more backbone cleavages and obtain more structural information, the charge state will need to be higher and it will be helpful to selectively modify the N-sulfate, the most fragile site.
De-N-Sulfation and N-Acetylation of HS dp6 and dp8
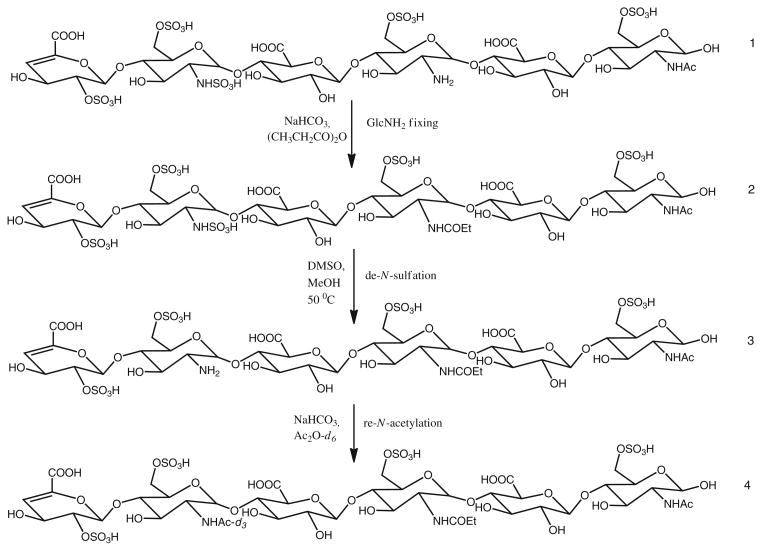
Heparan sulfate from porcine intestinal mucosa was digested with heparin lyase III to produce oligosaccharides that contain NS domains. These oligosaccharides were fractionated by size exclusion chromatography to generate fractions of oligosaccharides at different sizes, nominally dp6, dp8, dp10, and etc. Each fraction consists of a mixture of oligosaccharides of similar sizes but with different compositions. We selected two fractions, dp6 and dp8 oligosaccharides, as they mimic the interacting domains of HS with their protein binding partners [23, 52]. We used HILIC-MS to profile the compositions of dp6 and dp8, before and after each step of chemical modification (species 1, 2, 3, and 4 in Figure 5), so that the completeness and possible byproducts of these reactions modified from previous reports could be monitored [53, 54].
Figure 5.
Chemical modification schemes
As shown in Figure S5, the HS dp6 fraction consisted of hexasaccharides with 1 acetate and 3 to 6 sulfates, while HS dp8 fraction consists octasaccharides with both 1 or 2 acetates and 3 to 8 sulfates. Since there is a possibility of free glucosamine present in HS chains, to verify if the two remaining GlcN residues in the oligosaccharides contain N-sulfate or free GlcNH2, we first used propionic anhydride to fix any possible free glucosamine within the oligosaccharides. The condition of propionylation was tested with HS disaccharides D2H0 and D0H6, and the reactions were complete (a +56 D mass shift) and presented no side reactions. Both dp6 and dp8, before and after propionylation, exhibited identical HILIC LC/MS profiles, with no +56 D mass shift (+ m/z 28 shift in 2– charge state) found. The results indicate that there is no observable free glucosamine present for the dp6 and dp8 samples we studied, and if the free glucosamine did occur, it would be propionylated and its positional information would be retained.
Following propionylation, dp6 and dp8 were de-sulfated selectively at the NS positions. After this reaction, the newly generated free glucosamine was acetylated with acetic anhydride-d6. The HILIC LC/MS profiles showed very limited changes of the relative abundances of each composition, as summarized in Figures S5 and S6. The total yields of the entire reaction sequence are equal to or greater than 80 % when over 20 μg of starting material were used in this study. It should be noted that NS domains only constitute a small portion of typical HS chains. There are also multiple compositions and isomers for each size fraction of NS domains, which further spread mass spectrometric signals.
Charge Distribution and Tandem Mass Spectra Before and After Chemical Modification
The purpose of the chemical modification is to replace the most labile N-sulfate group with –COCD3, so that the overall sulfation degree of these HS oligosaccharides is decreased, and active protons also decreased, to the degree that precursor ions of these oligosaccharides experience minimal sulfate loss. As shown in Figure S7A, before modification, HS dp8 exhibits 2–, 3–, 4–, and some low abundances of 5– charge states. After the chemical modification, shown in Figure S7B, the overall charge states display a general decrease in absolute value. This is because, with the substitution of 2 or 3 N-sulfate groups with-COCD3, fewer sulfates entailed fewer charges during ionization. As a result, the decrease of the number of sulfates was compensated with lower charge state under our HILIC LC-ESI conditions, which did not effectively decrease FPI.
In our previous studies, we have demonstrated the make-up-flow pulse chip technology enabled the pulsing of charge-enhancing agent such as sulfolane at designated retention time during an LC-MS run. The post-column addition of sulfolane substantially increased the magnitude of the charge states of HS oligosaccharide negative ions and their ionization responses (Figure S7C). The charge state increase and, hence, the decrease of FPI for major compositions in HS dp6 and dp8 before modification, after modification, and after modification with sulfolane pulsing, are summarized in Table S1.
In our tandem mass spectrometric experiments, the lower the FPI of the precursor ions, the less sulfate loss occurred and the more backbone dissociation were observed. When FPI is equal to 0.5, at which all the sulfates are deprotonated, or adducted with metal ions, or in this study, selectively replaced with another functional group, sulfate loss will no longer be a significant dissociation pathway. It does happen, to a small degree, for larger precursor ions, presumably due to the presence of multiple charges bearing extra energy and causing the scrambling of protons from carboxylic acid to protonated sulfate energetically possible. Therefore, we selected FPI=0.5 as a benchmark when selecting precursor ions during targeted LC-MS/MS run. Avoiding ions with higher FPI makes the best use of tandem MS duty cycle and helps obtain the most structurally informative tandem mass spectra.
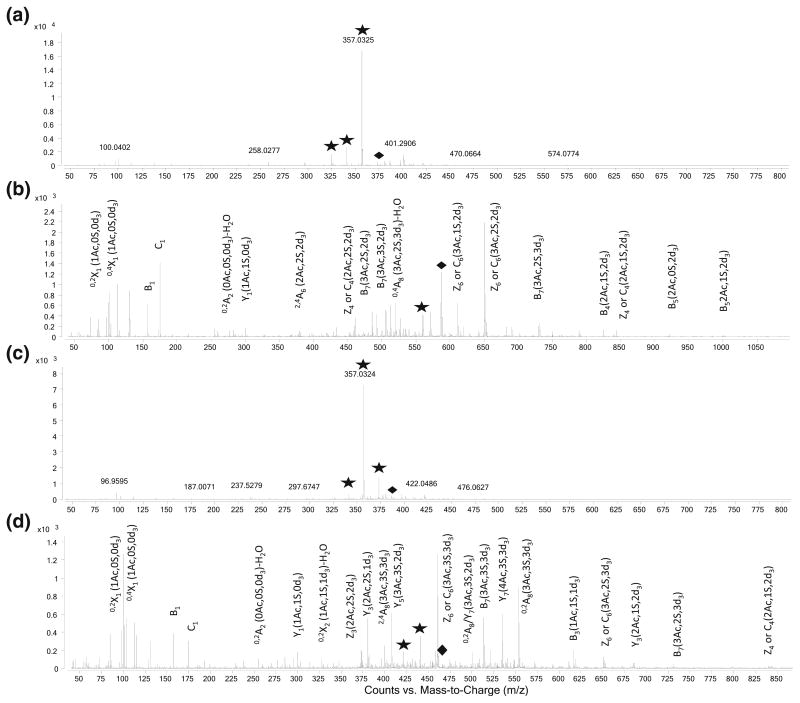
When we combined the chemical modification and pulsed-chip technology, we were able to obtain tandem mass spectra with the majority of the fragments being backbone cleavages, for oligosaccharides that are highly sulfated and thus would not produce such spectra in their native forms. Two sets of spectra are compared in Figure 6. Figure 6a is the tandem spectra of m/z 373, corresponding to [1,3,4,1,6]5–, and with FPI=0.625, whereas 6b is its modified product [1,3,4,4,3]-3d33– with m/z 560.8 and FPI =0.500. Although the native oligosaccharide carries 6 sulfates and was able to produce 5– charge state with sulfolane pulsing, the loss of sulfate is still substantial (59 %). After the modification, which replaced the three N-sulfates with deuterated acetates, the decrease in number of sulfates led to lower FPI and produced predominant backbone cleavage (95 %) The same occurred for dp8 [1,3,4,1,7]5– with m/z 389 and FPI=0.750 (Figure 6c) and its modified product [1,3,4,4,4]-3 d34– at m/z 460 (Figure 6d) and with FPI= 0.500. The major backbone cleavages were labeled according to the Domon-Costello convention followed by the number of sulfates, acetates, and deuterated acetates attached in the parenthesis. In order for these two octasaccharides, with 6 and 7 sulfates, respectively, to reach FPI=0.500 in their native forms, they would need to carry 6 and 7 negative charges. The high charge density in a limited-sized species will have severe charge-repulsion within the ion and cause the rupture of precursor ions during ion transmission and isolation. As a result, very high charge states (≥6) for the oligosaccharides we were studying are not achievable even with pulsing of sulfolane as they elute. This further demonstrates the need to chemically substitute the sulfate groups. A summary of the comparison of FPI and backbone cleavage percentage before and after the chemical modification is listed in Table 3. The percentage of backbone cleavages increased dramatically, in some cases closer to 100 %, after the chemical modification. These phenomena coincide with the decrease of FPI to about 0.5, highlighting the value of this index in selecting precursor ions during LC-MS/MS experiments to produce structurally meaningful spectra.
Figure 6.
Tandem spectra of (a) [1,3,4,1,6]5-, m/z 373, and its modified product (b) [1,3,4,4,3]-3d33- at m/z 560.8, (c) [1,3,4,1,7]5-, m/z 389 and its modified product [1,3,4,4,4]-3d34- at m/z 460. Major backbone fragments were assigned in Domon-Costello Convention in (b) and (d). Diamonds denote precursor ions, asterisks denote loss of sulfate ions
Table 3.
A Comparison of the Free Proton Index (FPI) and Backbone Cleavage Percentage, Defined as Glycosidic and Cross-Ring Cleavage, and Excluding the Neutral Sulfate Loss, for Some of the Oligosaccharides Studied. All LC-MS/MS Experiments were Performed with the Pulsing of Sulfolane to Produce Higher Charge States. The Last Compound is Arixtra
| Native composition, charge state | FPI | Backbone cleavage, % | Modified composition, charge state | FPI | Backbone cleavage, % |
|---|---|---|---|---|---|
| [1,2,3,1,3]3- | 0.500 | 96 | [1,2,3,3,1]-2d32- | 0.333 | 100 |
| [1,2,3,1,4]4- | 0.500 | 81 | [1,2,3,3,2]-2d33- | 0.333 | 99 |
| [1,2,3,1,5]4- | 0.667 | 24 | [1,2,3,3,3]-2d33- | 0.500 | 72 |
| [1,3,4,1,5]5- | 0.500 | 68 | [1,3,4,4,2]-3d33- | 0.375 | 100 |
| [1,3,4,1,6]5- | 0.625 | 41 | [1,3,4,4,3]-3d33- | 0.500 | 95 |
| [1,3,4,1,7]5- | 0.750 | 31 | [1,3,4,4,4]-3d34- | 0.500 | 93 |
| [1,3,4,1,8]5- | 0.875 | N.D. | [1,3,4,4,5]-3d35- | 0.500 | 99 |
| [0,2,3,0,8]-OMe4- | 1.200 | 22 | [0,2,3,3,5]-OMe-3 d35- | 0.400 | 96 |
N.D. = fragments that were of too low abundances to give definite assignments.
The oligosaccharides we studied were generated from heparin lyase digestion of HS from natural sources that consisted of many possible isomers for one particular composition. Therefore, the fragmentation ions alone are not sufficient to deduce the exact structure of the precursor ion without knowing if the precursor consists of a single structural isomer. To demonstrate the utility of the approach combing chemical modification and sulfolane pulsing in providing the complete structural information of a pure compound, we applied this strategy to Arixtra, a synthetic pentasaccharide with eight sulfates, including three N-sulfates. Figure S8A is the tandem mass spectrum of the 4– charge state of Arixtra (FPI=1.200), in which substantial sulfate loss and very limited backbone cleavages were observed. After the chemical modification, the sulfates number decreased to 5 and FPI dropped to 0.400 at 5–charge state. The tandem mass spectra yielded significantly more backbone cleavages (Figure S8B). In addition, major glycosidic bond cleavages, particularly those of B and Y ions, covered the entire sequence of the sugar chains, which in turn helped the assignment of sulfates to individual sugar rings. It should be noted that there is some degree of secondary loss of sulfate after dissociation of precursor ions. It is also difficult to assign of the 3-O sulfation due to the lack of cross-ring cleavage in the time-of-flight instrument we used in this study. We speculate that the secondary dissociation can be minimized and the differentiation of 6-O and 3-O-sulfation made possible if sulfolane pulsing is coupled with a hybrid ion trap-high resolution mass spectrometer, which will provide multi-stage tandem mass spectrometry capabilities.
After propionylation and de-N-sulfation, the N-sulfate groups have been removed and will not contribute to FPI. However, the charge states appear to be very low in absolute value for these oligosaccharides with multiple glucosamines (Figure S9, A). With the pulsing of sulfolane, there was very limited elevation of charge state (Figure S9, B). In contrast, the re-N-acetylation product responded much better with the pulsing of sulfolane (Figure S9, C). This phenomenon is probably due to the basicity of the amine groups, which will be protonated under HILIC conditions (pH=4.4). The carriage of the extra protons by the amine groups diminishes the effect of sulfolane pulsing.
Conclusions
Using MS3 experiments, we showed that N-sulfate groups were more prone to neutral loss than the O-sulfate groups. Through computational studies, it became clear that sulfate loss from a protonated sulfate is much more favored energetically than one from a deprotonated site, and metal adduction can also help stabilize sulfates. N-sulfate loss has a lower transition state barrier by 3 to 8 kcal/mol than either 2-O or 6-O sulfate loss, and the rate constant differences between N-sulfate and O-sulfate are enough to make the yield of N-sulfate loss markedly higher than O-sulfate loss. It is also revealed by calculation that carboxylic acid groups will be protonated over sulfate groups by about 27 kcal/mol and retro-Alder-Diels reaction at the non-reducing end proceeds through a transition state barrier similar to that of sulfate loss. We proposed the FPI concept that included all the factors that influence CID behaviors of HS negative ions, and aimed to reduce the FPI of precursor ions, minimize sulfate loss, and maximize backbone dissociation. We developed a procedure to selectively replace N-sulfate with acetate-d3. When combined with pulsing a charge-enhancing agent, the modified HS oligosaccharides possessed low FPI and produced abundant backbone cleavages that would not be possible without the chemical modification.
In this study, we investigated the fundamental thermodynamics and kinetics of sulfate loss at protonated, deprotonated, and metal-adducted sites. We demonstrated that N-sulfate is the most labile, and it is valuable to replace them with inert groups. The chemical modification developed in this work is selective, of high yield, amenable to low sample quantity, and complex mixtures. This study sheds light on the mechanisms of GAG ion dissociation and introduces a useful method to maximize abundances of structurally informative dissociations in LC-MS/MS experiments of HS oligosaccharides.
Supplementary Material
Acknowledgments
The authors acknowledge funding from NIH grants P41RR10888, P41GM104603, R01HL098950, and a research agreement with Bruker Daltonics Corporation. The authors thank Douglas Sondak at the Scientific Computational Facility at Boston University and Dr. Yiqun Huang of Boston University Center for Biomedical Mass Spectrometry for helpful discussions with the computational studies. They also thank Professor Irina Svir of Kharkov National University of Radio Electronics, Ukraine, and Professor Matthew Platz of Ohio State University for the use of KinFitSim software.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-012-0429-4) contains supplementary material, which is available to authorized users.
References
- 1.Esko JD, Kimata K, Lindahl U. Proteoglycans and Sulfated Glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. p. 229. [PubMed] [Google Scholar]
- 2.Roughley PJ, Lee ER. Cartilage proteoglycans: structure and potential functions. Microsc Res Tech. 1994;28:385–397. doi: 10.1002/jemt.1070280505. [DOI] [PubMed] [Google Scholar]
- 3.Conrad HE. Heparin Binding Proteins. Academic Press; New York: 1998. p. 183. [Google Scholar]
- 4.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 7.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 8.Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 9.Li JP. Heparin, heparan sulfate and heparanase in cancer: remedy for metastasis? Anti Cancer Agents Med Chem. 2008;8:64–76. doi: 10.2174/187152008783330824. [DOI] [PubMed] [Google Scholar]
- 10.Aikawa T, Whipple CA, Lopez ME, Gunn J, Young A, Lander AD, Korc M. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. J Clin Invest. 2008;118:89–99. doi: 10.1172/JCI32412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Rao G, Quiros RM, Kim AW, Miao HQ, Brunn GJ, Platt JL, Gattuso P, Prinz RA. In vivo and in vitro degradation of heparan sulfate (HS) proteoglycans by HPR1 in pancreatic adenocarcinomas. Loss of cell surface HS suppresses fibroblast growth factor 2-mediated cell signaling and proliferation. J Biol Chem. 2007;282:2363–2373. doi: 10.1074/jbc.M604218200. [DOI] [PubMed] [Google Scholar]
- 12.Escobar Galvis ML, Jia J, Zhang X, Jastrebova N, Spillmann D, Gottfridsson E, van Kuppevelt TH, Zcharia E, Vlodavsky I, Lindahl U, Li JP. Transgenic or tumor-induced expression of heparanase up-regulates sulfation of heparan sulfate. Nat Chem Biol. 2007;3:773–778. doi: 10.1038/nchembio.2007.41. [DOI] [PubMed] [Google Scholar]
- 13.Huntington JA, McCoy A, Belzar KJ, Pei XY, Gettins PGW, Carrell RW. The conformational activation of antithrombin—a 2.85-Å structure of a fluorescein derivative reveals an electrostatic link between the hinge and heparin binding regions. J Biol Chem. 2000;275:15377–15383. doi: 10.1074/jbc.275.20.15377. [DOI] [PubMed] [Google Scholar]
- 14.Jastrebova N, Vanwildemeersch M, Rapraeger AC, Gimenez-Gallego G, Lindahl U, Spillmann D. Heparan sulfate-related oligosaccharides in ternary complex formation with fibroblast growth factors 1 and 2 and their receptors. J Biol Chem. 2006;281:26884–26892. doi: 10.1074/jbc.M600806200. [DOI] [PubMed] [Google Scholar]
- 15.Goerges AL, Nugent MA. Regulation of vascular endothelial growth factor binding and activity by extracellular pH. J Biol Chem. 2003;278:19518–19525. doi: 10.1074/jbc.M211208200. [DOI] [PubMed] [Google Scholar]
- 16.Kreuger J, Spillmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spillmann D, Witt D, Lindahl U. Defining the interleukin-8-binding domain of heparan sulfate. J Biol Chem. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- 18.Feyzi E, Lustig F, Fager G, Spillmann D, Lindahl U, Salmivirta M. Characterization of heparin and heparan sulfate domains binding to the long splice variant of platelet-derived growth factor A chain. J Biol Chem. 1997;272:5518–5524. doi: 10.1074/jbc.272.9.5518. [DOI] [PubMed] [Google Scholar]
- 19.Walker A, Gallagher JT. Structural domains of heparan sulphate for specific recognition of the C-terminal heparin-binding domain of human plasma fibronectin (HEPII) Biochem J. 1996;317(Pt 3):871–877. doi: 10.1042/bj3170871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staples GO, Shi X, Zaia J. Extended NS domains reside at the non-reducing end of heparan sulfate chains. J Biol Chem. 2010;285:18336–18343. doi: 10.1074/jbc.M110.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem. 2004;279:5604–5611. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- 22.Safaiyan F, Lindahl U, Salmivirta M. Structural diversity of N-sulfated heparan sulfate domains: distinct modes of glucuronyl C5 epimerization, iduronic acid 2-O-sulfation, and glucosamine 6-O-sulfation. Biochemistry. 2000;39:10823–10830. doi: 10.1021/bi000411s. [DOI] [PubMed] [Google Scholar]
- 23.Naimy H, Leymarie N, Zaia J. Screening for anticoagulant heparan sulfate octasaccharides and fine structure characterization using tandem mass spectrometry. Biochemistry. 2010;49:3743–3752. doi: 10.1021/bi100135d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cravatt BF, Simon GM, Yates JR., III The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 25.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 26.Tissot B, North SJ, Ceroni A, Pang PC, Panico M, Rosati F, Capone A, Haslam SM, Dell A, Morris HR. Glycoproteomics: past, present and future. FEBS Lett. 2009;583:1728–1735. doi: 10.1016/j.febslet.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14:401–418. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol. 2010;5:105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 30.Whelan SA, Hart GW. Identification of O-GlcNAc Sites on Proteins. Methods Enzymol. 2006;415:113–133. doi: 10.1016/S0076-6879(06)15008-9. [DOI] [PubMed] [Google Scholar]
- 31.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 32.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff JJ, Leach FE, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ. Negative electron transfer dissociation of glycosaminoglycans. Anal Chem. 2010 doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19:1449–1458. doi: 10.1016/j.jasms.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adamson JT, Hakansson K. Electron detachment dissociation of neutral and sialylated oligosaccharides. J Am Soc Mass Spectrom. 2007;18:2162–2172. doi: 10.1016/j.jasms.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Mallis LM, Wang HM, Loganathan D, Linhardt RJ. Sequence analysis of highly sulfated, heparin-derived oligosaccharides using fast atom bombardment mass spectrometry. Anal Chem. 1989;61:1453–1458. doi: 10.1021/ac00188a030. [DOI] [PubMed] [Google Scholar]
- 37.Juhasz P, Biemann K. Utility of noncovalent complexes in the matrix-assisted laser desorption ionization mass spectrometry of heparin-derived oligosaccharides. Carbohydr Res. 1995;270:131–147. doi: 10.1016/0008-6215(94)00012-5. [DOI] [PubMed] [Google Scholar]
- 38.Saad OM, Leary JA. Delineating mechanisms of dissociation for isomeric heparin disaccharides using isotope labeling and ion trap tandem mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1274–1286. doi: 10.1016/j.jasms.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Saad OM, Leary JA. Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 40.Huang R, Pomin VH, Sharp JS. LC-MSn analysis of isomeric chondroitin sulfate oligosaccharides using a chemical derivatization strategy. J Am Soc Mass Spectrom. 2011;22:1577–1587. doi: 10.1007/s13361-011-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heiss C, Wang Z, Azadi P. Sodium hydroxide permethylation of heparin disaccharides. Rapid Commun Mass Spectrom. 2011;25:774–778. doi: 10.1002/rcm.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat Methods. 2008;5:291–292. doi: 10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, Shi X, Yu X, Leymarie N, Staples GO, Yin H, Killeen K, Zaia J. Improved liquid chromatography-MS/MS of heparan sulfate oligosaccharides via chip-based pulsed makeup flow. Anal Chem. 2011;83:8222–8229. doi: 10.1021/ac201964n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C.02. Gaussian, Inc; Wallingford, CT: 2004. [Google Scholar]
- 45.Curtissa LA, Raghavacharib K, Redferna PC, Pople JA. Investigation of the use of B3LYP zero-point energies and geometries in the calculation of enthalpies of formation. Chem Phys Lett. 1997;270:419–426. [Google Scholar]
- 46.Zaia J, Costello CE. Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal Chem. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 47.Naggar EF, Costello CE, Zaia J. Competing fragmentation processes in tandem mass spectra of heparin-like glycosaminoglycans. J Am Soc Mass Spectrom. 2004;15:1534–1544. doi: 10.1016/j.jasms.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 48.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790–798. doi: 10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLuckey SA, Cameron D, Cooks RG. Proton affinities from dissociations of proton-bound dimers. J Am Chem Soc. 1981;103:1313–1317. [Google Scholar]
- 50.Schnier PD, Jurchen JC, Williams ER. The effective temperature of peptide ions dissociated by sustained off-resonance irradiation collisional activation in Fourier transform mass spectrometry. J Phys Chem B. 1999;103:737–745. doi: 10.1021/jp9833193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morsa D, Gabelica V, De Pauw E. Effective temperature of ions in traveling wave ion mobility spectrometry. Anal Chem. 2011;83:5775–5782. doi: 10.1021/ac201509p. [DOI] [PubMed] [Google Scholar]
- 52.Naimy H, Buczek-Thomas JA, Nugent MA, Leymarie N, Zaia J. Highly sulfated nonreducing end-derived heparan sulfate domains bind fibroblast growth factor-2 with high affinity and are enriched in biologically active fractions. J Biol Chem. 2011;286:19311–19319. doi: 10.1074/jbc.M110.204693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumann H, Scheen H, Huppertz B, Keller R. Novel regio- and stereoselective O-6-desulfation of the glucosamine moiety of heparin with N-methylpyrrolidinone-water or N,N-dimethylformamide-water mixtures. Carbohydr Res. 1998;308:381–388. doi: 10.1016/s0008-6215(98)00097-4. [DOI] [PubMed] [Google Scholar]
- 54.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydr Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.