SUMMARY
Autophagy, a homeostatic process whereby eukaryotic cells target cytoplasmic cargo for degradation, plays a broad role in health and disease states. Here we screened the TRIM family for roles in autophagy and found that half of TRIMs modulated autophagy. In mechanistic studies we show that TRIMs associate with autophagy factors and act as platforms assembling ULK1 and Beclin 1 in their activated states. Furthermore, TRIM5α acts as a selective autophagy receptor. Based on direct sequence-specific recognition, TRIM5α delivered its cognate cytosolic target, a viral capsid protein, for autophagic degradation. Thus, our study establishes that TRIMs can function both as regulators of autophagy and as autophagic cargo receptors, and reveals a new basis for selective autophagy in mammalian cells.
INTRODUCTION
Autophagy is a eukaryotic homeostatic mechanism whereby cells remove from their cytoplasm toxic aggregates, damaged and surplus organelles, invading pathogens or utilize bulk cytosol for and metabolic needs (Mizushima et al., 2011). The key morphological presentation of autophagy is the appearance of autophagosomes, the organelles that carry out cytoplasmic cargo sequestration, driven by Atg factors (Mizushima et al., 2011; Yang and Klionsky, 2010). The canonical pathway leading to formation in the cytosol of the autophagic isolation membrane is under the control of the Ser/Thr protein kinase ULK1 (Atg1 in yeast), positioned downstream of mTOR and AMPK, which integrate nutritional and other signals (Mizushima et al., 2011). The mTOR and AMPK kinases phosphorylate ULK1 resulting in its inactivation or activation, respectively (Egan et al., 2011; Kim et al., 2011). ULK1 (Mizushima et al., 2011) and Beclin 1 (Liang et al., 1999) cooperate in the control of autophagy. A signaling cascade between ULK1 and Beclin 1 systems has been established via an activating phosphorylation of Beclin 1 by ULK1 (Russell et al., 2013). The subsequent stages of the pathway are controlled by the mammalian paralogues of the yeast Atg8. One of them, LC3B, is the most commonly used autophagosomal marker (Kabeya et al., 2000), whereas the role of other mammalian Atg8s is only beginning to be appreciated (von Muhlinen et al., 2012; Weidberg et al., 2010). At the end of a conjugation cascade initiated by Atg7, LC3s are lipidated at their C-termini as a defining event in the building of autophagic membranes (Mizushima et al., 2011). A phagophore sequesters the captured cytoplasmic cargo destined for autophagic disposal, which requires fusion of the autophagosome with lysosomes (Mizushima et al., 2011).
In contrast to bulk autophagy, targets of selective autophagy are recognized by autophagy receptors (Johansen and Lamark, 2011; Kirkin et al., 2009a; Li et al., 2013; Thurston et al., 2012; von Muhlinen et al., 2010; Wild et al., 2011) including p62/sequestosome 1 (p62) (Bjorkoy et al., 2005). The targets, earmarked by specific tags such as ubiquitin (Perrin et al., 2004) and galectins (Thurston et al., 2012), are delivered via cognate receptors to nascent autophagosomes (Johansen and Lamark, 2011; Kirkin et al., 2009b). In addition to placing ubiquitin tags on autophagic targets (Huett et al., 2012), E3 ligases have been implicated in autophagy activation of key regulatory factors (Nazio et al., 2013) downstream of signaling from TLR4, a pattern recognition receptor (PRR) (Shi and Kehrl, 2010). The engagement of PRRs in autophagy, as exemplified by TLR4 above, extends to nearly all major classes of PRRs (Deretic et al., 2013).
TRIMs represent a large family of proteins typically consisting of three motifs: an N-terminal RING domain, a B-box, and a coiled-coil domain (Kawai and Akira, 2011). Additionally, most TRIMs possess a variable C-terminal domain, which has a role in substrate binding (Kawai and Akira, 2011). Although TRIMs represent a large family of PRRs (Jefferies et al., 2011; Kawai and Akira, 2011; Ozato et al., 2008; Reymond et al., 2001) a systematic analysis of their involvement in autophagy has not been carried out. Since there are indications that TRIMs may be of relevance for autophagy (Barde et al., 2013; Khan et al., 2013; Niida et al., 2010; Pizon et al., 2013; Tomar et al., 2012; Yang et al., 2013), here we tested the hypothesis that, akin to other PRRs, TRIMs may play a general role in autophagy. We performed an siRNA screen examining the effects of TRIM knockdown on punctate LC3 and uncovered that a large number of TRIMs affect autophagy. We show here that TRIMs interact with ULK1, Beclin 1, and mammalian Atg8s. Further, we show that TRIM5α (Reymond et al., 2001) also acts as a receptor for selective autophagy.
RESULTS
TRIM proteins affect autophagy
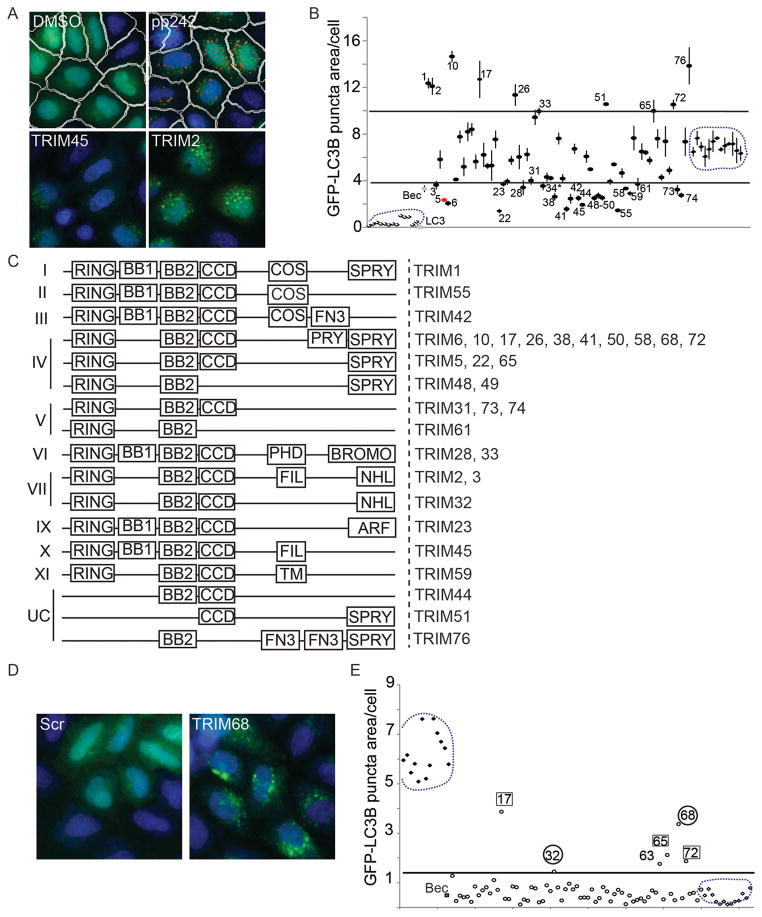
We employed a high-content image analysis (Figure 1A and Suppl. Figure S1A) with the autophagosomal marker LC3 (Kabeya et al., 2000; Mizushima et al., 2010) to screen the effects on autophagy of TRIM knockdowns (Figure 1B,C and Suppl. Figure S1A,B). Two conditions were examined, autophagy induced with the mTOR inhibitor pp242 (Figure 1A,B) and basal autophagy (Figure 1D,E). Automated image collection of >500 cells per siRNA were machine-analyzed using preset scanning parameters and object mask definition (iDEV software). Autophagy induction with pp242 resulted in a 17-fold induction of GFP-LC3B puncta area. TRIMs whose mean total area of GFP-LC3 per cell in three separate siRNA screen experiments (autophagy induced with pp242) differed by > 3 standard deviations above and below the mean of pp242-treated controls were reported as hits. Out of the 67 human TRIMs tested, knockdown of 21 different TRIMs reduced GFP-LC3B puncta area (Figure 1B) or puncta numbers (Suppl. Figure S1B) per cell under induced conditions, to an extent comparable to or exceeding the effect of Beclin 1 knockdown. Ten TRIMs showed a converse effect. Additional TRIMs affected basal autophagy (Figure 1D,E). Thus, a large fraction of TRIMs affect autophagy.
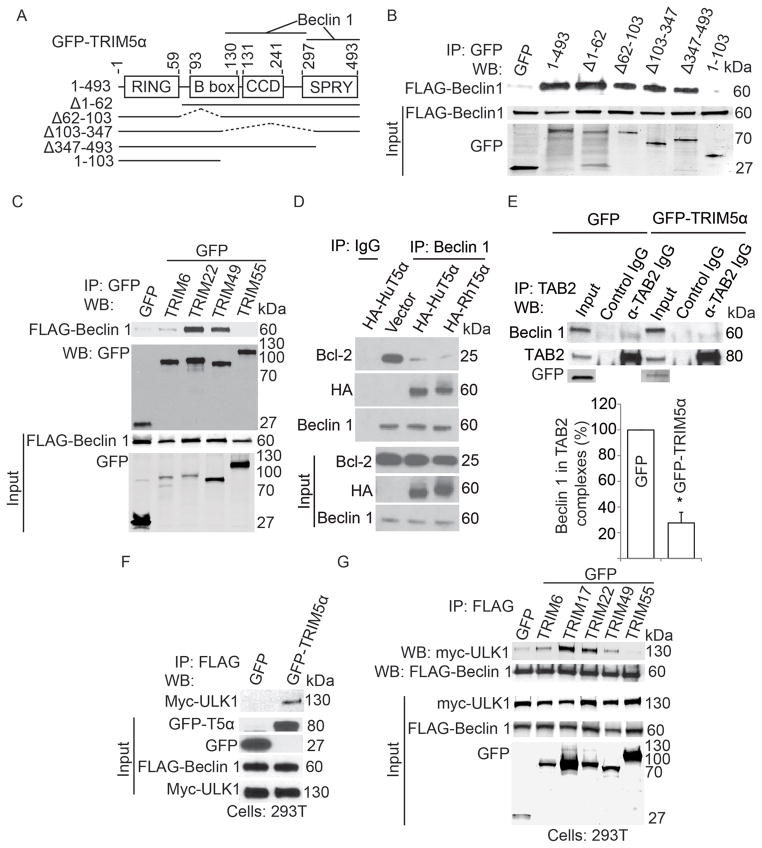
Figure 1. TRIM proteins regulate autophagy.
(A) HeLa cells stably expressing mRFP-GFP-LC3B were subjected to TRIM knockdowns, treated with pp242, and high content image analysis performed using a Cellomics HCS scanner and iDEV software. Shown are images (epifluorescence) with nuclear stain (blue) and GFP signal (green). Top, non-targeting siRNA-transfected cells treated with carrier (DMSO) or pp242. White lines, cell borders. Red, LC3B puncta borders. Bottom, representative images of cells subjected to knockdown of TRIM45 and TRIM2, both treated with pp242. (B) Average area of GFP-LC3B puncta per cell from cells treated as in (A) (data from multiple 96-well plates with identical siRNA arrangements represent means ± SE). Encircled are pp242-induced wells (pp242, right) and wells with vehicle controls (DMSO, bottom left). TRIM knockdowns that reduced or increased LC3B puncta readout by 3 SD (horizontal lines) from pp242-treated controls are indicated by corresponding TRIM numbers. Gray point (Bec), Beclin 1 knockdown; red point (numeral 5), TRIM5α. (C) Domain organization of TRIM sub-families (I–XI; UC, unclassified). Right, TRIM hits (LC3 puncta area >3 SD ± cutoff). (D) Representative images of TRIM knockdown cells under basal autophagy conditions. (E) High content image analysis (TRIM siRNA screen) under basal conditions (full medium). Encircled are scrambled siRNA controls: group on the left, pp242-induced wells; group on the bottom right, DMSO vehicle. Bec, Beclin 1 knock-down. TRIM knockdowns with GFP-LC3 puncta area >3 SD (horizontal bar) above unstimulated controls are indicated by corresponding TRIM numbers. Data, mean of two experiments. Numbers in squares, TRIMs scored as hits under both basal and induced conditions. Circled numbers, hits scored only under basal conditions. Cells treated with TRIM63 siRNA showed signs of apoptosis and were excluded from consideration. See also Figure S1.
TRIM5α interacts with Sequestosome 1/p62 and mammalian Atg8s
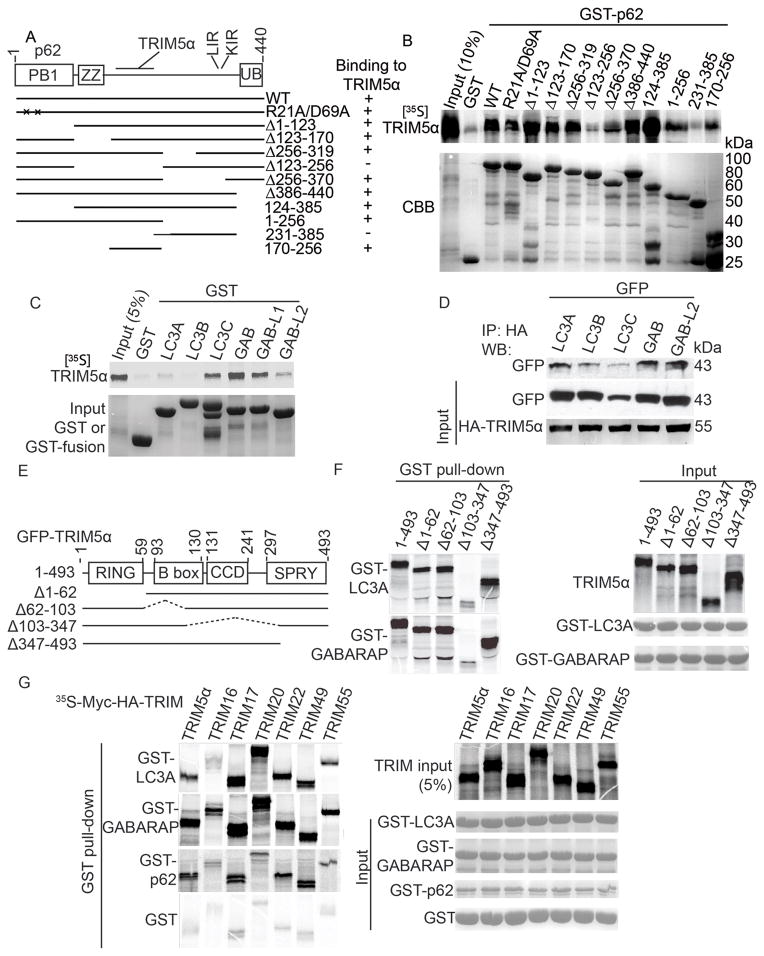
For detailed analysis of how a TRIM participates in autophagy, we chose to study TRIM5α. The rationale for focusing initially on TRIM5α was three-fold: (i) TRIM5α is physiologically highly relevant in cell-autonomous retroviral restriction (Stremlau et al., 2004; Stremlau et al., 2006); (ii) prior observations have indicated that TRIM5α may associate with the autophagy receptor p62 (O’Connor et al., 2010); and (iii) despite association with p62, no connections with autophagy have been previously suggested in the removal of TRIM5α’s cognate target, the retroviral capsid protein p24. We first confirmed the effects of TRIM5α knockdowns on LC3B puncta (Suppl. Figure S1C–E) and LC3-II levels in response to autophagy induction (Suppl. Figure S1F,G). Although the effects on LC3-II conversion were modest, they were in keeping with the similarly mild effects in this assay reported for Atg6 and Beclin 1 (Matsui et al., 2007; Suzuki et al., 2004). We next mapped the TRIM5α-binding domain on p62 to the region demarcated by residues 170–256 (Figure 2A,B). The biochemical analysis of p62-TRIM5α interaction was corroborated by immunofluorescence microscopy analysis, with p62 and TRIM5α co-localizing in puncta that were heterogeneous in size and distribution (Suppl. Figure S2A).
Figure 2. TRIMs interact with Sequestosome 1/p62 and mammalian Atg8s.
(A,B) Mapping of the Sequestosome 1/p62 region interacting with RhTRIM5α. (A) Domain organization of p62 and deletion/point mutation constructs employed to analyze interactions with TRIM5α as shown in (B). LIR, LC3-interacting region; KIR, KEAP1-interacting region. (B) Myc-TRIM5α was radiolabeled with [35S] methionine by in vitro translation and analyzed by GST pull-down assays with GST-p62 fusion proteins. Top, autoradiogram of pull-down products. Bottom, Coomassie Brilliant Blue (CBB)-stained SDS-polyacrylamide gel with GST-p62 proteins. Note TRIM5α input in first lane. (C) GST pull-down analysis of interactions between radiolabeled TRIM5α and GST-tagged mammalian Atg8 paralogues. (D) Co-immunoprecipitation analysis of interactions between HA-TRIM5α and GFP-tagged mammalian Atg8s in lysates from cells expressing the indicated constructs. (E) Domain organization of TRIM5α and schematic of deletion mutants used for mapping experiments in (F). (F) Analysis of TRIM5α domains interacting with LC3A, and GABARAP. (G) GST-pulldown analysis of binding between indicated radiolabeled TRIM proteins and GST-LC3A, GST-GABARAP, and GST-p62. See also Figure S2.
Sequestosome 1/p62 plays a role in autophagy, but also has other functions (Moscat and Diaz-Meco, 2009), and thus we tested whether TRIM5α connected with any additional autophagy factors. One of the best defined motifs for interactions between autophagy factors is the LC3-interacting region (LIR). Using the LIR consensus algorithm ([DEST]x(0,1)[WFY]{RKGP}{RKGP}[LIV]), which was defined based on alignment of 26 validated LIR sequences and mutation analysis of the ULK1 and ATG13 LIRs interacting with GABARAP (Alemu et al., 2012), we searched TRIM5α for the presence of putative LIRs. The best match was the sequence 186..DFEQL..190. We tested TRIM5α interactions with the full complement of mammalian Atg8 paralogs, of which LC3B is one member. LC3B showed minimal or no signal in the GST pull-down experiment with TRIM5α (Figure 2C). However, we detected robust interactions in vitro with other mammalian Atg8 paralogs (mAtg8s) GABARAP and GABARAPL1, and to a lesser extent with LC3A, LC3C, and GABARAPL2 (Figure 2C). Similar relationships with a subset of the mAtg8s were seen in cells as determined by co-immunoprecipitation experiments (Figure 2D). Whereas LC3B signal in TRIM5α GST pull-down assays was negligible (compared to the GST control; Figure 2C), the co-immunoprecipitation studies in cell lysates suggested that LC3B may nonetheless be in protein complexes in vivo with TRIM5α (Figure 2D). TRIM5α co-localized with punctate LC3B (Suppl. Figure S2B,C) and co-fractionated with the membrane-associated LC3B-II form (Suppl. Figure S2D). A p62 knockdown reduced the levels of LC3B in co-immunoprecipitates with TRIM5α (Suppl. Figure S2E), suggesting that LC3B detected in vivo in TRIM5α complexes was directly or indirectly affected by p62 levels. We next mapped the region of TRIM5α responsible for interactions with the mAtg8s LC3A and GABARAP through GST pull-down experiments (Figure 2E,F). We found that loss of the TRIM5α region encompassed by amino acids 103–347 ablated these interactions (Figure 2F). Thus, TRIM5α interacts directly with a subset of mAtg8s and p62.
We next asked, using a subset of TRIMs from our screen, whether other TRIMs interacted with p62 and mAtg8s. We included representative TRIMs based on whether they modulated (TRIMs 17, 22, 49 and 55) or showed no significant effect (TRIMs 16 and 20) on LC3B puncta. All TRIMs tested interacted with GABARAP (Figure 2G). TRIMs also interacted with LC3A and p62 (Figure 2G) albeit with exceptions (TRIM16 and TRIM20; Figure 2G). Thus, binding to GABARAP is a common feature among TRIMs, whereas other mAtg8s as well as p62 show variable but still prominent association with the TRIMs tested.
TRIMs interact with ULK1, an early regulator of autophagy initiation
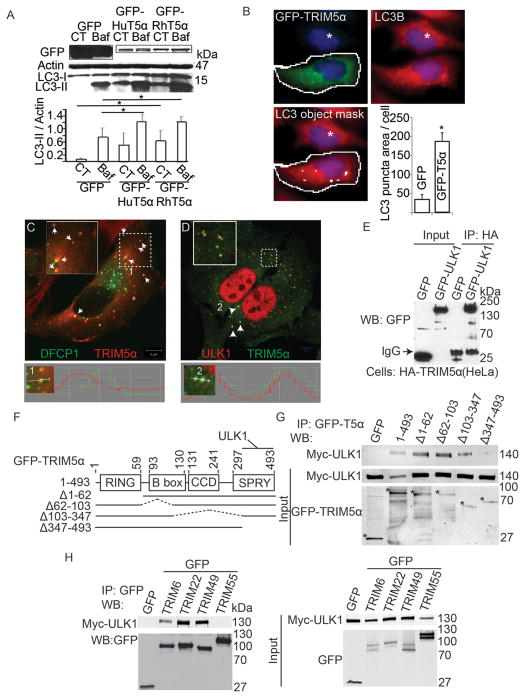
Overexpression of TRIM5α induced autophagy (Figure 3A,B). In these experiments we used TRIM5α clones from two different species, human (HuTRIM5α) and rhesus macaque (RhTRIM5α). Although HuTRIM5α and RhTRIM5α show differences associated with their binding to viral capsid proteins (Stremlau et al., 2006), overexpression of GFP-tagged TRIM5α from either source increased the abundance of LC3-II (Figure 3A). This indicated that TRIM5α from either species could be used interchangeably in autophagy activation experiments. Using the standard bafilomycin A1 flux assay (Mizushima et al., 2010), we established that TRIM5α overexpression induced autophagy rather than blocked autophagic maturation (Figure 3A). Overexpression of GFP-TRIM5α also increased LC3 puncta relative to cells over-expressing GFP alone (Figure 3B, Suppl. Figure S3A) in an ATG7-dependent manner (Suppl. Figure S3B).
Figure 3. TRIM5α and additional TRIMs interact with ULK1.
(A) Top, 293T cells were transfected with GFP-tagged human (HuT5α) or rhesus (RhT5α) TRIM5α or GFP alone and treated or not with bafilomycin A1 (Baf) and levels of LC3B and actin were assayed by immunoblot. Bottom, quantitation of LC3B-II/actin ratios. CT, control without Baf. (B) High content analysis of endogenous LC3 puncta in HeLa cells (full media) transfected with GFP or GFP-TRIM5α. Object masks: White contour line, gating for primary objects (GFP-positive cells). White internal small object masks, LC3B puncta. White asterisk, GFP-negative cell (manually entered; excluded from analysis by iDEV software). Graph, quantification of LC3B puncta area per green fluorescent (GFP+ or GFP-TRIM5α+) cell. Data, means ± SE, n ≥ 3 experiments, *, P bold> 0.05 (t test). (C) Confocal microscopy of HeLa cells stably expressing HA-TRIM5α and transiently expressing GFP-DFCP1. Numeral 1, a punctum displayed in line tracing profile below. (D) Confocal microscopy analysis of HA-tagged TRIM5α (green) and endogenous ULK1 (red) in HeLa cells. Numeral 2, two puncta displayed in line tracing profile below. (E) Lysates from HeLa cells stably expressing HA-tagged TRIM5α and transiently over-expressing either GFP-ULK1 or GFP alone were subjected to immunoprecipitation with anti-HA and immunoblots probed with anti-GFP. (F) Domain organization of TRIM5α and deletion constructs used in mapping experiments. (G) Co-immunoprecipitation analysis of full-length or deletion variants of TRIM5α (as GFP fusions; asterisks denote fusion products on the bottom blot) with Myc-ULK1 transiently expressed in 293T cells. (H) Co-immunoprecipitation analysis of interactions between indicated TRIMs (as GFP fusions) and Myc-ULK1 in 293T cell extracts. See also Figure S3.
Early autophagosomal structures in mammalian cells form in the vicinity of an endoplasmic reticulum-derived structure termed the omegasome (Axe et al., 2008). We examined the intracellular localization of TRIM5α relative to the omegasome marker DFCP1 (Figure 3C, Suppl. Figure S3C). Stably transfected HeLa cells expressing HA-TRIM5α showed a punctate cytoplasmic distribution of TRIM5α as previously described (Reymond et al., 2001). TRIM5α showed morphological linkage and spatial proximity with DFCP1 (Figure 3C). A similar juxtaposition has previously been noted for DFCP1 and the early autophagy factors of the ULK1 complex during characterization of the mammalian autophagosome formation sites (Itakura and Mizushima, 2010). We thus tested whether TRIM5α associated with ULK1 and found that TRIM5α and ULK1 co-localized (Figure 3D, Suppl. Figure S3D).
Based on their co-localization, we tested whether the two proteins interacted and found that HA-TRIM5α co-immunoprecipitated with GFP-ULK1 (Figure 3E) and endogenous ULK1 (Suppl. Figure S3E). We next mapped the ULK1 interacting region on TRIM5α (Figure 3F,G and Suppl. Figure S3F). Deletion mutants of TRIM5α lacking the C-terminus of the protein (amino acids 347–493) lost association with Myc-ULK1 in co-immunoprecipitation studies, indicating that the SPRY domain of TRIM5α was required for ULK1 inclusion in the complex (Figure 3G). When a panel of TRIMs that behaved similarly to TRIM5α in the screen was tested, they too co-immunoprecipitated with ULK1 (Figure 3H). One exception was TRIM55, a TRIM that lacks a SPRY domain. These findings indicate that TRIM5α and additional TRIMs act early in the autophagy pathway and that TRIMs’ SPRY domains are required to engage ULK1.
TRIM5α interacts with activated ULK1 and affects its intracellular distribution
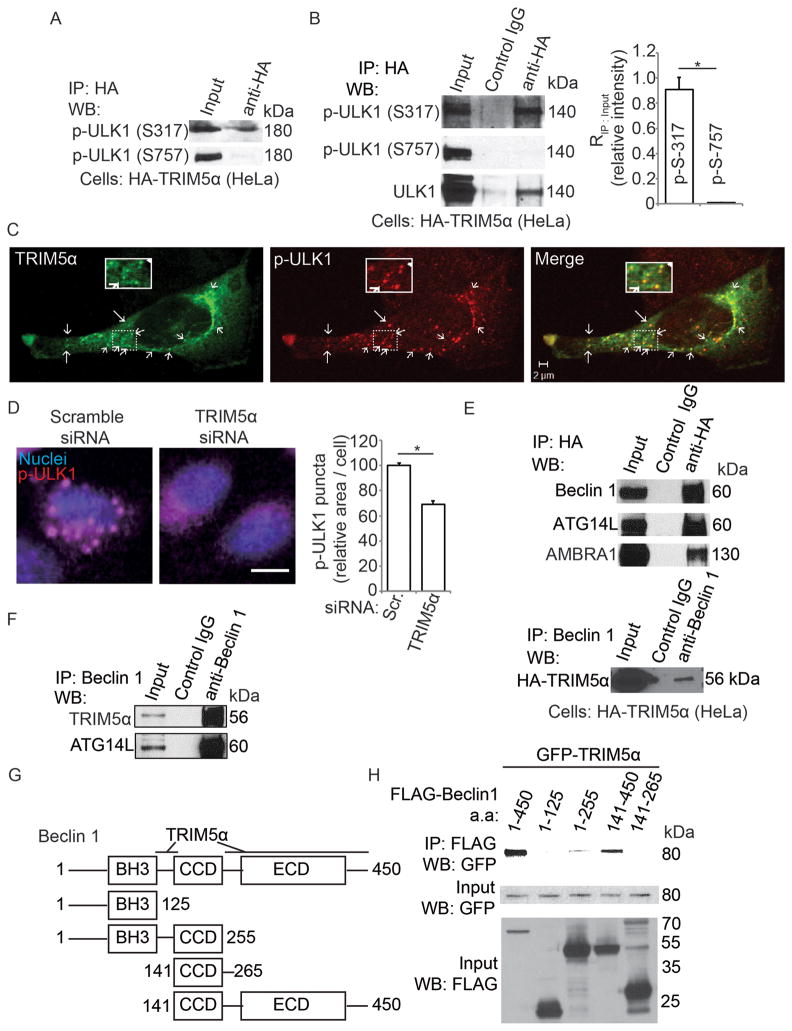
ULK1 activity is regulated by its phosphorylation status (Egan et al., 2011; Kim et al., 2011), with phosphorylations at Ser-317 activating and at Ser-757 inactivating ULK1 (Kim et al., 2011). We found that active p-ULK1 (Ser-317) co-immunoprecipitated with TRIM5α (Figure 4A,B). In contrast, inactive p-ULK1 (Ser-757) was dis-enriched in TRIM5α complexes (Figure 4A,B). We also observed substantial co-localization between TRIM5α and p-ULK1 (Ser-317) (Figure 4C). Of note, p-ULK1 (Ser-317) displayed a punctate pattern (Figure 4C). Thus, we considered a model in which TRIM5α affected cytoplasmic distribution of p-ULK1 (Ser-317). For this we developed a p-ULK1 assay (PULKA) based on high content microscopy to determine the abundance of p-ULK1 puncta. Using the PULKA approach, we found that cells knocked down for TRIM5α had fewer p-ULK1 puncta (Figure 4D). This indicates that TRIM5α may play a role in autophagy by concentrating active p-ULK1, possibly at sites of autophagy initiation.
Figure 4. TRIM5α interacts with activated ULK1 and with Beclin 1.
(A) Lysates from HeLa cells stably expressing HA-TRIM5α and transiently transfected with GFP-ULK1 were immunoprecipitated with anti-HA and blots probed with antibodies against Ser-317 or Ser-757 phospho-ULK1. (B) Lysates from HeLa cells stably expressing HA-TRIM5α were immunoprecipitated with anti-HA and blots probed as in A for endogenous ULK1. Graph, ratio of immunoprecipitated phospho-ULK1 to phospho-ULK1 in the input. (C) Confocal microscopy of cells stably expressing HA-tagged TRIM5α (green) and stained to detect p-ULK1 (p-Ser 317; red). Arrows, overlaps between p-ULK1 and HA-TRIM5α. (D) High content analysis of endogenous p-ULK1 (p-Ser 317; red) in control HeLa cells or cells subjected to TRIM5α knock-down. Blue, nuclei. Graph, area of phospho-ULK1 (Ser-317) puncta per cell. (E) Top, lysates from cells stably expressing HA-tagged TRIM5α were immunoprecipitated with anti-HA and immunoblots probed as indicated. Bottom, lysates as above were subjected to immunoprecipitated with anti-Beclin 1 and immunoblots probed for HA-TRIM5α. (F) Lysates from rhesus epithelial cells (FRhK4) were immunoprecipitated with anti-Beclin 1 and immunoblots probed with the indicated antisera. (G,H) Mapping of Beclin 1 regions interacting with TRIM5α. Schematic (G) of Beclin 1 constructs used in immunoprecipitation experiments in H. Lysates of 293T cells co-expressing GFP-tagged TRIM5α and the indicated FLAG-tagged Beclin 1 constructs in D were immunoprecipitated with anti-GFP and immunoblots probed as indicated. Data, means ± SE, n ≥ 3 experiments, *, P < 0.05 (t test). See also Figure S4.
TRIMs interact with the autophagy regulator Beclin 1
In addition to ULK1, autophagy initiation requires a second regulatory system centered on the Beclin 1 (Liang et al., 1999; Mizushima et al., 2011), which itself is a target of ULK1 (Russell et al., 2013). Thus, we considered whether TRIM5α action may engage Beclin 1. Beclin 1 co-immunoprecipitated with TRIM5α (Figure 4E,F; Suppl. Figure S4A). Furthermore, ATG14L (Itakura et al., 2008; Sun et al., 2008) and AMBRA1 (Fimia et al., 2007), components of the Beclin 1 complex engaged in autophagic initiation, were found in complexes with TRIM5α (Figure 4E,F; Suppl. Figure S4A). TRIM5α and Beclin 1 interaction was confirmed by proximity ligation assay (PLA; Suppl. Figure S4B–C), which reports direct protein-protein interactions in situ (Suppl. Figure S4C, scheme) (Soderberg et al., 2006). The positive PLA results with Beclin 1-TRIM5α were comparable to those with proteins known to be in complexes with TRIM5α, i.e. p62 and TAB2 (O’Connor et al., 2010; Pertel et al., 2011) (Suppl. Figure S4B,C), but not TAK1, which nevertheless co-localized with TRIM5α (Pertel et al., 2011) (Suppl. Figure S4D).
To map Beclin 1 domains required for interactions with TRIM5α, co-immunoprecipitation experiments were carried out with deletion mutants of Beclin 1. TRIM5α (HuTRIM5α and RhTRIM5α) bound Beclin 1 at regions defined by residues 1–255 (encompassing the BH3 and CCD domains) and 141–450 (encompassing the CCD and ECD domains) (Figure 4G,H; Suppl. Figure S4E). The CCD domain region (residues 141–265) was insufficient for TRIM5α binding (Figure 4H). Thus, TRIM5α directly interacts with Beclin 1, likely at two sites (Figure 4G). We also mapped the Beclin 1 interacting region on TRIM5α (Figure 5A,B). Only the TRIM5α deletion mutant lacking the amino acids 104–493 (consisting of part of the B box domain and all of the CCD and SPRY domains) lost the ability to interact with Beclin 1, whereas all other TRIM5α deletion mutants retained Beclin 1 binding (Figure 5A). These data suggest the presence of two separate TRIM5α-interacting regions on Beclin 1 and a longer TRIM5α region required for interactions with Beclin 1.
Figure 5. TRIM5α interacts with activated Beclin 1 and assembles ULK-1 with Beclin 1.
(A) Domain organization of TRIM5α and deletion constructs used in mapping experiments in B. (B) Co-immunoprecipitation analysis of full-length or deletion mutants of GFP-TRIM5α with FLAG-tagged Beclin 1. (C) Co-immunoprecipitation analysis of interactions between indicated TRIMs (as GFP fusions) and FLAG-Beclin 1 in 293T cell extracts. (D) Bcl-2-Beclin 1 complexes assessed by co-immunoprecipitation from control cells or cells expressing HA-HuTRIM5α or HA-RhTRIM5α. (E) Abundance of TAB2-Beclin 1 complexes assessed by co-immunoprecipitation from 293T expressing GFP-TRIM5α or GFP alone. Transfected cells were subjected to TAB2 immunoprecipitation and intensities of Beclin 1 bands in the precipitates normalized to TAB2 in the same samples. Differences in the normalized values (set at 100% for GFP-expressing cells) were assessed between cells expressing GFP or GFP-TRIM5α. Data, means ± SE, n = 3 experiments, *, P < 0.05 (t test). (F) Assessment of TRIM5α effects on ULK1 presence in Beclin 1 complexes. 293T cells were transiently transfected with Myc-ULK1, FLAG-Beclin 1, and either GFP-TRIM5α or GFP alone. Lysates were immunoprecipitated with anti-FLAG, and immunoblots probed as indicated. (G) Lysates from 293T cells expressing GFP or the indicated GFP-TRIMs, Myc-ULK1, and FLAG-Beclin 1 were treated as in F. See also Figure S5.
We tested other TRIMs (TRIMs 6, 22, 49, 55) for Beclin 1 binding. As was the case for ULK1, all TRIMs tested bound to Beclin 1 with the exception of TRIM55 (Figure 5C). These findings indicate that TRIM5α, as well as additional SPRY domain-containing TRIMs that behaved similarly to TRIM5α in the screen, interacted with Beclin 1, another key regulator of autophagy.
TRIM5α activates Beclin 1
We next tested whether TRIM5α affected Beclin 1 activation states. Beclin 1 is negatively regulated by two principal inhibitory binding partners, Bcl-2 (Wei et al., 2008) and TAB2 (Criollo et al., 2011; Takaesu et al., 2012). A dissociation of these factors is necessary to initiate autophagy (Criollo et al., 2011; Takaesu et al., 2012; Wei et al., 2008). Overexpression of TRIM5α (HuTRIM5α or RhTRIM5α) caused a dissociation of Bcl-2 from Beclin 1 as evidenced by diminished amounts of Bcl-2 that co-immunoprecipitated with Beclin 1 (Figure 5D). Overexpression of TRIM5α also caused dissociation of Beclin 1 from TAB2, evidenced by lower amounts of Beclin 1 found in TAB2 complexes (Figure 5E). We confirmed this using PLA methodology, and observed TRIM5α-dependent reduction in the PLA signal representing Beclin 1-TAB2 interactions when cells expressing GFP-TRIM5α were compared to cells expressing GFP (Suppl. Figure S5A). Thus, TRIM5α de-represses Beclin 1 by causing release of its negative regulators.
TRIMs serve as platforms to assemble ULK1 and Beclin 1 into a complex
Because TRIM5α binds both ULK1 and Beclin 1, we tested the possibility that it could act as a platform upon which Beclin 1 and ULK1 assembled together. ULK1 and Beclin 1 co-immunprecipitated in cells expressing GFP-TRIM5α but not GFP alone (Figure 5F). Furthermore, we detected the form of Beclin 1 phosphorylated by ULK1 (phospho-Beclin 1 at Ser-15; (Russell et al., 2013)) in complexes with TRIM5α (Suppl. Figure S5B). Thus, TRIM5α acts as a platform for ULK1 and Beclin 1 assembly and activation.
We next tested whether the TRIM5α capacity for bringing together ULK1 and Beclin 1 extended to other TRIMs (Figure 5G). We included the same TRIM panel (TRIMs 6, 22, 49 and 55) employed for the analysis of binding of ULK1 or Beclin 1, and found that those TRIMs (TRIMs 6, 22 and 49) which bound ULK1 or Beclin 1 individually also brought these molecules together into a complex. Co-expression of these TRIMs enriched ULK1 in the immunoprecipitated Beclin 1 complexes. In contrast, TRIM55, which lacks a SPRY domain and does not associate with either ULK1 or Beclin 1, had no such effect. Thus, the tested SPRY-endowed TRIMs acted as platforms for the assembly of autophagy regulators ULK1 and Beclin 1 into joint complexes.
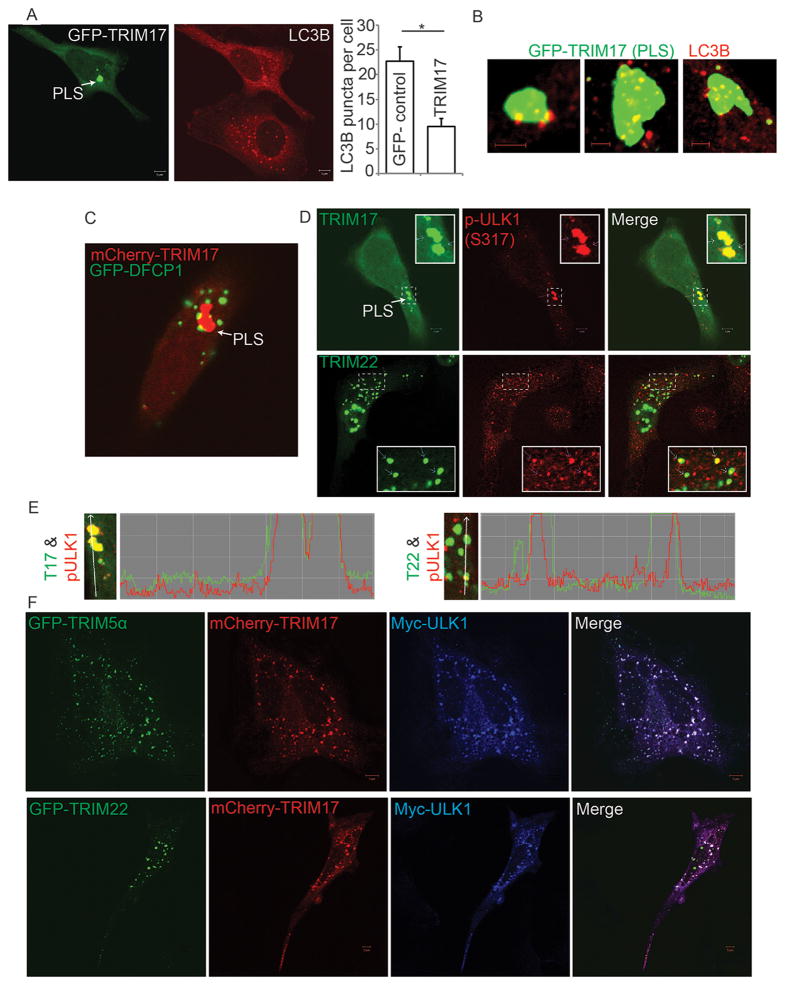
TRIM17 focuses autophagic machinery to a localized domain in the cell
A knockdown of TRIM17 in the siRNA screen increased LC3 puncta under both inducing and basal conditions. Accordingly, overexpression of TRIM17 reduced the number of LC3B puncta per cell (Figure 6A). When we examined these cells for TRIM17 localization, it became evident that TRIM17 formed one to two prominent large structures per cell (PLS; Figure 6A), observed also with antibody against endogenous TRIM17 (Suppl. Figure S6A). Endogenous LC3B puncta coalesced with TRIM17 PLS profiles in GFP-TRIM17-overexpressing cells (Figure 6A,B). Similarly, DFCP1 puncta, indicators of active autophagosome formation (Axe et al., 2008), associated with TRIM17 PLS (Figure 6C). TRIM17 bound ULK1 and Beclin 1 (Suppl. Figure S6B,C) and furthermore enriched ULK1 in Beclin 1 complexes (Figure 5G), just like most other TRIMs acting as inducers of autophagy. In cells with over-expressed TRIM17, p-ULK1 dots were reduced in total area per/cell (Suppl. Figure S6D) and changed their normal pattern of dispersed small puncta (italic>1.3 μm) within the cytoplasm, to coalescence at the PLS (> 3 μm) (Figure 6D,E). In contrast, neither TRIM5α (Suppl. Fig. S6E,F) nor TRIM22 (Figure 6D,E) overexpression caused this p-ULK1 phenotype. The unique features of TRIM17 in organizing active autophagic machinery at the PLS also extended to its recruitment of Exo84, an exocyst component previously associated with active autophagic complexes, to ULK1 complexes (Bodemann et al., 2011), contrasting with TRIM5α (Suppl. Fig. S6G). Thus, TRIM17 organizes active autophagy complexes but focuses them at the PLS.
Figure 6. TRIM17 focuses active autophagy machinery whereas TRIM5α and TRIM22 can redirect its localization.
(A) Effect of GFP-TRIM17 over-expression on LC3B puncta abundance in HeLa cells. Images, representative confocal micrographs showing GFP-TRIM17 and LC3B (red) under starvation conditions. PLS, prominent large structure containing GFP-TRIM17. Graph, number of LC3B puncta per cell; *, P < 0.05 (t test); n ≥ 30 cells. (B) Confocal micrographs showing association between TRIM17 PLS profile (green) with endogenous LC3B (red) in HeLa cells under starvation conditions. Scale bar, 2 μm. (C) Confocal micrograph of the association between PLS structure (labeled with mCherry-TRIM17, red) and omegasome marker GFP-DFCP1. (D) Intracellular distribution of endogenous p-ULK1 (p-Ser 317) in HeLa cells expressing GFP-TRIM17 or GFP-TRIM22. (E) Fluorescence intensity line tracings show phospho-ULK1 localization relative to GFP-tagged TRIMs. (F) Confocal micrograph of HeLa cells co-expressing mCherry-TRIM17 with Myc-ULK1 and GFP-TRIM5α (top) or with Myc-ULK1 and GFP-TRIM22 (bottom). See also Figure S6.
The above analysis indicates that although TRIM17 appeared as a negative regulator of autophagy in the initial siRNA screen, its effect on LC3 puncta distribution is due to the recruitment of autophagic factors to a localized, limited area of the cell. We next asked whether and how other TRIMs can overcome this effect. When TRIM5α and TRIM17 were co-expressed in HeLa cells, TRIM5α exerted dominance over TRIM17 (Figure 6F) by altering the localization pattern of ULK1 and TRIM17 away from forming a PLS and towards a dispersed cytoplasmic overall distribution that is typical of TRIM5α. Moreover, TRIM17 and ULK1 now co-localized strongly with TRIM5α (Figure 6F). TRIM22 showed the same effect, acting dominantly over TRIM17 (Figure 6F). Thus, the co-distribution of autophagic machinery with various TRIMs in the cell determines the localization of autophagosome formation.
A role for TRIM5α as an autophagic receptor
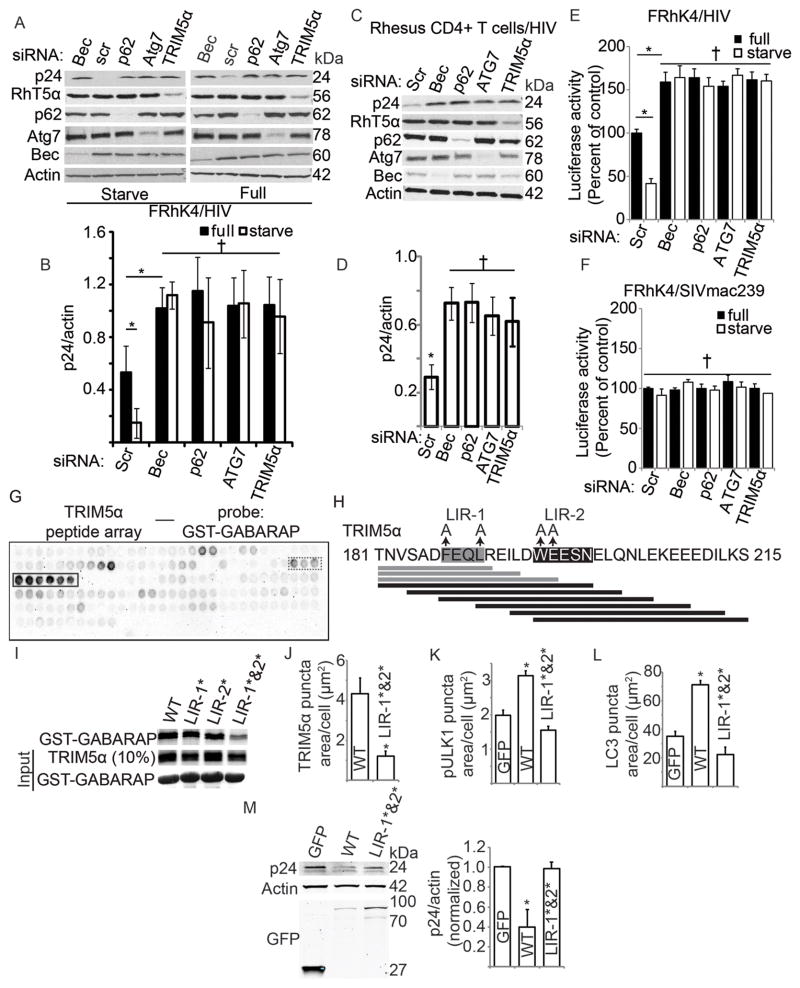
The above studies indicate a regulatory role for TRIMs in autophagy. Because TRIM5α is known to bind to a retroviral capsid as a part of its anti-retroviral activity (Stremlau et al., 2006), we wondered whether TRIM5α plays an additional role in autophagy by targeting its cognate cargo for autophagic degradation. The RhTRIM5α SPRY domain binds to human immunodeficiency virus 1 (HIV-1) capsid composed of the viral protein p24 (Stremlau et al., 2006) (Suppl. Figure S7A). We thus tested whether HIV-1 capsid protein p24 can be a substrate for lysosomal degradation in rhesus cells, FRhK4, which express endogenous RhTRIM5α. The experiments were carried out using VSVG-pseudotyped HIV-1 core viral particles. The experiments in Suppl. Figure S7B,C indicated that inhibition of lysosomal proteases protected HIV-1 p24 from degradation in rhesus cells. We next tested whether the observed lysosomal degradation of p24 was through autophagy. The data in Figure 7A,B indicated that p24 degradation was dependent on the autophagy factors Atg7, Beclin 1, p62, and TRIM5α. Induction of autophagy by starvation increased degradation of p24 in control cells but not in FRhK4 cells subjected to Atg7, Beclin 1, p62 or TRIM5α knockdowns (Figure 7A,B). Furthermore, ALFY/WDFY3, a potentiator of p62-dependent autophagy (Filimonenko et al., 2010), co-localized with TRIM5α (Suppl. Figure S7D), and a knock-down of ALFY in FRhK4 cells protected p24 from degradation, abrogating the effect of starvation (Suppl. Figure S7E,F). Collectively, these data are consistent with the interpretation that p24 is a target for autophagic degradation and that this process is directed by TRIM5α in cooperation with p62 and ALFY. A physiological relevance of the autophagic action of TRIM5α was indicated by the role that TRIM5α and autophagic factors played in diminishing the p24 levels in rhesus primary CD4+ T cells upon infection (Figure 7C,D).
Figure 7. Autophagy degrades protein target of TRIM5α in a manner requiring direct cargo recognition and mAtg8-interacting motifs in TRIM5α.
(A,B) Levels of intracellular p24 were determined by immunoblotting from rhesus cells (FRhK4) that had been subjected to knockdown of autophagy factors (Scr, scrambled siRNA) or TRIM5α and then exposed to pseudotyped HIV-1 for 4 h under full media or starvation conditions. (C,D) Immunoblot-based assessment of HIV-1 p24 in primary rhesus CD4+ T cells subjected to indicated knockdowns, infected with VSVG-pseudotyped HIV-1, and induced for autophagy by starvation for 4 h. (E,F) Luciferase activity of fed or starved FRhK4 cells subjected to indicated knockdowns and infected with luciferase-expressing pseudotyped HIV-1 (panel E) or SIVmac239 (panel F). (G,H) Binding of GST-GABARAP to a TRIM5α peptide array. A 20-mer peptide array corresponding to the entire TRIM5α sequence scanned in 3 amino acid residue shifts was subjected to a dot blot with GST-GABARAP as a probe. (H) Identification of a GABARAP interacting region on TRIM5α. Bars indicate peptide spots on the arrays with black bars indicating strongest binding (corresponding to the charcoal-framed dots in G) and gray bars to weaker binding (dashed-framed dots in G). Highlighted sequences, canonical LIR (LIR-1) and alternative LIR (LIR-2) involved in GST-GABARAP binding. (I) Mutational analysis of RhTRIM5α LIR motifs for effects on TRIM5α binding to GABARAP by GST pull-down. (J) High-content imaging analysis of the punctate distribution (vs. diffuse cytoplasmic) of GFP-tagged WT or double-LIR mutant (LIR-1*&2*) RhTRIM5α. (K) High-content analysis of the abundance of punctate p-ULK1 (Ser-317) in HeLa cells expressing GFP alone or GFP-RhTRIM5α (WT or LIR-1*&2*). (L) High-content analysis of the abundance of endogenous LC3B puncta in cells as in K. (M) Transfected 293T cells transiently expressing GFP alone or GFP-RhTRIM5α (WT or LIR-1*&2*) were infected with VSVG-pseudotyped HIV-1, induced for autophagy by starvation for 4 h, and levels of intracellular p24 were determined by immunoblotting. Data, means ± SE; n ≥ 3 experiments; *, P < 0.05; †, P ≥ 0.05 (t test or ANOVA). See also Figure S7.
Since upon viral entry TRIM5α recognizes the capsid protein only in the specific tertiary structure of the viral capsid (Stremlau et al., 2006), we considered for further study the use of a viral infection output assay. This was possible, since knocking down TRIM5α or autophagy factors resulted in an increase of the abundance of proviral DNA (Suppl. Figure S7G) and reverse transcriptase (Suppl. Figure S7H) in accordance with the results from the p24 assay described above. Having established that autophagy can lead to elimination of a portion of the incoming HIV-1 viral particles in cells expressing RhTRIM5α, we next sought to determine if this function was dependent on the specific interaction between TRIM5α as an autophagic receptor and its viral protein target. To do this, we utilized a well-characterized feature of the RhTRIM5α SPRY domain that recognizes HIV-1 capsid but is unable to recognize the equivalent simian immunodeficiency virus (SIV) capsid (Stremlau et al., 2004; Stremlau et al., 2006). A prediction based on this property of RhTRIM5α is that rhesus TRIM5α and autophagy can act upon HIV but cannot affect SIV. To test this hypothesis, we employed an assay with viral infection measured by luciferase outputs. Autophagy, induced by starvation, as in the p24 assays above, resulted in a reduced output of virally encoded luciferase only with HIV but not with SIV (Figure 7E,F). Moreover, Atg7, Beclin 1, p62 and TRIM5α were all required for optimal effects, again affecting luciferase outputs only in the case of HIV but not SIV (Figure 7E,F). These studies establish that mobilization of the viral target for degradation by the autophagic apparatus directly correlates with the target’s amino acid sequence recognized by TRIM5α. Collectively, these findings establish TRIM5α as an autophagic receptor.
Interaction of TRIM5α with mAtg8s is necessary for its action as an autophagic regulator and a receptor
As established in previous sections, TRIM5α interacts with mAtg8s, with the most prominent interaction being with GABARAP. We used GABARAP association with TRIM5α to delimit the sequence required for binding (Figure 7G,H). A 20-mer peptide array representing the entire TRIM5α sequence in increments staggered by 3 amino acid residues was subjected to a dot blot with GST-GABARAP as a probe (Figure 7G). Several peptides showed positive signals. We focused on the positive peptides with at least three consecutive binding signals. One such series containing exclusively the canonical LIR sequence 186-DFEQL-190 (Figure 7G, dashed framed dots) conferred only a weak GST-GABARAP binding. However, those peptides (Figure 7G, solid line framed dots) that contained an adjacent sequence, WEESN, located at positions 196–200, conferred strong GABARAP binding. Mutational analysis of key residues (LIR-1*: AEQA, instead of FEQL; LIR-2*: AAESN, instead of WEESN; Figure 7I and Suppl. Fig. S7I) indicated that GABARAP and other mAtg8s tested utilized both sequences. These experiments identified LIR-1 and LIR-2 in TRIM5α as the key motifs interacting with mammalian Atg8s, with LIR-2 showing similarity to LIR variants reported to bind strongly to GABARAP (Knaevelsrud et al., 2013; Ma et al., 2013).
We employed the above information to test whether TRIM5α’s interactions with mAtg8s were important for its autophagic functions. RhTRIM5α mutant with altered LIR-1 and LIR-2 motifs (RhTRIM5α LIR-1*&2*) showed diffuse cytoplasmic appearance instead of punctate distribution (Figure 7J, Suppl. Figure S7J). Whereas expression of the wild type RhTRIM5α resulted in increased p-ULK1 puncta (Ser-317), mutation of LIR-1 and LIR-2 (RhTRIM5α LIR-1*&2*) abrogated this effect (Figure 7K, Suppl. Figure S7K). RhTRIM5α’s ability to increase LC3B puncta, as a read-out of autophagy induction, was abrogated by LIR-1 and LIR-2 mutations (Figure 7L, Suppl. Figure S7L). Mutations of LIR-1 and LIR-2, which caused the above phenotypes, also precluded RhTRIM5α from targeting HIV-1 p24 for degradation (Figure 7M). Thus, LIR-1 and LIR-2, the newly identified motifs in TRIM5α, are essential for its regulatory effects on autophagy and for its ability to act as an autophagic receptor.
DISCUSSION
This study shows that TRIM proteins as a family broadly affect autophagy. The interactions between autophagy factors and TRIMs (e.g. TRIMs 5α, 6, 16, 17, 20, 22, 49, and 55) are extensive and involve both the regulators and effectors (ULK1, Beclin1, mAtg8s, and p62/sequestosome 1). In their regulatory role, TRIMs 5α, 6, 17, 22, and 49 act as platforms to bring together ULK1 and Beclin 1 into a single complex. By affecting the distribution of phospho-ULK1 (p-Ser 317) these TRIMs position the autophagy machinery. Our data with TRIM5α also reveal that TRIMs can play a role of cargo receptors for selective autophagy: TRIM5α targets a retroviral capsid protein for autophagic degradation by direct recognition of the target’s amino acid sequence in a manner dependent on interactions with mAtg8s. Based on these features, we propose a model in which TRIMs embody within one entity several aspects of selective autophagy such as target recognition, recruitment of the autophagy initiation machinery, and execution of the autophagic degradation of the cargo.
Our screen reveals a broad engagement of TRIMs in autophagy. Additional work supports TRIM connections with autophagy: TRIM13 overexpression can induce autophagy (Tomar et al., 2012); TRIM28 may regulate autophagy (Barde et al., 2013; Yang et al., 2013); several TRIMs interact with p62 (Fusco et al., 2012; Khan et al., 2013; Kim and Ozato, 2009; Pizon et al., 2013; Tomar et al., 2012); the expression of TRIM55 correlates with that of autophagy factors (Perera et al., 2011; Pizon et al., 2013); TRIMs are on lists in genome-wide autophagy screens (Behrends et al., 2010; Lipinski et al., 2010; McKnight et al., 2012); and TRIM21 and murine TRIM30α induce lysosomal degradation of certain targets (Niida et al., 2010; Shi et al., 2008).
The assembly of autophagic machinery (ULK1, Becln 1 and mATg8s) on TRIMs leads us to propose the concept of the TRIMosome (Suppl. Figure S7M). TRIMosome acts as a platform to focus selective autophagy on highly specific targets. However, additional processes may also be at play. For instance, TRIM28 was reported to influence expression of autophagy genes via miRNAs (Barde et al., 2013). Furthermore, TRIM55 in our experiments did not bind ULK1 and Beclin 1 (albeit it bound mAtg8s) but still strongly affected autophagy.
TRIM17 and TRIM22, while both binding to ULK1 and Beclin 1, distribute differently the activated form of ULK1 (p-Ser 317) in the cell, possibly to accomplish differential physiological tasks or to capture different cognate cargoes. TRIM17 focuses active autophagic machinery in a very restricted part of the cell, of as yet to be determined function. This can explain why in the siRNA screen TRIM17 knockdown increased generic LC3 puncta as a read-out of nonselective (pp242-induced) autophagy. In competition experiments, TRIM5α and TRIM22 both redirected and dispersed the autophagic machinery from the TRIM17 foci. Since TRIM expression responds to cytokines (Carthagena et al., 2009) and other signals (Bodine et al., 2001), this may differentially regulate levels of individual TRIMs and positioning of TRIMosomes in selective autophagy.
In its role as an autophagic cargo receptor, TRIM5α directly recognizes viral capsid sequences via its SPRY domain (Stremlau et al., 2004; Stremlau et al., 2006). This is an example of selective autophagy in mammalian cells, which we propose occurs via direct substrate recognition by TRIMs. We note that TRIMs contain SPRY or other types of C-terminal domains with the potential to recognize diverse protein targets or other molecular patterns (Kawai and Akira, 2011): COS - microtubule binding; FN3 - DNA or heparin binding; PHD - histone binding; BROMO - acetylated Lys residues binding; FIL - actin crosslinking; and NHL – protein interactions. Thus, we propose that TRIM proteins, as a group, comprise a new class of broad-repertoire high-fidelity selective autophagic receptors. These receptors may directly recognize their cognate targets without a need for ubiquitin tagging, which is the currently prevailing model of how autophagy targets its cargo (Shaid et al., 2013). The receptor function of TRIM5α shown here is reminiscent of selective autophagy in yeast where this process is independent of ubiquitin tags including the Cvt pathway (Lynch-Day and Klionsky, 2010), mitophagy (Kanki et al., 2009; Okamoto et al., 2009), and pexophagy (Farre et al., 2008). Should other TRIMs act as receptors that directly bind to their targets via cargo-recognition domains, this would confer exclusivity of autophagic targeting that a generic tagging with ubiquitin lacks.
In conclusion, our study reports the recognition of a global control of autophagy by TRIMs. TRIM proteins interact with components of the autophagic apparatus including mAtg8s, p62, and activated ULK1 and Beclin 1. TRIM5α acts both as a regulator of autophagy by providing a platform for the assembly of activated ULK1 and Beclin 1, and as a receptor for selective autophagy. Thus, TRIM5α embodies in one core entity two essential aspects of selective autophagy – recognition of the cargo and initiation of autophagy. Since TRIMs play diverse physiological roles (Barde et al., 2013; Cavalieri et al., 2011; Chen et al., 2012; Jefferies et al., 2011; Kawai and Akira, 2011; Ozato et al., 2008; Pertel et al., 2011; Versteeg et al., 2013) and have been linked to human inflammatory diseases and cancer (Hatakeyama, 2011; Jefferies et al., 2011; Kawai and Akira, 2011), our study invites explorations of the broad connections revealed here between TRIMs and autophagy.
METHODS
See also supplementary experimental procedures.
TRIM family screen
HeLa cells stably expressing mRFP-GFP-LC3B were cultured in 96-well plates containing siRNA smart pools and transfection reagent (Dharmacon). 48 h after plating, cells were treated with pp242 (as inducer of autophagy) or DMSO (control) for 2 h, fixed, and stained with Hoechst 33342. Plates with cells were subjected high content analysis for image acquisition and data processing. Three separate siRNA screens for induced autophagy and two separate siRNA screens for basal autophagy were carried out with the same cutoff (>3 SD above the mean of stimulated or unstimulated controls, respectively) for hits.
High content image analysis
High content imaging analysis was performed using a Cellomics HCS scanner and iDEV software (Thermo) Automated epifluorescence image collection was carried out until a minimum of 500 cells per well per siRNA knockdown per plate was acquired. Epifluorescent images were machine-analyzed using preset scanning parameters and object mask definitions. Cells were identified based on nuclear staining and cell outlines defined by background staining of the cytoplasm, and the mean per cell total area of GFP puncta or number of GFP puncta per cell were reported. Autophagy induction with pp242 resulted in a Z′ value of 0.52. When results were expressed as puncta area per cell, the units corresponded to μm2/cell.
High content analysis in subpopulations of transfected cells
HeLa cells were transfected with GFP or GFP-TRIM5α plasmids with or without siRNA, and cultured in full media for 48 h. Cells were then immunostained to detect endogenous LC3. High content image analysis was used in a mode discriminating transfected (GFP-positive cells) from untransfected cell subpopulations. High content imaging analysis, discriminating transfected cell subpopulations, was performed using a Cellomics HCS scanner and iDEV software (Thermo); >200 cells were analyzed per treatment in quadruplicate per experiment. Cell outlines were automatically determined based on background nuclear staining, and the mean total area of punctate LC3, p-ULK1 (Ser-317), or TRIM5α per cell was determined within the sub-population of cells that were successfully transfected as determined by having above background GFP fluorescence.
Confocal microscopy
Fluorescence confocal microscopy was carried out using a Zeiss META microscope.
Antibodies for immunoblotting, immunolabeling for microscopy and co-immunoprecipitation
Antibodies used were: ATG7 (Santa Cruz), ATG14L (MBL), Beclin 1 (Novus and Santa Cruz), Flag (Sigma), HA (Sigma and Roche), p62 (Abcam), TAB2 (Santa Cruz), TAK1 (Abcam), TRIM5α (Abcam), ULK1 (Sigma), ULK1 p-Ser 317 & pSer 757(Cell Signaling), Beclin 1 p-Ser15 (Abbiotec), FIP200 (provided by J._L. Guan, University of Cincinnati), Exo84 (Abcam), c-Myc (Santa Cruz). All other antibodies and methods are defined in supplementary experimental procedures.
Peptide array overlay assay and GST pull-downs
Peptide arrays were synthesized on cellulose membranes using a MultiPep automated peptide synthesizer (INTAVIS Bioanalytical Instruments AG, Germany). Peptide interactions were probed by overlaying the membranes with 1 μg/ml of recombinant protein for 2 h. Bound proteins were detected with HRP-conjugated anti-GST antibody (1:5000; clone RPN1236; GE Healthcare). GST pulldowns are described in supplementary experimental procedures.
Viral infection
Viral detection and yield measurements are detailed in supplementary materials. HIV-1 or SIVmac239 viruses for single cycle infection were generated by co-transfection of plasmids encoding VSV-G protein and the NL43 or SIVmac239 clones lacking the env gene into 293T cells.
Supplementary Material
Highlights.
The TRIM family of proteins regulate autophagy and direct selective autophagy
TRIMs serve as platforms for the assembly of the autophagic initiation machinery
TRIM5α is an autophagic receptor governing selective autophagy of HIV-1 capsid
Acknowledgments
We thank George Kyei, Manohar Pilli, Nicolas Dupont, Eliseo Castillo, Steven Bradfute, Michal Mudd and J. L. Guan. This work was supported by grants AI042999 and AI111935 from NIH and a Bill and Melinda Gates Foundation. M. Mandell and C. Dinkins were supported by NIH training grant T32AI007538. F. Kirchhoff acknowledges German Research Foundation (DFG) and the Zeiss foundation. G. Silvestri was supported by OD P51OD011132 to the Yerkes National Primate Research Center of Emory University. B. Levine was supported by NIH grant CA109618 and CPRIT RP120718-P1. T. Johansen was supported by grant 196898 from the Norwegian Research Council and grant 71043-PR-2006-0320 from the Norwegian Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Overvatn A, Kirkin V, Johansen T. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. RalB and the Exocyst Mediate the Cellular Starvation Response by Direct Activation of Autophagosome Assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, Mothes W, Hazan U, Transy C, Pancino G, et al. Human TRIM gene expression in response to interferons. PLoS One. 2009;4:e4894. doi: 10.1371/journal.pone.0004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri V, Guarcello R, Spinelli G. Specific expression of a TRIM-containing factor in ectoderm cells affects the skeletal morphogenetic program of the sea urchin embryo. Development. 2011;138:4279–4290. doi: 10.1242/dev.066480. [DOI] [PubMed] [Google Scholar]
- Chen L, Chen DT, Kurtyka C, Rawal B, Fulp WJ, Haura EB, Cress WD. Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J Biol Chem. 2012;287:40106–40118. doi: 10.1074/jbc.M112.380865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, et al. Inhibition of autophagy by TAB2 and TAB3. The EMBO journal. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nature reviews Immunology. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Developmental cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Fusco C, Micale L, Egorov M, Monti M, D’Addetta EV, Augello B, Cozzolino F, Calcagni A, Fontana A, Polishchuk RS, et al. The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome. PLoS One. 2012;7:e40440. doi: 10.1371/journal.pone.0040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ. The LRR and RING Domain Protein LRSAM1 Is an E3 Ligase Crucial for Ubiquitin-Dependent Autophagy of Intracellular Salmonella Typhimurium. Cell host & microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6 doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies C, Wynne C, Higgs R. Antiviral TRIMs: friend or foe in autoimmune and autoinflammatory disease? Nat Rev Immunol. 2011;11:617–625. doi: 10.1038/nri3043. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO molecular medicine. 2011;3:513–527. doi: 10.1002/emmm.201100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, Reischl M, Carnio S, Labeit D, Sandri M, et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 2013;10 doi: 10.4161/auto.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Ozato K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. Journal of immunology. 2009;182:2131–2140. doi: 10.4049/jimmunol.0802755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009a;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009b;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Soreng K, Raiborg C, Haberg K, Rasmuson F, Brech A, Liestol K, Rusten TE, Stenmark H, Neufeld TP, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202:331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wandel MP, Li F, Liu Z, He C, Wu J, Shi Y, Randow F. Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Science signaling. 2013;6:ra9. doi: 10.1126/scisignal.2003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS letters. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Schwarten M, Schneider L, Boeske A, Henke N, Lisak D, Weber S, Mohrluder J, Stoldt M, Strodel B, et al. Interaction of Bcl-2 with the autophagy-related GABAA receptor-associated protein (GABARAP): biophysical characterization and functional implications. J Biol Chem. 2013;288:37204–37215. doi: 10.1074/jbc.M113.528067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 2012;31:1931–1946. doi: 10.1038/emboj.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nature cell biology. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- Niida M, Tanaka M, Kamitani T. Downregulation of active IKK beta by Ro52-mediated autophagy. Molecular immunology. 2010;47:2378–2387. doi: 10.1016/j.molimm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor C, Pertel T, Gray S, Robia SL, Bakowska JC, Luban J, Campbell EM. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J Virol. 2010;84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera S, Holt MR, Mankoo BS, Gautel M. Developmental regulation of MURF ubiquitin ligases and autophagy proteins nbr1, p62/SQSTM1 and LC3 during cardiac myofibril assembly and turnover. Developmental biology. 2011;351:46–61. doi: 10.1016/j.ydbio.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V, Rybina S, Gerbal F, Delort F, Vicart P, Baldacci G, Karsenti E. MURF2B, a Novel LC3-Binding Protein, Participates with MURF2A in the Switch between Autophagy and Ubiquitin Proteasome System during Differentiation of C2C12 Muscle Cells. PLoS One. 2013;8:e76140. doi: 10.1371/journal.pone.0076140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nature cell biology. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell death and differentiation. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Noda T, Ohsumi Y. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast. 2004;21:1057–1065. doi: 10.1002/yea.1152. [DOI] [PubMed] [Google Scholar]
- Takaesu G, Kobayashi T, Yoshimura A. TGFbeta-activated kinase 1 (TAK1)-binding proteins (TAB) 2 and 3 negatively regulate autophagy. Journal of biochemistry. 2012;151:157–166. doi: 10.1093/jb/mvr123. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar D, Singh R, Singh AK, Pandya CD. TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochimica et biophysica acta. 2012;1823:316–326. doi: 10.1016/j.bbamcr.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Versteeg Gijs A, Rajsbaum R, Sánchez-Aparicio Maria T, Maestre Ana M, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, García-Sastre A. The E3-Ligase TRIM Family of Proteins Regulates Signaling Pathways Triggered by Innate Immune Pattern-Recognition Receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. LC3C, Bound Selectively by a Noncanonical LIR Motif in NDP52, Is Required for Antibacterial Autophagy. Molecular cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N, Thurston T, Ryzhakov G, Bloor S, Randow F. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy. 2010;6:288–289. doi: 10.4161/auto.6.2.11118. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. The EMBO journal. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6841–6846. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.