Abstract
We investigate the differences in molecular signature and clinical outcomes between multiple lesion glioblastoma (GBM) and single focus GBM in the modern treatment era. Between August 2000 and May 2010, 161 patients with GBM were treated with modern radiotherapy techniques. Of this group, 33 were considered to have multiple lesion GBM (25 multifocal and 8 multicentric). Patterns of failure, time to progression and overall survival were compared based on whether the tumor was considered a single focus or multiple lesion GBM. Genomic groupings and methylation status were also investigated as a possible predictor of multifocality in a cohort of 41 patients with available tissue for analysis. There was no statistically significant difference in overall survival (p < 0.3) between the multiple lesion tumors (8.2 months) and single focus GBM (11 months). Progression free survival was superior in the single focus tumors (7.1 months) as compared to multi-focal (5.6 months, p = 0.02). For patients with single focus, multifocal and multicentric GBM, 81, 76 and 88 % of treatment failures occurred in the 60 Gy volume (p < 0.5), while 54, 72, and 38 % of treatment failures occurred in the 46 Gy volume (p < 0.4). Out of field failures were rare in both single focus and multiple foci GBM (7 vs 3 %). Genomic groupings and methylation status were not found to predict for multifocality. Patterns of failure, survival and genomic signatures for multiple lesion GBM do not appreciably differ when compared to single focus tumors.
Keywords: Glioblastoma, Multifocal, Multicentric
Introduction
Multiple lesion glioblastoma (GBM) represent a population of GBM patients in which there exist multiple distinct foci of tumor enhancement. These multiple focus tumors represent between 0.5 and 20 % of all GBMs diagnosed, though some more recent reports indicate a higher incidence [1–3]. The population of multiple lesion GBM has previously been subdivided into multifocal and multicentric GBM. Multifocal GBM exhibit a contiguity or clear pathway of spread between foci. Multicentric tumors, on the other hand, do not exhibit a clear connection between the difference foci of disease and are often found in distinct lobes or hemispheres of the brain, and thus appear metastatic. The current literature regarding clinical outcomes of multiple lesion GBM has not distinguished between multiple lesion and single focus GBM, despite some literature that suggests that multiple lesion GBM may have worse outcomes.
The current study represents a single institution retrospective comparison of the clinical outcomes of single focus and multiple lesion GBM in which overall survival, progression-free survival and patterns of failure were assessed. The study aimed to determine if there were any differences in these outcomes between single focus and multiple lesion GBM in the modern treatment era of 3D conformal radiotherapy and temozolomide-based chemotherapy. In addition this study investigates potential correlation between multiple lesion GBM and previously determined clinically relevant genomic groupings and methylation states (Fig. 1).
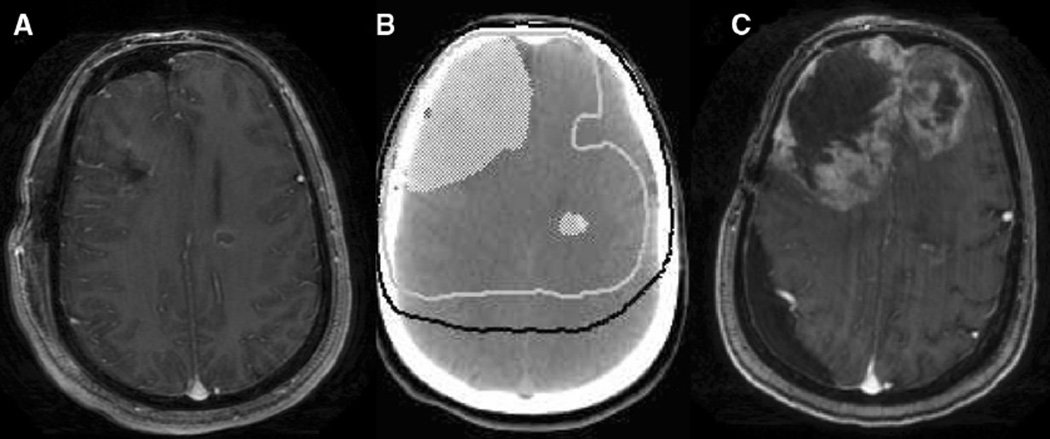
Fig. 1.
a T1 axial contrast-enhanced MRI done in the post-operative for multicentric GBM. There is a resection cavity in the right frontal lobe and a residual ring-enhancing lesion in the left frontal lobe. 1b Treatment planning axial non-contrast-enhanced CT demonstrating post-operative tumor volume in white colorwash, 60 Gy isodose line represented as white solid line, and 46 Gy isodose line represented as black solid line. 1c T1 axial contrast-enhanced MRI done at time of tumor progression showing tumor progression predominantly in the highest dose radiation volume
Materials and methods
Data acquisition and patient characteristics
This study was approved by the Wake Forest University Institutional Review Board. The Wake Forest University Radiation Oncology Database was searched for patients with diagnosis of GBM who were treated at our institution with radiation therapy. Between August 2000 and May 2010, 161 patients with GBM were treated with radiotherapy using modern treatment planning techniques. Patient characteristics for the study are summarized in Table 1. Patients with multifocal and multicentric tumors were identified by review of the diagnostic (pre-surgical) MRI. Tumors were classified as multifocal or multicentric based on whether there was a clear pathway of spread between foci of disease. Tumors were also classified as having multiple enhancing lesions, or whether any of the foci of tumor were non-enhancing tumors detected on T2 or fluid attenuated inversion recovery (FLAIR) sequences.
Table 1.
Patient characteristics
| Characteristic | Single focus | Multifocal | Multicentric |
|---|---|---|---|
| Age at diagnosis, years (median, range) | 61 (14–84) | 62 (45–83) | 64 (14–72) |
| Sex | |||
| Female | 47 | 13 | 0 |
| Male | 81 | 12 | 8 |
| Resection type | |||
| Gross total resection (GTR) | 71 (55 %) | 6 (24 %) | 1 (12.5 %) |
| Sub-total resection (STR) | 28 (22 %) | 12 (48 %) | 2 (25 %) |
| Biopsy (bx) | 29 (23 %) | 7 (28 %) | 5 (63.5 %) |
| RT modality | |||
| 3D Conformal | 96 (75 %) | 19 (76 %) | 8 (100 %) |
| IMRT | 32 (25 %) | 6 (24 %) | 0 (0 %) |
| Chemotherapy | |||
| Temodar-based | 83 (65 %) | 19 (76 %) | 6 (75 %) |
| Non-temodar-based | 5 (4 %) | 2 (8 %) | 1 (12.5 %) |
| None | 40 (31 %) | 4 (16 %) | 1 (12.5 %) |
Radiotherapy
Patients were treated at the Wake Forest University Comprehensive Cancer Center with non-whole brain radiotherapy fields using a shrinking field technique. The clinical target volume (CTV) margins used for individual patients were based upon physician discretion unless the patient was enrolled on a clinical trial, in which case the CTV margins were dictated by the trial guidelines. CTV margins for this analysis varied from 0.5 to 2.0 cm. These margins were determined using archived plans from the pinnacle treatment planning system (Philips Healthcare, Andover, MA). Planned treatment volume (PTV) margins were 5 mm.
Patient follow-up and response assessment
Patients were followed clinically by a multidisciplinary team and with serial MRI. Imaging was generally performed in the immediate post-operative setting, 1 month after completion of radiation therapy, and then every 2 months for 3 consecutive scans. Subsequent imaging was performed every 3 months, unless patients developed new or progressive symptoms warranting a scan. Determination of a gross total resection (GTR) was made based upon post-operative imaging, and required that all enhancing tumor be removed. Surgery was considered a subtotal resection (STR) if greater than 10 % of the tumor mass was debulked, but if the surgery was not a GTR. Surgery was considered a biopsy if less than 10 % of the tumor was debulked.
Electronic medical records and imaging review were used to retrospectively determine date of initial failure using the response assessment in neuro-oncology (RANO) criteria published by Wen et al. [4]. Treatment planning data were fused to MRI at date of failure using the pinnacle treatment planning system. Location of treatment failure was categorized as within the 60 Gy volume, within the 46 Gy volume or as distant failure (beyond the 46 Gy volume). Patients were considered to have failure in both the 60 and 46 Gy volumes if MRI on the date of failure demonstrated new or worsening areas of enhancement that were both enclosed by the 60 Gy isodose line, and between the 60 and 46 Gy isodose lines. Patterns of failure, time to progression and overall survival were compared based on whether the patient was considered a multiple lesion or single focus GBM.
Genomic analysis
Of the 161 patients identified above, fresh-frozen tumor tissues from 51 subjects were available for genomic analyses using Affymetrix Human Exon 1.0 ST GeneChips (Affymetrix, Santa Clara, CA) and Infinium HumanMethylation (HM) 450 BeadChip from Illumina (Illumina, San Diego, CA). Tissue banking was standardized at our institution in 2008, and tissue obtained prior to that was done so more sporadically and was based upon neurosurgeon participation in the banking program. Tissue banking was generally not performed on samples obtained from stereotactic biopsy because of insufficient tissue quantity. Of 51 patients for which sample existed in the tumor bank, 41 patients had sufficient tissue for analysis.
Genomic DNA and total RNA were isolated from these tumor samples using a QIAGEN DNA purification kit and TRIzol Reagent from Invitrogen, respectively, according to manufacturer’s instructions. The quality of genomic DNA and total RNA was evaluated as described previously [5]. We purchased all reagents and carried out transcriptome and genome-wide methylation assays following manufacturer’s instructions. Raw data generated from the exon GeneChips was first analyzed and evaluated using Affymetrix Expression Console. The data sets passed QCs were then imported into Partek Genomics Suite 6.6 software and Quantile-normalized. Pre-background was adjusted for GC content and for probe sequence. Log 2 probe intensities were finally analyzed for association between gene expression and clinical outcomes.
These 41 tumors were clustered into clinically relevant gene expression-based molecular classifications using hierarchical clustering with average linkage and a panel of 840 genes with high predictive values as previously described by The Cancer Genomic Atlas (TCGA) project [6]. Raw data generated from the HM450 BeadChip were obtained using GenomeStudio software (Illumina). Color balance adjustment, simple scaling normalization and data analysis were performed using the Bioconductor lumi package [7]. The β (methylation) value was calculated as described previously [8]. DNA methylation profiles for these 41 samples were evaluated for methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene, and for G-Cimp methylation phenotype [9, 10].
Statistics
Kaplan–Meier analysis was used to generate survival curves and chi square contingency analysis was used to determine differences between failure patterns between single focus and multiple lesion populations. Fisher’s exact test was used to determine differences between genomic groupings and methylation states. Multivariate analysis was performed using stepwise Cox proportional hazard regression to determine the relative value of factors that independently predicted for endpoints of survival and progression free survival. Multivariable models were built by a priori consideration of the factors for which data were gathered. Only variables with p significance of ≤0.2 were entered into the multivariable model, and if factors did not reach significance of 0.2 on stepwise regression, they were subsequently removed from the model. All statistics were performed using the SPSS program.
Results
Prevalence of multifocal and multicentric tumors
Of the 161 patients with GBM treated at our institution between August 2000 and May 2010, 33 patients (21 %) had multiple lesion GBM. Of these, 25 patients (16 %) had multifocal GBM and 8 patients (5 %) had multicentric GBM. Six of 33 patients (18 %) with multiple lesion GBM had foci of non-enhancing tumor.
Survival
Median overall survival of patients was statistically equivalent in patients who had single focus tumors and those with multiple lesion GBM (11 vs. 8.2 months, p = 0.3). Median time to progression was improved in the patients with single focus GBMs when compared to multiple lesion tumors (7.1 vs 5.6 months, p = 0.02).
Median survival was equivalent between multicentric GBM and multifocal GBM (8.2 months each, p = 0.90). Median time to progression was also equivalent between multicentric and multifocal GBM (4.0 vs 5.6 months, p = 0.3). A multivariate analysis was performed for all multiple lesion GBM which demonstrated no significant predictors amongst variables of age, performance status, gender, multicentricity, and degree of resection. If patients had tumors with at least one focus of non-enhancing FLAIR tumor, there were trends towards improved survival (HR 2.0, p = 0.13) and progression free survival (HR = 2.4, p = 0.08). Table 2 shows a univariate analysis for overall survival and univariate and multivariate analyses for progression free survival. A multivariate analysis was not performed for the endpoint of overall survival because only a single variable met the criteria to be entered into a multivariate model.
Table 2.
Univariate and multivariate analyses for survival and progression free survival in multiple lesion GBM
| Overall survival (Univariate) | Progression free survival (Univariate) |
Progression free survival (multivariate) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| P | Hazard ratio | 95 % CI | P | Hazard ratio | 95 % CI | P | Hazard ratio | 95 % CI | |
| Age | 0.6 | 1.0 | 0.97–1.0 | 0.9 | 1.0 | 0.96–1.0 | |||
| Gender (female) | 0.9 | 1.0 | 0.5–2.0 | 0.5 | 1.3 | 0.6–2.6 | |||
| KPS (≥80) | 0.9 | 1.0 | 0.5–2.0 | 0.002 | 3.4 | 1.5–7.8 | |||
| Multicentricity | 0.6 | 0.8 | 0.4–1.8 | 0.2 | 0.4 | 0.2–1.4 | |||
| Presence of non-enhancing tumor on FLAIR | 0.1 | 2.0 | 0.8–5.0 | 0.04 | 2.7 | 1–6.7 | 0.07 | 2.4 | 0.9–6.3 |
| Gross total resection | 0.6 | 1.22 | 0.5–2.8 | 0.7 | 0.8 | 0.4–2.0 | |||
Patterns of failure
Failure patterns were not significantly different between single focus and multiple focus GBM. For patients with single focus, multifocal and multicentric GBM, 81, 76 and 88 % of treatment failures occurred in the 60 Gy volume (p = 0.5), while 54, 72, and 38 % of treatment failures occurred in the 46 Gy volume (p = 0.4). Out of field failures were rare in both single focus and multiple foci GBM (7 % vs 3 %).
Value of surgical debulking
Of the patients with multiple lesion GBM, gross total resection was performed in 8 patients (24 %), subtotal resection was performed in 13 patients (39 %), and biopsy alone was performed in 12 patients (36 %). Of the single focus GBM, gross total resection was performed in 71 patients (55 %), subtotal resection was performed in 28 (22 %), and biopsy alone was performed in 29 (23 %).
Kaplan–Meier analysis was performed to determine whether surgery (either gross total or subtotal resection) was beneficial over biopsy alone for patients with multiple lesion GBM. Median survival for patients with multiple lesion GBM was equivalent between patients undergoing resection vs ones undergoing biopsy alone (8.4 vs 7.9 months, p = 0.2).
Genomic analyses
Of the 161 patients initially identified, 41 had genomic expression and methylation data available. Of the 41 patients with genomic data, 8 patients (20 %) had multiple lesion GBMs. Table 3 shows results of the genomic and methylation analyses. In summary, none of the TCGA genomic groups (proneural, neural, mesenchymal, or classical) predicted for multiple lesion GBMs (p = 0.6). In addition, MGMT methylation (p = 0.4) and G-CIMP methylation phenotype (p = 0.3) were not statistically different for multiple lesion versus single focus GBM.
Table 3.
Genomic characteristics of tumors
| Characteristic | Single focus | Multifocal |
|---|---|---|
| TCGA group (p = 0.575) | ||
| Classical | 4 (66 %) | 2 (33 %) |
| Mesenchymal | 15 (75 %) | 5 (25 %) |
| Neural | 7 (100 %) | 0 (0 %) |
| Proneural | 6 (86 %) | 1 (14 %) |
| MGMT methylation (p = 0.698) | ||
| Yes | 16 (76 %) | 5 (24 %) |
| No | 16 (84 %) | 3 (16 %) |
| G-Cimp phenotype (p = 0.563) | ||
| Yes | 5 (100 %) | 0 (0 %) |
| 27 (77 %) | 8 (23 %) | |
Discussion
While several reports of multiple lesion GBM have been published, it has been unclear whether these tumors represent distinct biologic variants of GBM or whether multifocal progression is an inevitable step in the natural history of GBM. Recent evidence suggests that there are several molecular subclasses of GBM with distinct molecular signatures [11]. A recent study from Cedars-Sinai Medical Center analyzed molecular markers from both multiple lesion GBM and single focus GBM showing no expression difference in a molecular panel including phosphorylated MAPK, PTEN, MGMT, laminin β1 and β2, and EGFR amplification between the tumor types [3]. While it remains unknown as to whether any particular subclasses of GBM or specific molecular pathways may influence multifocality of GBM, clinical trials have begun to target this possibility. As the glutamatergic system is considered important in the migration of glioblastoma cells and may play a role in the development of multiple lesion tumors, a recent phase II study was conducted in the NABTT consortium assessing the efficacy of talampanel, an AMPA receptor blocker, with standard chemoradiotherapy [12]. Radiation therapy fields, too, have the potential to be affected by a molecular signature. It has been demonstrated that tumors that experience progression outside of the highest dose region have a better prognosis [13, 14]. If a signature could be defined prior to treatment that predicts for out-of-field failure, the radiation field could be tailored to account for failure pattern.
The present report offers preliminary results on the distribution of single focus and multiple lesion GBM across the TCGA genomic subclasses and currently represents the only series in the scientific literature to have attempted this comparison. However, due to the small number of multiple lesion observations in the neural subclass and G-CIMP methylation phenotype groups and the small number of microarrays done in total, no definitive conclusions can be drawn concerning the impact of these genetic traits on whether GBM can form multiple lesions as the study was underpowered to answer the question. An additional concern is that our analysis does not account for the possibility of heterogeneity between tumors within the same patient, or even the possibility of intra-tumoral heterogeneity.
Multiple lesion GBM have been reported to have worse clinical outcomes than their single focus counterparts. The reason for this worsening has been hypothesized to be because of (1) higher burden of disease, (2) genetically more aggressive phenotype, and (3) inability for gross tumor resection given the multiple tumors. The present series shows that patients with multiple glioblastoma lesions had a worsened progression free survival than patients with single focus tumors, though there was no difference in overall survival. However, amongst subgroups, our data suggests that patients with tumors that have foci of non-enhancing FLAIR abnormality have improved progression free survival. This may be due to the presence of secondary GBM in this subgroup, as secondary GBM derive from lower grade gliomas that commonly have FLAIR abnormalities on imaging. Furthermore, secondary GBM are known to have improved outcomes [15].
The patterns of failure also do not appear to be significantly different between single focus and multiple lesion GBM, and the current series represents the first in the temozolomide treatment era to demonstrate this. There has been an increase of out-of-field treatment failures since the advent of temozolomide as evidenced by a series by Brandes et al. [14] that demonstrated this phenomenon. Prior to the era of temozolomide, multiple lesion GBM had a similar local failure pattern as single focus lesions. A retrospective study of 50 patients from Thomas Jefferson University demonstrated that the pattern of progression of these multiple-lesion GBMs was overwhelmingly local and no recurrences were seen distant from the original foci without significant local progression [1]. In the current series, regardless of the tumor type, the overwhelming majority of tumors experienced progression within the high-dose radiation volume (81 % for unifocal tumors, 76 % for multifocal, and 88 % for multicentric), while out of field failures were rare in all populations. Our data mimics what has been reported in the scientific literature as out of field failures are generally found in fewer than 10 % of cases [13, 16]. Because of the predominantly local treatment failure, use of standard non-whole brain treatment fields appears to have the potential to produce regional control while limiting toxicity in this population as in the population presenting with a single GBM focus.
The role of surgical de-bulking for multiple lesion GBM is controversial. There are several series in the literature that suggest that a greater degree of resection (from biopsy to subtotal resection to gross total resection) imparts a survival advantage to patients with GBM [17]. This may also be the case for multiple lesion tumors [17, 18]. A recent series from MD Anderson suggested that gross total resection of multiple lesions can impart a survival advantage to patients with even multicentric tumors [19]. Any advantage of surgical resection of multiple lesion GBM must be weighed, however, against the potential for toxicity. In the current series, gross total resection was performed in significantly fewer patients with multiple lesion GBM (21 vs. 55 %) because of a perceived risk by the operating surgeon. Performance status remains a dominant prognostic factor in GBM patients, and this is particularly relevant in multiple lesion GBM for which the therapeutic ratio may be narrower.
While our series represents a single institution retrospective series, it does represent one of the larger series of patients with multiple lesion GBM in the scientific literature, and the first to directly compare clinical outcomes and genomic characteristics of single versus multiple lesion GBM. From our series, it appears that multiple lesion GBM do not differ significantly in response to standard therapies when compared to their single focus counterparts. This is clinically relevant because in the current era of stratification of GBM by genetic signature, it appears that an imaging signature of detectable multifocality or multicentricity probably does not need to be included in this stratification, unless the relevant endpoint is the complete resectability of the tumor. Because of the retrospective nature of our report, these data need to be validated in a prospective manner.
Conclusion
Multiple lesion GBM do not differ significantly with regard to survival or patterns of failure when compared to single focus GBM. Differences in progression-free survival may be due to increased likelihood of gross total resection in single focus GBM.
Supplementary Material
Acknowledgements
The investigators responsible for this manuscript would like to sincerely thank Marlene Pratto and the Pratto Fund for Brain Tumor Research for the generous gift which helped to fund this research. We would like to thank Dr. Thomas L. Ellis, who contributed significantly to this work until his untimely death in 2012. We would also like to thank our patients as they are the inspiration behind our work.
Funding Pratto Fund for Brain Tumor Research; Dalton McMichael Fund in Cancer Research (to A.M.P); Comprehensive Cancer Center of Wake Forest University supported by the National Cancer Institute Cancer Center Support Grant [P30CA012197].
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-014-1515-1) contains supplementary material, which is available to authorized users.
This work has previously been presented at the ASTRO 2012 National Meeting in Boston, MA.
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Anna K. Paulsson, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA
Jordan A. Holmes, Department of Radiation Oncology, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA
Ann M. Peiffer, Department of Radiation Oncology, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA Brain Tumor Center of Excellence, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
Lance D. Miller, Department of Cancer Biology, Wake Forest Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA
Wennuan Liu, Center for Genomics and Personalized Medicine Research, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
Jianfeng Xu, Center for Genomics and Personalized Medicine Research, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
William H. Hinson, Department of Radiation Oncology, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA
Glenn J. Lesser, Brain Tumor Center of Excellence, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA Department of Medicine (Hematology & Oncology), Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
Adrian W. Laxton, Department of Neurosurgery, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA
Stephen B. Tatter, Brain Tumor Center of Excellence, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA Department of Neurosurgery, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
Waldemar Debinski, Brain Tumor Center of Excellence, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Department of Neurosurgery, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
Michael D. Chan, Email: mchan@wfubmc.edu, Department of Radiation Oncology, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA; Brain Tumor Center of Excellence, Wake Forest School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157, USA.
References
- 1.Showalter TN, Andrel J, Andrews DW, Curran WJ, Jr, Daskalakis C, Werner-Wasik M. Multifocal glioblastoma multiforme: prognostic factors and patterns of progression. Int J Radiat Oncol Biol Phys. 2007;69:820–824. doi: 10.1016/j.ijrobp.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 2.Thomas RP, Xu LW, Lober RM, Li G, Nagpal S. The incidence and significance of multiple lesions in glioblastoma. J Neurooncol. 2013;112:91–97. doi: 10.1007/s11060-012-1030-1. [DOI] [PubMed] [Google Scholar]
- 3.Patil CG, Yi A, Elramsisy A, Hu J, Mukherjee D, Irvin DK, Yu JS, Bannykh SI, Black KL, Nuno M. Prognosis of patients with multifocal glioblastoma: a case-control study. J Neurosurg. 2012;117:705–711. doi: 10.3171/2012.7.JNS12147. [DOI] [PubMed] [Google Scholar]
- 4.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, Tsien C, Mikkelsen T, Wong ET, Chamberlain MC, Stupp R, Lamborn KR, Vogelbaum MA, van den Bent MJ, Chang SM. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 5.Liu W, Lindberg J, Sui G, Luo J, Egevad L, Li T, Xie C, Wan M, Kim ST, Wang Z, Turner AR, Zhang Z, Feng J, Yan Y, Sun J, Bova GS, Ewing CM, Yan G, Gielzak M, Cramer SD, Vessella RL, Zheng SL, Gronberg H, Isaacs WB, Xu J. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2012;31:3939–3948. doi: 10.1038/onc.2011.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 8.Kim JW, Kim ST, Turner AR, Young T, Smith S, Liu W, Lindberg J, Egevad L, Gronberg H, Isaacs WB, Xu J. Identification of new differentially methylated genes that have potential functional consequences in prostate cancer. PLoS ONE. 2012;7:e48455. doi: 10.1371/journal.pone.0048455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, Dietrich PY, Weller M, Mariani L, Heppner FL, McDonald DR, Lacombe D, Stupp R, Delorenzi M, Hegi ME. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Grossman SA, Ye X, Chamberlain M, Mikkelsen T, Batchelor T, Desideri S, Piantadosi S, Fisher J, Fine HA. Talampanel with standard radiation and temozolomide in patients with newly diagnosed glioblastoma: a multicenter phase II trial. J Clin Oncol. 2009;27:4155–4161. doi: 10.1200/JCO.2008.21.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulsson AK, McMullen KP, Peiffer AM, Hinson WH, Kearns WT, Johnson AJ, Lesser GJ, Ellis TL, Tatter SB, Debinski W, Shaw EG, Chan MD. Limited margins using modern radiotherapy techniques does not increase marginal failure rate of glioblastoma. Am J Clin Oncol. 2014;37:177–181. doi: 10.1097/COC.0b013e318271ae03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandes AA, Tosoni A, Franceschi E, Sotti G, Frezza G, Amista P, Morandi L, Spagnolli F, Ermani M. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation with MGMT promoter methylation status. J Clin Oncol. 2009;27:1275–1279. doi: 10.1200/JCO.2008.19.4969. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobelbower MC, Burnett Iii OL, Nordal RA, Nabors LB, Markert JM, Hyatt MD, Fiveash JB. Patterns of failure for glioblastoma multiforme following concurrent radiation and temozolomide. J Med Imaging Radiat Oncol. 2011;55:77–81. doi: 10.1111/j.1754-9485.2010.02232.x. [DOI] [PubMed] [Google Scholar]
- 17.Simpson JR, Horton J, Scott C, Curran WJ, Rubin P, Fischbach J, Isaacson S, Rotman M, Asbell SO, Nelson JS, et al. Influence of location and extent of surgical resection on survival of patients with glioblastoma multiforme: results of three consecutive radiation therapy oncology group (RTOG) clinical trials. Int J Radiat Oncol Biol Phys. 1993;26:239–244. doi: 10.1016/0360-3016(93)90203-8. [DOI] [PubMed] [Google Scholar]
- 18.Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen HJ. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7:392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- 19.Hassaneen W, Levine NB, Suki D, Salaskar AL, de Moura Lima A, McCutcheon IE, Prabhu SS, Lang FF, DeMonte F, Rao G, Weinberg JS, Wildrick DM, Aldape KD, Sawaya R. Multiple craniotomies in the management of multifocal and multicentric glioblastoma: clinical article. J Neurosurg. 2011;114:576–584. doi: 10.3171/2010.6.JNS091326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.