Abstract
Mitochondrial DNA (mtDNA) is replicated by DNA polymerase γ in concert with accessory proteins such as the mitochondrial DNA helicase, single-stranded DNA binding protein, topoisomerase, and initiating factors. Defects in mtDNA replication or nucleotide metabolism can cause mitochondrial genetic diseases due to mtDNA deletions, point mutations, or depletion, which ultimately cause loss of oxidative phosphorylation. These genetic diseases include mtDNA depletion syndromes such as Alpers or early infantile hepatocerebral syndromes, and mtDNA deletion disorders, such as progressive external ophthalmoplegia, ataxia-neuropathy, or mitochondrial neurogastrointestinal encephalomyopathy. This review focuses on our current knowledge of genetic defects of mtDNA replication (POLG, POLG2, C10orf2, and MGME1) that cause instability of mtDNA and mitochondrial disease.
Keywords: Alpers syndrome, ataxia-neuropathy, DNA polymerase γ, mitochondrial DNA depletion syndrome, mitochondrial DNA replication, POLG, progressive external ophthalmoplegia
Introduction
The mitochondrial genome is a closed circular genome of 16 569 bp that codes for 37 genes, all of which are directly or indirectly involved in the production of adenosine triphosphate. Thirteen of these genes encode protein subunits involved in electron transport to carry out oxidative phosphorylation. The remaining 24 genes encode the transfer RNAs (22 genes) and ribosomal RNAs (2 genes) required for mitochondrial protein synthesis of the 13 polypeptides. Animal tissue cells can contain several thousand copies of mitochondrial DNA (mtDNA) spread out over hundreds of mitochondria.1
Mitochondrial DNA is replicated and repaired by DNA polymerase γ (pol γ) only known DNA polymerase to be found in animal cell mitochondria. The holoenzyme of pol γ in humans consists of a catalytic subunit (encoded by POLG at chromosomal locus 15q25) and a dimeric form of its accessory subunit (encoded by POLG2 at chromosomal locus 17q24.1). The catalytic subunit is a 140 kDa enzyme (p140) that has DNA polymerase, 3’–5’ exonuclease, and 5’ dRP lyase activities.2,3 The accessory subunit is a 55 kDa protein (p55) required for tight DNA binding and processive DNA synthesis.4 The pol γ holoenzyme functions in conjunction with the mtDNA helicase, Twinkle or C10orf2, and mitochondrial single-stranded binding protein (mtSSB) to form the minimal replication apparatus.5,6 A recently identified 5’-3’ exonuclease, MGME1, also appears crucial in the replication of mtDNA.
Mitochondrial diseases can be caused by genetic defects in mtDNA or in nuclear genes that encode proteins that function in the mitochondria.7 During the past 10 years, researchers have defined a list of genes linked to instability of mtDNA, either due to mitochondrial DNA depletion syndrome or disorders characterized by multiple deletions, with or without pointmutations. Table 1 lists the current genes associated with mitochondrial DNA depletion syndrome and other deletion syndromes. Disorders associated with deletions and point mutations include commonly known disorders such as progressive external ophthalmoplegia and ataxia-neuropathy syndromes, but also some very rare disorders of tricarboxylic acid cycle abnormalities.8 Mitochondrial DNA depletion syndrome includes early childhood disorders such as Alpers syndrome, hepatocerebral syndromes, myocerebrohepatopathy spectrum disorders, and fatal myopathies.9,10 Gene mutations in POLG, POLG2, C10orf2, and MGME1 have been implicated in mitochondrial disease and are discussed below.
Table 1.
Nuclear Genes That Affect the Stability of Mitochondrial DNA
| Gene | Disorder | Chromosomal locus |
Function |
|---|---|---|---|
| POLG | PEO / Alpers / ataxia | 15q25 | Mitochondrial DNA polymerase |
| POLG2 | PEO | 17q23–24 | Pol γ accessory subunit |
| PEO1 (Twinkle) | PEO / ataxia | 10q24 | Mitochondrial DNA helicase |
| MGME1 | PEO, MtDNA depletion | 20p11.23 | RecB type exonuclease |
| ANT1 | PEO | 4q34–35 | Adenine nucleotide translocator |
| TP | MNGIE | 22q13.32 | Thymidine phosphorylase |
| DGUOK | MtDNA depletion | 2p13 | Deoxyguanosine kinase |
| TK2 | MtDNA depletion | 16q22 | Mitochondrial thymidine kinase |
| MPV17 | MtDNA depletion | 2p21–23 | Mt inner membrane protein |
| SUCLA2 | MtDNA depletion | 13q12.2–13.3 | ATP-dependent Succinate-CoA ligase |
| SUCLG1 | MtDNA depletion | 2p11.2 | GTP-dependent Succinate CoA ligase |
| RRM2B | MtDNA depletion | 8q23.1 | p53-Ribonucleotide reductase, small subunit |
| OPA1 | Dominant optic atrophy | 3q28–29 | Dynamin-related GTPase |
| FBXL4 | MtDNA depletion, Encephalopathy | 6q16.1–16.3 | Mitochondrial LRR F-Box protein |
Abbreviations: ATP, adenosine- 5’-triphosphate; CoA, coenzyme A; GTP, guanosine-5'-triphosphate; LLR, leucine-rich repeats; PEO, progressive external ophthalmoplegia; MNGIE, mitochondrial neurogastrointestinal encephalomyopathy; mtDNA, mitochondrial DNA.
POLG, the Catalytic Subunit of the Human DNA Polymerase γ
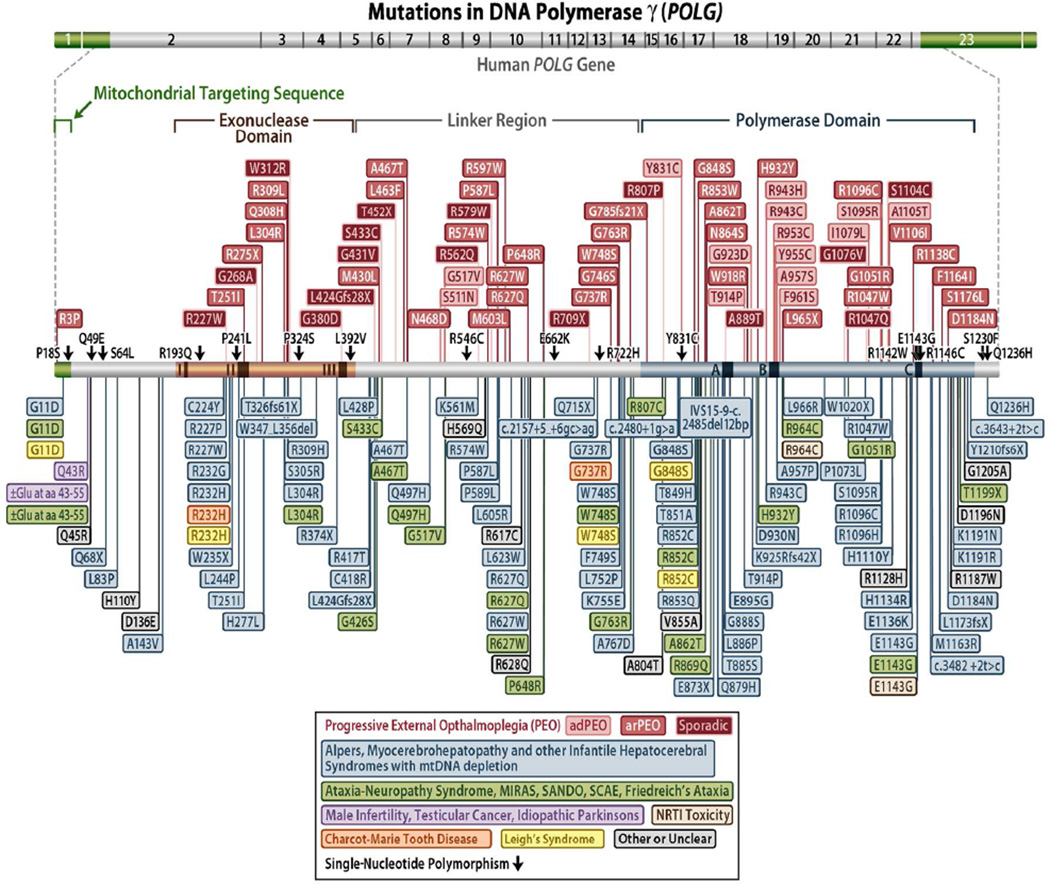
In 2001, Van Goethem and colleagues published a paper describing 4 mutations in the POLG gene associated with progressive external ophthalmoplegia.11 This was the first of many papers to follow identifying disease mutations in the POLG gene. To date, there are nearly 250 pathogenic mutations in POLG (http://tools.niehs.nih.gov/polg/)8,12–15) (Figure 1). POLG-related disorders are currently defined by at least 6 major phenotypes of neurodegenerative disease that include Alpers-Huttenlocher syndrome, childhood myocerebrohepatopathy spectrum, myoclonic epilepsy myopathy sensory ataxia, the ataxia neuropathy spectrum, autosomal recessive progressive external ophthalmoplegia, and autosomal dominant progressive external ophthalmoplegia.9,10,16,17 Also, alteration of the (CAG)10 repeat in the second exon of POLG has been implicated in male infertility, testicular cancer, and Parkinsonism, and has been reviewed by Saneto and Naviaux.10 The POLG gene is unique in the number of pathogenic mutations spread out over the gene and by the variety of diseases they cause, some of which follow an autosomal dominant pattern of inheritance (although the majority are autosomal recessive).
Figure 1.
Schematic diagram of the human DNA polymerase γ gene and protein illustrating the positions of disease mutations.
Progressive external ophthalmoplegia is a mitochondrial disorder associated with mtDNA deletions and point mutations.11,18–20 It is characterized by late-onset (between 18 and 40 years of age) bilateral ptosis and progressive weakening of the external eye muscle, resulting in blepharoptosis and ophthalmoparesis, proximal muscle weakness and wasting, and exercise intolerance. The disease is often accompanied by cataract, hypogonadism, dysphagia, and hearing loss, and may, within several years, lead to development of neuromuscular problems.18,21 Neurological problems may include depression or avoidant personality.22 Skeletal muscles of patients with progressive external ophthalmoplegia present red ragged fibers and lowered activity of respiratory chain enzymes. Progressive external ophthalmoplegia can be inherited in an autosomal dominant or recessive manner. Autosomal dominant progressive external ophthalmoplegia mutations in POLG are generally found in very conserved residues within the active site of the DNA polymerase domain,23 whereas recessive progressive external ophthalmoplegia mutations are spread throughout the gene.
Alpers syndrome is an autosomal recessive mitochondrial DNA depletion disorder that affects children and young adults. It is a devastating disease characterized by psychomotor retardation, hepatic failure, and intractable seizures, as well as tissue-specific mtDNA depletion. Alpers patients rarely survive past 10 years of age.
Childhood myocerebrohepatopathy spectrum includes myopathy or hypotonia, developmental delay or dementia, and liver dysfunction.9 In addition, patients can have either a liver biopsy that excluded classical Alpers hepatopathy,24 or at least 2 of the following 8 findings: (1) neuropathy, (2) seizures, (3) elevated blood or cerebrospinal fluid lactic acid, (4) dicarboxylic aciduria, (5) renal tubular dysfunction with aminoaciduria, glucosuria, or bicarbonaturia, (6) hearing loss, (7) abnormal MRI with either cerebral volume loss, delayed myelination, or white matter disease, and (8) deficiency of either Complex IV (cytochrome c oxidase, [COX]) in isolation, or 2 or more electron transport complexes in skeletal muscle or liver biopsy.9 In some cases, patients came to diagnosis without, or before, the onset of liver dysfunction. In these cases, at least 3 of the 8 supportive diagnostic findings were required. Patients with POLG mutations meeting the diagnostic features for childhood myocerebrohepatopathy spectrum were first described by Ferrari et al25 and de Vries et al26.
Myoclonus epilepsy myopathy sensory ataxia is an overlapping spectrum of disorders of myopathy, epilepsy, and ataxia in the absence of ophthalmoplegia with or without ragged red fibers.27 A subclinical sensory polyneuropathy leading to ataxia is usually the first sign of the disease. Usually seen later in the disease course, myoclonic seizures are often focal, frequently involving an arm, and can become generalized over time and comorbid with a progressive encephalopathy.
Ataxia-neuropathy resulting from mutations in POLG is an autosomal recessive disorder affecting patients in their mid-teens to later years, usually resulting in premature death. The disease is accompanied mainly by mtDNA deletions. The ataxia usually occurs in combination with various central nervous system features, including myoclonus, epilepsy, cognitive decline, nystagmus, dysarthria, thalamic and cerebellar white matter lesions on MRI, and evidence of neuronal loss in discrete gray nuclei.
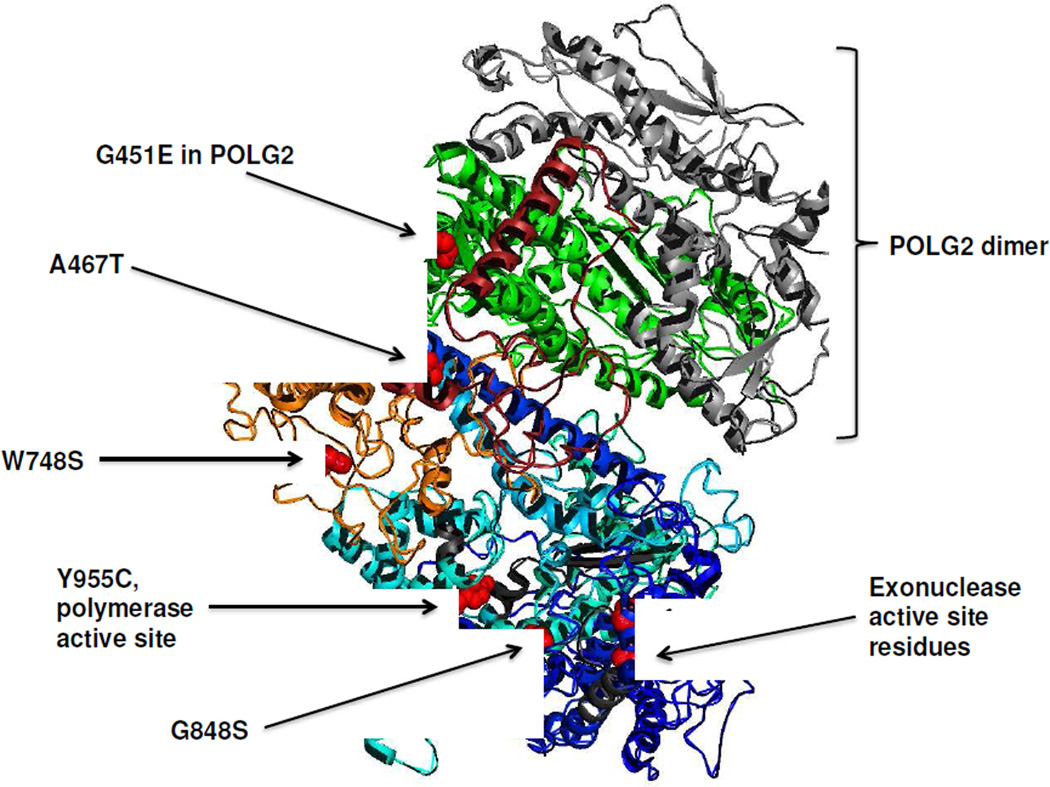
Biochemical and genetic analysis of POLG mutations has provided a useful understanding of the biochemical defect as well as prediction of the recessive or dominant nature of the mutation. A 3.2 Å structure of the human holoenzyme has been solved by Whitney Yin and coworkers (Figure 2).28 The most valuable benefit of this structure is the understanding of the subunit interface. The interaction of p140 with p55 is unique because of the asymmetric binding of the accessory subunit dimer with the catalytic subunit. The catalytic subunit extends an ‘arm’ partially around the p55 dimer using the first half of the linker region between the exonuclease and polymerase active sites. The structure also illuminates a subdomain composed of residues 1050–1090 that partially blocks the modeled DNA binding channel. The availability of this structure helps to predict the consequence of POLG mutations on function. Analysis of structure-function relationship of Alpers mutations has revealed that recessive mutations cluster within 5 distinct functional modules in the POLG catalytic subunit.29 This clustering can serve as a diagnostic tool to evaluate the consequence of new POLG mutations.
Figure 2.
Three-dimensional structure of the human DNA polymerase γ holoenzyme.28 The color scheme of the catalytic subunit is: light blue for the polymerase domain, dark blue for the exonuclease region, red for the accessory interacting domain, and orange for the intrinsic processivity domain. The 2 accessory subunits are colored in green for the proximal subunit and grey for the distal subunit. Common disease mutations and the active site residues in the polymerase and exonuclease are colored red and labeled accordingly.
The A467T mutation is the most common disease mutation in POLG and is estimated to occur in 36% of all alleles associated with POLG disease.9,24–26,30 In the general population, the frequency of the A467T mutation has been found to exist in 0.2%–1% of asymptomatic European populations.11,30–32 Ataxic patients who are homozygous for the A467T mutation present with symptoms in their early to late teens;31,33,34 however, patients have often been reported to be compound heterozygous with A467T and at least one other POLG disease-associated mutation. We found that the A467T mutant enzyme retains only 4% polymerase activity compared with wild-type enzymes and is also compromised for its interaction with the accessory subunit.35 In 2004, Naviaux and colleagues reported the genetic cause of Alpers syndrome by identification of the in trans recessive A467T with E873stop mutations.36 The POLG messenger RNAs (mRNAs) that contain the Glu873 nonsense mutation are degraded from the pool of mRNAs by nonsense mediated decay, resulting in mono-allelic expression of the other allele containing the A467T mutation.37
Haplotype analysis of the Finnish population demonstrates a carrier frequency of 1:125 for a common polymerase domain mutation, W748S.38 We showed that the W748S mutation caused the polymerase to have a low catalytic activity and a severe DNA-binding defect.39 The W748S mutation is often found in combination with E1143G in ataxia patients31,40 and was originally identified as a single-nucleotide polymorphism in 4% of the general population.33,41 However, biochemical evidence shows that the E1143G substitution partially reduces the deleterious effects of the W748S mutation, suggesting that E1143G may modulate the effects of disease mutations in POLG.39
With the exception of one mutation, all dominant POLG mutations responsible for developing progressive external ophthalmoplegia are mapped to the polymerase domain of pol γ. Three of the substitutions, H932Y, R943H, and Y955C, change side chains that interact directly with the incoming deoxynucleoside triphosphates (dNTP).23,42 These enzymes retain less than 1% of the wild-type polymerase activity and display a severe decrease in processivity,23 characteristics that likely cause the severe clinical presentation in R943H and Y955C heterozygotes. In addition, in vitro assays showed that the Y955C substitution also increases nucleotide misinsertion errors 10- to 100-fold in the absence of exonucleolytic proofreading.43 Alteration of this conserved tyrosine in the mitochondrial polymerase gene of Saccharomyces cerevisiae, mip1, enhanced mtDNA damage, increased mutagenesis, and resulted in cells with dysfunctional mitochondria.44,45
Similar to dominance seen in human patients, heterozygous yeast that alter the conserved tyrosine show a dominant increase in mitochondrial dysfunction, suggesting that the mutant enzymes compete with the wild-type polymerases for binding at the replication fork.45 Additionally, mouse transgenic model with the Y955C POLG allele targeted to the heart resulted in cardiomyopathy, loss of mtDNA, and enlarged hearts.46 These models strongly suggest that large reductions in pol γ polymerase activity are sufficient to cause mitochondrial dysfunction that is central to POLG-related disease. Lastly, the Y955C pol γ displays relaxed discrimination during incorporation of 8-oxo-dGTP or translesion synthesis opposite 8-oxo-dG.47
Other POLG disease-associated mutations have been assayed for polymerase activity to determine the potential to cause disease. The polymerase active site mutations G923D and A957S found in patients with progressive external ophthalmoplegia exhibited 21% and 23% polymerase activity, respectively.23 Analysis of a cluster of Alpers mutations in the thumb domain has shown a striking correlation with the severity of the defect and the degree of conservation of amino acid sequences among various eukaryotes.48 Mutations in the most conserved sites represented by G848S, T851A, R852C, and R853Q exhibited less than 1% wild-type enzyme activity.48 Mutations in codons for less conserved amino acids (Gln879 and Thr885) resulted in only moderate reduction in activity.48
To better understand the consequences of POLG disease-associated mutations in vivo, 31 mutations in the conserved regions of the S. cerevisiae MIP1 gene were altered to the disease mutation found in humans.42 Twenty mip1 mutant enzymes were shown to disrupt mtDNA replication and were sufficient to cause disease. Previously uncharacterized sporadic mutations, Q308H, R807C, G1076V, R1096H, and S1104C, caused decreased polymerase activity leading to mtDNA depletion and mitochondrial dysfunction. Despite mutations residing in the exonuclease region, it was found that point mutagenesis by these mutations plays a limited role in mitochondrial dysfunction and disease progression. Instead, most mitochondrial defective mip1 mutants displayed reduced or depleted mtDNA. In general, the severity of the phenotype of the mip1 mutant strain correlated with the age of onset of disease associated with the human ortholog.
Role of the Exonuclease in Polymerase Gamma
The intrinsic 3' to 5' exonuclease activity that contributes to replication fidelity can be completely eliminated by substitution of alanine for Asp198 and Glu200 in the conserved ExoI motif of human pol γ.49 Comparing the in vitro error rates for the exonuclease proficient and deficient forms of pol γ indicates that exonucleolytic proofreading contributes at least 20-fold to the fidelity of mtDNA synthesis.50 The exonuclease function is also necessary to suppress mtDNA deletions between direct repeats.51
Several groups have constructed mouse models with disrupted pol γ exonuclease activity by creating knock-in mice homozygous for mutations that disrupted exonuclease function.52,53 These mice exhibit premature aging between 6 and 9 months, characterized by graying hair, loss of hair and hearing, curvature of the spine, enlarged hearts, and decreased body weight and bone density.52,53 While the progeroid phenotype of mice lacking pol γ exonuclease activity implied a causative role of mtDNA mutations in aging,52,53 asymptomatic exonuclease deficient heterozygotic mice accumulate 500-fold more point mutations than aged wild-type mice,54 suggesting that mtDNA point mutagenesis does not limit life span and that mitochondria are rather tolerant of point mutagenesis up to a point, as the homozygous mouse progeroid phenotype was associated with 2000-fold more point mutations. This also demonstrates that disruption of exonuclease activity is not embryonic lethal, as compare to the knockout of the entire POLG gene function.55
POLG2, the gene for the p55 Accessory Subunit of Pol γ
Using a partial human POLG2 clone identified during the cloning of the Drosophila accessory subunit,56 the full-length cDNA of the human accessory subunit was isolated, overexpressed, and protein purified from E. coli.4 The function of the accessory subunit was demonstrated to impart high processivity on the pol γby increasing the affinity to DNA.4 The mammalian accessory subunit is a 55 kDa protein (p55) that forms a homodimer.57,58 The C-terminal region of one monomer (proximal subunit) in the dimer makes the majority of contact with the catalytic subunit, primarily with the linker region of the catalytic subunit.28
Mutation of the POLG2 gene is rarely described as compared with mutations in the gene for the catalytic subunit. The first mutation described (c.1352G>A; G451E) was in a patient with late-onset autosomal dominant progressive external ophthalmoplegia with multiple mtDNA deletions in muscle and ptosis.59 The G451E substitution was in a region of the p55 protein that interacts with the p140 subunit (Figure 2), and caused a decreased in processivity of the enzyme complex due to a compromised p55–p140 subunit interaction.59 The second case also involved a patient with late-onset autosomal dominant progressive external ophthalmoplegia with mtDNA deletions who harbored a c.1207–1208ins24 mutation, causing mis-splicing and skipping of exon 7, thus impairing the C-terminal domain required for enzyme processivity.13
In an exploration of nuclear genes for mtDNA maintenance linked to mitochondrial disease, 8 heterozygous mutations (7 novel) in POLG2 were identified in a cohort of 112 patients suspected of POLG involvement but absent of POLG mutations.60 These 7 novel mutations (G103S, L153V, P205R, R369G, D386E, S423Y, and L475DfsX2) encode amino acid substitutions that map throughout the protein, including the p55 dimer interface and the C-terminal domain that interacts with the catalytic subunit. Recombinant proteins harboring these alterations were assessed for stimulation of processive DNA synthesis, binding to the p140 catalytic subunit, binding to double-stranded DNA and self-dimerization. Whereas the G103S, L153V, D386E, and S423Y proteins displayed wild-type behavior, the P205R and R369G p55 variants had reduced stimulation of processivity and decreased affinity for the catalytic subunit.60 The R369G mutation was subsequently found to be associated with autosomal dominant progressive external ophthalmoplegia in a patient displaying multiple mtDNA deletions and COX-deficient muscle fibers.61 The L475DfsX2 variant, which possesses a C-terminal truncation, was unable to bind the p140 catalytic subunit and unable to bind dsDNA, and formed aberrant oligomeric complexes. The failure to enhance processivity in the catalytic subunit by these mutant variants would cause the complex to stall during DNA replication and is consistent with the accumulation of mtDNA deletions detected in progressive external ophthalmoplegia.
Although the catalytic subunit has been shown to be essential for embryo development,55 genetic data regarding the accessory subunit has been lacking in mammalian systems. To address the role of POLG2, we generated heterozygous (Polg2(+/−)) and homozygous (Polg2(−/−)) knockout (KO) mice.62 Polg2(+/−) mice are haplosufficient and developed normally with no discernible difference in mitochondrial function through 2 years of age. In contrast, the Polg2(−/−) was embryonic lethal at day 8.0–8.5 p.c. with concomitant loss of mtDNA and mtDNA gene products. This finding was similar to the mouse knockout of the POLG catalytic subunit.55 Electron microscopy showed severe ultrastructural defects and loss of organized cristae in mitochondria of the Polg2(−/−) embryos as well as an increase in lipid accumulation compared with both wild-type (WT) and Polg2(+/−) embryos. This data indicates that Polg2 function is crucial to mammalian embryogenesis and mtDNA replication, but that a single copy of Polg2 is sufficient to sustain life.
Twinkle Helicase, C10orf2, the Mitochondrial DNA Helicase
The mitochondrial helicase, referred to as the Twinkle helicase, and encoded by the C10orf2 gene (sometimes referred as PEO1) was originally identified by Spelbrink and coworkers in 2001.63 The protein shows striking homology to the T7 helicase-primase sequence, but lacks specific primase motifs. Missense mutations in the human C10orf2 gene co-segregate with mitochondrial diseases such as adult-onset progressive external ophthalmoplegia, hepatocerebral syndrome with mtDNA depletion syndrome, and infantile-onset spinocerebellar ataxia. Screening of the C10orf2 gene in individuals with autosomal dominant progressive external ophthalmoplegia, associated with multiple mtDNA deletions, identified 11 different mutations that co-segregated with the disorder in 12 affected families.63 At least 23 additional missense mutations in C10orf2 associated have been reported in autosomal dominant progressive external ophthalmoplegia.64,65 Although mutations in C10orf2 are mainly associated with autosomal dominant progressive external ophthalmoplegia, several reports have described recessive mutations as a cause of either epileptic encephalopathy with mtDNA depletion or infantile-onset spinocerebellar ataxia.66–68
Expression of this protein in baculovirus, purification, and characterization has verified that the gene functions as a DNA helicase.69 Further studies with pol γ and single-stranded binding protein demonstrated that these 3 protein complexes act in concert to carry out DNA replication, lending support for the role of Twinkle as the mtDNA replicative helicase.5 Overexpression of dominant disease variants of the mtDNA helicase in cultured human or Schneider cells results in stalled mtDNA replication or depletion of mtDNA,70–72 which emulates the disease state. Two of 5 autosomal dominant progressive external ophthalmoplegia mutants exhibited a dominant negative phenotype with mtDNA depletion in Schneider cells.71 Disease mutations in the linker region were shown to disrupt protein hexamerization and abolish DNA helicase activity.73 Four mutations in the N-terminal domain demonstrated a dramatic decrease in ATPase activity.74
A comprehensive study of recombinant disease variants overproduced and purified from E. coli has revealed that all of the disease variants display some level of activity where mild to moderate defects were seen variably over 20 different variants with defects in helicase activity, adenosine triphosphate hydrolysis and stability.12 All 20 mutant variants retain helicase function under optimized in vitro conditions despite partial reductions in DNA binding affinity, nucleotide hydrolysis, or thermal stability for some mutants. Such partial defects are consistent with the delayed presentation of mitochondrial diseases associated with mutation of C10orf2.
A mouse model of Twinkle deficiency has been produced by transgenic expression of Twinkle cDNA with the autosomal dominant disease mutation found in patients.75,76 The mouse developed progressive respiratory chain deficiency at 1 year of age in skeletal muscle, cerebellar Pukinje cells, and hippocampal neurons. The affected cells accumulated multiple mtDNA deletions. This ‘Deletor’ mouse model recapitulates many of the symptoms associated with progressive external ophthalmoplegia and provides a useful model for further study.
MGME1, a New Player on the Block
Two families exhibiting a severe, recessive multi-systemic mitochondrial disorder were described in a recent report.77 Causative mutations in mtDNA and known nuclear genes associated with mitochondrial disease were excluded by targeted conventional sequencing, prompting deep exome sequencing of affected families. Homozygous nonsense mutations in the orphan C20orf72 gene were identified in affected members of 2 families with heterozygous carrier parents and unaffected siblings. Renamed MGME1, the C20orf72 gene encodes a mitochondrial RecB-type exonuclease of the PD-(D/E)XK nuclease superfamily.
Cellular fractionation indicated mitochondrial localization and protease-resistance for the native protein, and confocal microscopy convincingly demonstrated mitochondrial localization of a green-fluorescent protein (GFP)-tagged recombinant form. Patient samples exhibited partial deletion and depletion of mtDNA, and the postulated direct involvement of MGME1 in maintenance of mtDNA and turnover of prematurely terminated 7S DNA replication intermediates is quite compelling. Indeed, patient fibroblasts (MGME1 null) depleted of mtDNA by continuous culture in the presence of 2’, 3’-dideoxycytidine failed to repopulate their mtDNA upon release from ddC, whereas wild-type fibroblasts were able to do so. The accumulation of mtDNA replication intermediates in HeLa cells subjected to MGME1 small interfering RNA (siRNA) was clearly demonstrated by 2D native agarose gel electrophoresis, further supporting a role for MGME1 in maintenance of mtDNA replication in vivo. Full-length, strep-tagged MGME1 protein was produced in HEK293 cells and purified by biotin-affinity chromatography. Preliminary qualitative characterization revealed the recombinant enzyme cleaves DNA but not RNA, requires a free 5’-end to a nucleic acid substrate, and prefers single-stranded DNA over double-stranded DNA in vitro.77 The MGME1 protein is postulated to have direct involvement in maintenance of mtDNA and turnover of prematurely terminated 7S mtDNA replication intermediates.
Conclusion
Mutations in nuclear genes that function to maintain our mitochondrial DNA are clearly common disease alleles in mitochondrial disease. Diseases of mtDNA stability are found in core proteins of mtDNA replication or in genes involved in supplying the mitochondrial nucleotide precursors needed for DNA replication (Table 1). With our awareness of disease mutations in these genes, the number of mitochondrial patients identified as having mutation in these genes will continue to increase. As an example, the number of individuals harboring a recessive pathogenic mutation in POLG has been estimated to approach 2% of the population.78 However, the varied polymorphic nature of these diseases as well as the age of presentation due to these gene mutations stumps our understanding and challenges clinician and researchers. Continued in vitro biochemistry and yeast models along with animal models are essential to understand the consequence of these disease mutations and to predict the in vivo consequences of newly identified mutations with these genes.
Acknowledgments
Funding
This work was supported by intramural funds from the National Institute of Environmental Health Sciences, National Institutes of Health (ES 065078 and ES 065080), and the National Institute of Neurological Disorders and Stroke (R13NS04925).
Footnotes
Presented at the Neurobiology of Disease in Children Symposium: Mitochondrial Disease, in conjunction with the 42nd Annual Meeting of the Child Neurology Society, Austin, TX, October 30, 2013. The author thanks Melanie Fridl Ross, MSJ, ELS, for editing this paper.
Declaration of Conflicting Interests
The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval
Not applicable
References
- 1.Miller FJ, Rosenfeldt FL, Zhang C, et al. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31(11):e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 3.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev. 2006;106(2):383–405. doi: 10.1021/cr040463d. [published online ahead of print December 31, 2005]. [DOI] [PubMed] [Google Scholar]
- 4.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274(53):38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 5.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J. 2004;23(12):2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 7.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 8.Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong LJ, Naviaux RK, Brunetti-Pierri N, et al. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum Mutat. 2008;29(9):E150–E172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saneto RP, Naviaux RK. Polymerase gamma disease through the ages. Dev Disabil Res Rev. 2010;16(2):163–174. doi: 10.1002/ddrr.105. [DOI] [PubMed] [Google Scholar]
- 11.Van Goethem G, Dermaut B, Lofgren A, et al. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28(3):211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 12.Longley MJ, Humble MM, Sharief FS, Copeland WC. Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis and helicase activity. J Biol Chem. 2010;285(39):29690–29702. doi: 10.1074/jbc.M110.151795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter MC, Czermin B, Muller-Ziermann S, et al. Late-onset ptosis and myopathy in a patient with a heterozygous insertion in POLG2. J Neurol. 2010;257(9):1517–1523. doi: 10.1007/s00415-010-5565-9. [DOI] [PubMed] [Google Scholar]
- 14.Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Tang S, Wang J, Lee NC, et al. Mitochondrial DNA polymerase {gamma} mutations: an ever expanding molecular and clinical spectrum. J Med Genet. 2011;48(10):669–681. doi: 10.1136/jmedgenet-2011-100222. [DOI] [PubMed] [Google Scholar]
- 16.Cohen BH, Chinnery PF, Copeland WC. GeneReviews at GeneTests: Medical Genetics Information Resource [database online] Seattle: University of Washington; 1997–2010. [March 16, 2010]. POLG-Related Disorders. Available at http://www.genetests.org. [Google Scholar]
- 17.Saneto RP, Lee IC, Koenig MK, et al. POLG DNA testing as an emerging standard of care before instituting valproic acid therapy for pediatric seizure disorders. Seizure. 2010;19(3):140–146. doi: 10.1016/j.seizure.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano M, Marti R, Ferreiro-Barros C, et al. Defects of intergenomic communication: autosomal disorders that cause multiple deletions and depletion of mitochondrial DNA. Semin Cell Dev Biol. 2001;12(6):417–427. doi: 10.1006/scdb.2001.0279. [DOI] [PubMed] [Google Scholar]
- 19.Zeviani M, Servidei S, Gellera C, et al. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339(6222):309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 20.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 21.Servidei S, Zeviani M, Manfredi G, et al. Dominantly inherited mitochondrial myopathy with multiple deletions of mitochondrial DNA: clinical, morphologic, and biochemical studies. Neurology. 1991;41(7):1053–1059. doi: 10.1212/wnl.41.7.1053. [DOI] [PubMed] [Google Scholar]
- 22.Suomalainen A, Majander A, Wallin M, et al. Autosomal dominant progressive external ophthalmoplegia with multiple deletions of mtDNA: clinical, biochemical, and molecular genetic features of the 10q-linked disease. Neurology. 1997;48(5):1244–1253. doi: 10.1212/wnl.48.5.1244. [DOI] [PubMed] [Google Scholar]
- 23.Graziewicz MA, Longley MJ, Bienstock RJ, et al. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat Struct Mol Biol. 2004;11(8):770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen KV, Sharief F, Chan SSL, et al. Molecular diagnosis of Alpers syndrome. J Hepatology. 2006;45:108–116. doi: 10.1016/j.jhep.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 25.Ferrari G, Lamantea E, Donati A, et al. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain. 2005;128(Pt4):723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- 26.de Vries MC, Rodenburg RJ, Morava E, et al. Multiple oxidative phosphorylation deficiencies in severe childhood multi-system disorders due to polymerase gamma (POLG1) mutations. Eur J Pediatr. 2007;166(3):229–234. doi: 10.1007/s00431-006-0234-9. [DOI] [PubMed] [Google Scholar]
- 27.Van Goethem G, Mercelis R, Lofgren A, et al. Patient homozygous for a recessive POLG mutation presents with features of MERRF. Neurology. 2003;61(12):1811–1813. doi: 10.1212/01.wnl.0000098997.23471.65. [DOI] [PubMed] [Google Scholar]
- 28.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139(2):312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Euro L, Farnum GA, Palin E, et al. Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol {gamma} Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath R, Hudson G, Ferrari G, et al. Phenotypic spectrum associated with mutations of the mitochondrial polymerase {gamma} gene. Brain. 2006;129(7):1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- 31.Winterthun S, Ferrari G, He L, et al. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology. 2005;64(7):1204–1208. doi: 10.1212/01.WNL.0000156516.77696.5A. [DOI] [PubMed] [Google Scholar]
- 32.Luoma PT, Luo N, Loscher WN, et al. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum Mol Genet. 2005;14(14):1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- 33.Van Goethem G, Luoma P, Rantamaki M, et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63(7):1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- 34.Tzoulis C, Engelsen BA, Telstad W, et al. The spectrum of clinical disease caused by the A467T and W748S POLG mutations: a study of 26 cases. Brain. 2006;129(7):1685–1692. doi: 10.1093/brain/awl097. [DOI] [PubMed] [Google Scholar]
- 35.Chan SSL, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 36.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers' syndrome and mitochondrial DNA depletion. Ann Neurol. 2004;55(5):706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 37.Chan SSL, Longley MJ, Naviaux RK, Copeland WC. Mono-allelic POLG expression resulting from nonsense-mediated decay and alternative splicing in a patient with Alpers syndrome. DNA Repair. 2005;4(12):1381–1389. doi: 10.1016/j.dnarep.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Hakonen AH, Heiskanen S, Juvonen V, et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am J Hum Genet. 2005;77(3):430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan SS, Longley MJ, Copeland WC. Modulation of the W748S mutation in DNA polymerase gamma by the E1143G polymorphism in mitochondrial disorders. Hum Mol Genet. 2006 Dec 1;15(23):3473–3483. doi: 10.1093/hmg/ddl424. Epub 2006 Nov 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakonen AH, Davidzon G, Salemi R, et al. Abundance of the POLG disease mutations in Europe, Australia, New Zealand, and the United States explained by single ancient European founders. Eur J Hum Genet. 2007;15(7):779–783. doi: 10.1038/sj.ejhg.5201831. [DOI] [PubMed] [Google Scholar]
- 41.Di Fonzo A, Bordoni A, Crimi M, et al. POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions. Hum Mutat. 2003;22(6):498–499. doi: 10.1002/humu.9203. [DOI] [PubMed] [Google Scholar]
- 42.Stumpf JD, Bailey CM, Spell D, Stillwagon M, Anderson KS, Copeland WC. mip1 Containing mutations associated with mitochondrial disease causes mutagenesis and depletion of mtDNA in Saccharomyces cerevisiae. Hum Mol Genet. 2010;19(11):2123–2133. doi: 10.1093/hmg/ddq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponamarev MV, Longley MJ, Nguyen D, et al. Active site mutation in DNA polymerase gamma associated with progressive external ophthalmoplegia causes error-prone DNA synthesis. J Biol Chem. 2002;277(18):15225–15228. doi: 10.1074/jbc.C200100200. [DOI] [PubMed] [Google Scholar]
- 44.Baruffini E, Lodi T, Dallabona C, et al. Genetic and chemical rescue of the Saccharomyces cerevisiae phenotype induced by mitochondrial DNA polymerase mutations associated with progressive external ophthalmoplegia in humans. Hum Mol Genet. 2006;15(19):2846–2855. doi: 10.1093/hmg/ddl219. [DOI] [PubMed] [Google Scholar]
- 45.Stuart GR, Santos JH, Strand MK, et al. Mitochondrial DNA defects in Saccharomyces cerevisiae with mutations in DNA polymerase gamma associated with progressive external ophthalmolplegia. Hum Mol Genet. 2006;15(2):363–374. doi: 10.1093/hmg/ddi454. Epub 2005 Dec 20. [DOI] [PubMed] [Google Scholar]
- 46.Lewis W, Day BJ, Kohler JJ, et al. MtDNA depletion, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graziewicz MA, Bienstock RJ, Copeland WC. The DNA polymerase {gamma} Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2'-deoxyguanosine. Hum Mol Genet. 2007;16(22):2729–2739. doi: 10.1093/hmg/ddm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasiviswanathan R, Longley MJ, Chan SS, Copeland WC. Disease mutations in the human mitochondrial DNA polymerase thumb subdomain impart severe defects in MtDNA replication. J Biol Chem. 2009;284(29):19501–19510. doi: 10.1074/jbc.M109.011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longley MJ, Ropp PA, Lim SE, Copeland WC. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37(29):10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 50.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276(42):38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 51.Stumpf JD, Copeland WC. The exonuclease activity of the yeast mitochondrial DNA polymerase gamma suppresses mitochondrial DNA deletions between short direct repeats in Saccharomyces cerevisiae. Genetics. 2013;194(2):519–522. doi: 10.1534/genetics.113.150920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 53.Kujoth GC, Hiona A, Pugh TD, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 54.Vermulst M, Bielas JH, Kujoth GC, et al. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39(4):540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 55.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14(13):1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 56.Fan L, Sanschagrin PC, Kaguni LS, Kuhn LA. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc Natl Acad Sci U S A. 1999;96(17):9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell. 2001;7(1):43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 58.Fan L, Kim S, Farr CL, et al. A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase. J Mol Biol. 2006;358(5):1229–1243. doi: 10.1016/j.jmb.2006.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Longley MJ, Clark S, Yu Wai Man C, et al. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am J Hum Genet. 2006;78(6):1026–1034. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young MJ, Longley MJ, Li FY, et al. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20(15):3052–3066. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Craig K, Young MJ, Blakely EL, et al. A p.R369G POLG2 mutation associated with adPEO and multiple mtDNA deletions causes decreased affinity between polymerase gamma subunits. Mitochondrion. 2011;12(2):313–319. doi: 10.1016/j.mito.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Humble MM, Young MJ, Foley JF, et al. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum Mol Genet. 2013;22(5):1017–1025. doi: 10.1093/hmg/dds506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spelbrink JN, Li FY, Tiranti V, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28(3):223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 64.Van Hove JL, Cunningham V, Rice C, et al. Finding twinkle in the eyes of a 71-year-old lady: a case report and review of the genotypic and phenotypic spectrum of TWINKLE-related dominant disease. Am J Med Genet A. 2009;149A(5):861–867. doi: 10.1002/ajmg.a.32731. [DOI] [PubMed] [Google Scholar]
- 65.Fratter C, Gorman GS, Stewart JD, et al. The clinical, histochemical, and molecular spectrum of PEO1 (Twinkle)-linked adPEO. Neurology. 2010;74(20):1619–1626. doi: 10.1212/WNL.0b013e3181df099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson G, Deschauer M, Busse K, et al. Sensory ataxic neuropathy due to a novel C10Orf2 mutation with probable germline mosaicism. Neurology. 2005;64(2):371–373. doi: 10.1212/01.WNL.0000149767.51152.83. [DOI] [PubMed] [Google Scholar]
- 67.Hakonen AH, Isohanni P, Paetau A, et al. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain. 2007;130(Pt 11):3032–3040. doi: 10.1093/brain/awm242. [DOI] [PubMed] [Google Scholar]
- 68.Lonnqvist T, Paetau A, Valanne L, Pihko H. Recessive twinkle mutations cause severe epileptic encephalopathy. Brain. 2009;132(Pt 6):1553–1562. doi: 10.1093/brain/awp045. [DOI] [PubMed] [Google Scholar]
- 69.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5' ->3' DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278(49):48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 70.Wanrooij S, Goffart S, Pohjoismaki JL, et al. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35(10):3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsushima Y, Kaguni LS. Differential phenotypes of active site and human adPEO mutations in drosophila mitochondrial DNA helicase expressed in Schneider cells. J Biol Chem. 2007;282(13):9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goffart S, Cooper HM, Tyynismaa H, et al. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18(2):328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korhonen JA, Pande V, Holmlund T, et al. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol. 2008;377(3):691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 74.Holmlund T, Farge G, Pande V, et al. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim Biophys Acta. 2009;1792(2):132–139. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Tyynismaa H, Mjosund KP, Wanrooij S, et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102(49):17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 2009;10(2):137–143. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kornblum C, Nicholls TJ, Haack TB, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45(2):214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen BH, Naviaux RK. The clinical diagnosis of POLG disease and other mitochondrial DNA depletion disorders. Methods. 2010;51(4):364–373. doi: 10.1016/j.ymeth.2010.05.008. [DOI] [PubMed] [Google Scholar]