Abstract
Globally, hepatocellular carcinoma (HCC) accounts for 70–85% of primary liver cancers and ranks second in the leading cause of male cancer death. Serum alpha-fetoprotein (AFP), normally highly expressed in the liver only during fetal development, is reactivated in 60% of HCC tumors and associated with poor patient outcome. We hypothesize that AFP+ and AFP-tumors differ biologically. Multivariable analysis in 237 HCC cases demonstrates that AFP level predicts poor survival independent of tumor stage (p<0.043). Using microarray-based global microRNA profiling, we found that miR-29 family members were the most significantly (p<0.001) down-regulated miRNAs in AFP+ tumors. Consistent with miR-29’s role in targeting DNA methyltransferase 3A (DNMT3A), a key enzyme regulating DNA methylation, we found a significant inverse correlation (p<0.001) between miR-29 and DNMT3A gene expression suggesting that they might be functionally antagonistic. Moreover, global DNA methylation profiling reveals that AFP+ and AFP− HCC tumors have distinct global DNA methylation patterns and that increased DNA methylation is associated with AFP+ HCC. Experimentally, we found that AFP expression in AFP− HCC cells induces cell proliferation, migration and invasion. Over expression of AFP, or conditioned media from AFP+ cells, inhibits miR-29a expression and induces DNMT3A expression in AFP− HCC cells. AFP also inhibited transcription of the miR-29a/b-1 locus and this effect is mediated through c-MYC binding to the transcript of miR-29a/b-1. Further, AFP expression promotes tumor growth of AFP− HCC cells in nude mice.
Conclusion
our findings indicate that tumor biology differs considerably between AFP+ HCC and AFP− HCC and that AFP is a functional antagonist of miR-29, which may contribute to global epigenetic alterations and poor prognosis in HCC.
Keywords: DNMT3A, c-MYC, Epigenetics, Liver cancer, DNA methylation
Introduction
The worldwide incidence of HCC is currently estimated at nearly 750,000 new cases each year resulting in over 600,000 deaths annually and remains on the rise (1). Patients are typically diagnosed with late stage disease leading to poor survival rates. Two major risk factors are chronic hepatitis B (HBV) and hepatitis C virus (HCV) infections, which are responsible for 93% of cases in developing countries and 53% of cases in developed countries (1). Additional risk factors include chronic alcohol consumption, aflatoxin-B1 contaminated foods and other conditions that cause cirrhosis (2).
Patients at risk for HCC are screened and monitored for serum alpha-fetoprotein (AFP) levels. AFP is a molecular marker elevated (>1000ng/ml) in 60–75% of HCC patients making it the key biomarker used for HCC surveillance (3). Though it is used for surveillance and to assess patient risk, its low sensitivity makes it inadequate to detect all patients that will develop cancer (3). In fact, many cirrhotic patents develop HCC without any increase in AFP. In the cohort of HCC patients that we study, 40% have normal levels (<20ng/ml) of the protein. Currently, the function of AFP is not well understood and it is mainly thought of clinically as a diagnostic marker.
AFP is an oncofetal protein highly elevated during embryogenesis and detected mainly in the fetal liver and yolk sac (3). It is secreted through the cell membrane and part of the albuminoid gene family which also includes serum albumin, vitamin D binding protein and alpha-albumin (3–5). The synthesis of AFP decreases rapidly after birth and levels remain below 20 ng/mL in adults (3, 6). It has been shown that AFP binds and transports unsaturated fatty acids, estrogen, retinoids, steroids, flavanoids, heavy metals, dioxins, and bilirubin (4, 5). AFP also interacts with macrophages and inhibits natural killer cells (4). In addition, the oncofetal protein plays a role in the regulation of cell proliferation and tumor growth. However, evidence for both stimulatory and inhibitory effects on cell growth remains contradictory and may be estrogen dependent (4, 7).
Though the physical, chemical, and immunological properties of AFP have been well studied, the mechanisms underlying its biological function and its role in carcinogenesis remain unclear (3, 4). AFP is elevated in many HCC patients; however, levels are heterogeneous suggesting that the biology of AFP+ and AFP− tumors may be different (8). For example, AFP levels may be low in patients with early HCC but very high in patients who have cirrhosis without HCC (9).
Aberrant microRNA expression is a ubiquitous feature in an increasing number of cancers including HCC (10). Studies indicate that these microRNAs (miRNAs) are directly connected with epigenetic factors that regulate gene expression (11). MiRNAs are short, non-coding RNAs about 22 nucleotides in length that regulate the function of messenger RNA (mRNA) (12). Partial sequence homology allows miRNAs to bind to the 3’UTR of target mRNAs inhibiting translation or causing mRNA degradation (11, 12). Recently, a specific group of miRNAs have been designated “epi-miRNAs” as they target effectors of epigenetic machinery (13). In addition to their functional role, miRNAs show promise as biomarkers for early detection, prognosis, diagnosis and treatment subgroups (13–16).
In this study, we found that AFP+ and AFP− HCC cases were biologically different according to the miRNA, mRNA and methylation expression patterns. In addition, we reveal an important functional role of AFP in HCC. Not only is AFP inversely correlated to miR-29 in HCC tumor tissue, we demonstrate that AFP transcriptionally down regulates miR-29a through c-MYC. Identifying the molecular mechanisms underlying AFP+ tumors will help build an understanding of heterogeneity in HCC as well as our understanding of AFP’s functional role in HCC progression.
Experimental Procedures
Patient studies and tumor specimens
Paired tumor and non-tumor hepatic samples were obtained with informed consent and the collection of these samples for research were approved by the Institutional Review Board of the Liver Cancer Institute and Zhongshan Hospital (Fudan University, Shanghai, China) as described in previous studies (17, 18). Refer to Experimental Procedures in the Supplemental Material for details.
Xenograft study
The animal study protocol was approved by the NIH Animal Care and Use Committee and all animals received human care according to the NIH “Guide for Care and Use of Laboratory Animals.” Refer to Experimental Procedures in the Supplemental Material for details.
Plasmids, lentiviral vectors, siRNA, qRT-PCR, conditioned media study design, protein expression analysis, cell proliferation assays, ChiP assay, and all statistical analysis
Please refer to Experimental Procedures in the Supplementary Materials.
Results
AFP+ HCC patients have a distinct genomic profile that is linked to poor survival
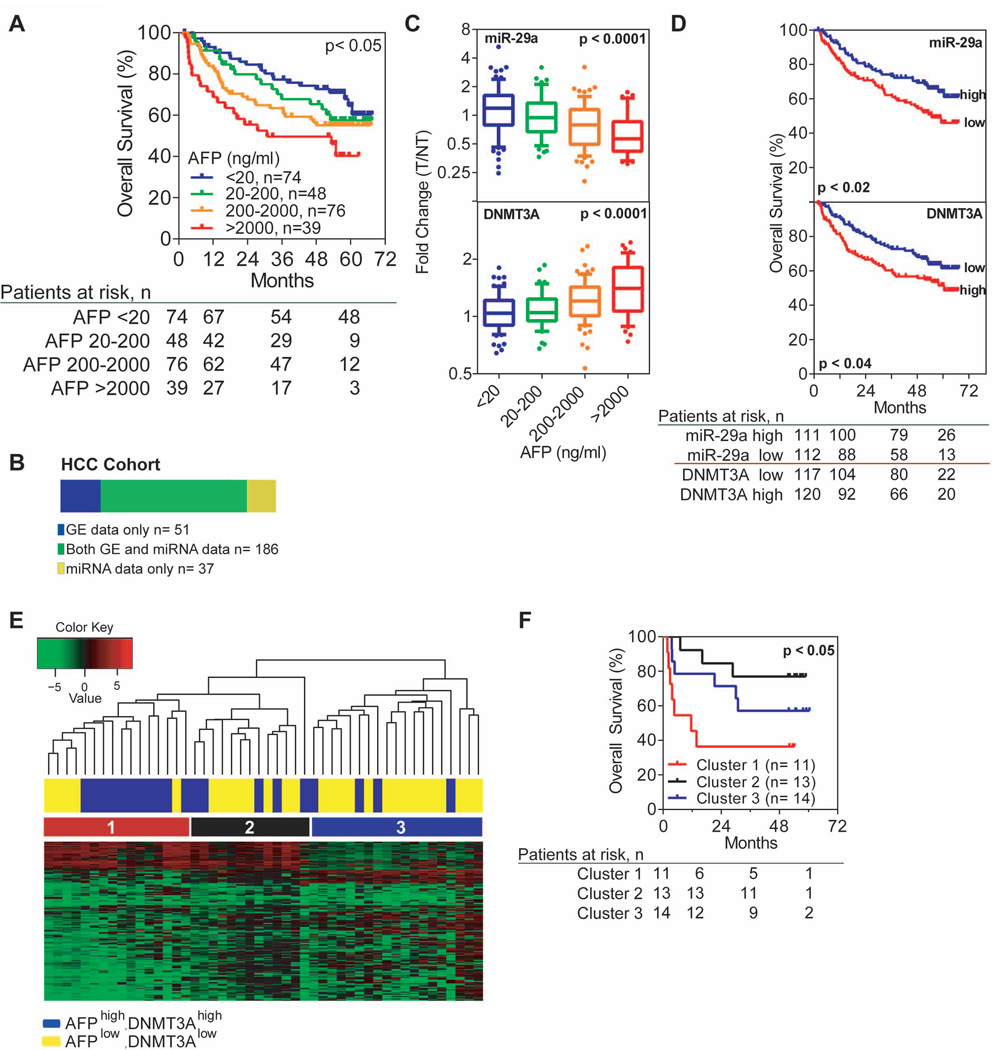
In our cohort, we subdivided HCC patients into four groups based on serum AFP levels starting at a normal level (<20ng/ml) and increasing ten-fold (i.e., 20–200, 200–2000, and >2000ng/ml) where patients with >2000ng/ml AFP were considered to be extremely high cases. Overall survival data confirms that poor clinical outcome is associated with increasing serum AFP suggesting that the biological make up of these HCCs differ from those that are AFP− (Figure 1A, Mantel-Cox p<0.05; log-rank p<0.005). Additional clinical characteristics of HCC cases in each subgroup can be found in Suppl. Table 1). To test whether AFP identifies a unique molecular subclass rather than late-stage tumors, we performed a multivariable analysis between AFP level and TNM staging, the only two variables from the univariable analysis that passed a stepwise selection process using both forward addition and backwards subtraction with a p-value cut-off of <0.05. Both AFP level and TNM staging were significant in the multivariable analysis indicating that AFP predicts poor overall survival independent of tumor stage (Table 1).
Figure 1.
AFP is inversely correlated with miR-29 and associated with increased DNA methyl transferase expression. A) HCC patients with high levels of serum AFP are associated with poor survival. Mantel-Cox p<0.05; log-rank p<0.05; n=237. B) The LCS cohort includes 274 HCC patients with matched tumor and non-tumor tissue samples. 186 patients (shown in green) have both mRNA expression and miRNA expression achieved by the Affymatrix gene expression array and OSU-CCC miRNA array, respectively. An additional 51 patients have gene expression data only (blue) and 37 patients have miRNA expression data only (yellow). C) miR-29a significantly decreases (top panel, n=223) and DNMT3A significantly increases (bottom panel, n=237) as serum AFP expression increases in HCC patients. D) Low expression of miR-29a (top panel, n=223) and increased DNMT3A (bottom panel, n=237) are associated with poor survival, respectively. E) Unsupervised hierarchical clustering of 48 HCC tumor samples reveals a unique methylation profile in patients with high AFP and DNMT3A gene expression. Manhattan clustering was used with a standard deviation cut-off of 2, which showed 211 probes to be differentially methylated. A median cut-off was used to determine high or low gene expression of AFP and DNMT3A for patient labeling. Fisher’s exact test confirms AFP high/DNMT3A high patients are enriched in cluster #1 (p<0.05). F) The three clusters observed in the unsupervised hierarchical clustering exhibit distinct differences in overall survival. Cluster #1, with predominantly AFP high and DNMT3A high patients, shows significantly worse overall survival than patients in cluster #2 or #3. Mantel-Cox p<0.05; n=48.
Table 1.
Univariable and multivariable analyses of factors associated with disease stage, aggressive HCC and survival (n= 237)
| Univariable analysisa | Multivariable analysisb | |||
|---|---|---|---|---|
| Clinical Variable | Hazard ratio (95% CI) |
p-value | Hazard ratio (95% CI) |
p-value |
| AFP (> 300 ng/mL vs ≤ 300 ng/mL) | 1.7 (1.1 – 2.5) | 0.011 | 1.6 (1.0 – 2.4) | 0.043 |
| Age (≥ 50 yr vs <50 yr) | 0.8 (0.5 – 1.2) | 0.26 | n.a. | |
| Sex (male vs female) | 1.8 (0.9 – 3.7) | 0.116 | n.a. | |
| Cirrhosis (yes vs no) | 5.2 (1.3 – 20.9) | 0.022 | n.a. | |
| Tumor size (> 3 cm vs ≤ 3 cm) | 2.4 (1.5 – 3.8) | < 0.001 | n.a. | |
| Multinodularity | 1.6 (1.0 – 2.5) | 0.046 | n.a. | |
| Vascular invasion | 1.8 (1.2 – 2.9) | 0.007 | n.a. | |
| TNM staging (II–III vs I)c | 2.9 (1.8 – 4.8) | < 0.001 | 2.9 (1.8 – 4.8) | < 0.001 |
Univariable analysis, Cox proportional hazards regression.
Multivariable analysis, Cox proportional hazards regression.
Stages II and III were combined because of the presence of vascular invasion at these stages.
CI, confidence interval.
n.a., not applicable.
Affymetrix mRNA expression data are available for 237 paired tumor and non-tumor tissue samples as described in earlier studies (17–19), while miRNA expression data are available on a subset of patients (n=223) in our cohort (Figure 1B). Global DNA methylation profiles were also obtained for a subset of HCC patients who have mRNA expression data.
We next determined if AFP+ HCC may have different miRNA expression. We performed a class comparison analysis between 74 patients with normal serum AFP (<20ng/ml) and 39 patients with extremely high AFP levels (>2000ng/ml) and found that 18 miRNAs were differentially expressed using a p-value cut-off of 0.001 and a false discovery rate of 5% (Table 2). Of the twelve down regulated miRNAs (ranked by fold change), seven were in the miR-29 family. All members of the miR-29 family were also associated with poor survival in HCC (Suppl. Table 2). Interestingly, when examining available array CGH data (20), none of the miR-29 family members are located in frequent genomic areas of loss suggesting that miR-29 repression in HCC may be at the epigenetic or transcriptional level (Table 2).
Table 2.
The miR-29 family is differentially expressed between patients with normal levels versus those with extremely high levels of serum AFP
| Raw Intensity Valueb | ||||||||
|---|---|---|---|---|---|---|---|---|
| miRNA Probea |
AFP >2000ng/ulc |
AFP <20ng/uld |
Fold Change |
Parametric p-value |
FDR | Chromosome Location |
Gain (%)e |
Loss (%)e |
| miR-29a (1) | 1230.14 | 2193.45 | 0.56 | < 0.00001 | 0.0001 | 7q32 | 6.6 | 1.3 – 2.6 |
| miR-29a (2) | 1542.24 | 2730.72 | 0.56 | < 0.00001 | 0.0004 | 7q32 | 6.6 | 1.3 – 2.6 |
| miR-29b-2 | 1756 | 3106.81 | 0.57 | < 0.00001 | 0.0001 | 1q32 | 34.2 | 1.3 |
| miR-29b-1 (1) | 1477.04 | 2545.82 | 0.58 | < 0.00001 | 0.0004 | 7q32 | 6.6 | 1.3 – 2.6 |
| miR-122a (1) | 16063.12 | 27088.34 | 0.59 | < 0.00001 | 0.0059 | 18q21 | 0 | 6.6 |
| miR-29b-1 (2) | 1377.04 | 2242.49 | 0.61 | < 0.00001 | 0.0100 | 7q32 | 6.6 | 1.3 – 2.6 |
| miR-29b-1 (2) | 960.32 | 1559.23 | 0.62 | < 0.00001 | 0.0040 | 7q32 | 6.6 | 1.3 – 2.6 |
| miR-29c | 1418.1 | 2247.65 | 0.63 | < 0.00001 | 0.0059 | 1q32 | 34.2 | 1.3 |
| miR-125b-1 (1) | 1613.31 | 2482.49 | 0.65 | 0.00110 | 0.0328 | 11q24 | 1.3 | 6.6 |
| miR-125b-1 (2) | 1448.15 | 2194.68 | 0.66 | 0.00120 | 0.0315 | 11q24 | 1.3 | 6.6 |
| miR-122a (2) | 29564.15 | 40839.46 | 0.72 | 0.00070 | 0.0250 | 18q21 | 0 | 6.6 |
| miR-let-7d | 1116.67 | 1429.47 | 0.78 | 0.00030 | 0.0233 | 9q22 | 2.6 | 13.2 |
| miR-let-7f | 679.58 | 862.59 | 0.79 | 0.00060 | 0.0250 | 9q22 | 2.6 | 13.2 |
| miR-181b-2 (1) | 2216.1 | 1622.01 | 1.37 | 0.00010 | 0.0059 | 9q33 | 5.3 | 10.5 |
| miR-181c | 1279.19 | 930.72 | 1.37 | 0.00010 | 0.0093 | 19q13 | 9.2 – 10.5 | 3.9 |
| miR-181b-1 | 1807.52 | 1193.32 | 1.51 | 0.00010 | 0.0044 | 1q32 | 34.2 | 1.3 |
| miR-181b-2 (2) | 2289.77 | 1470.03 | 1.56 | < 0.00001 | 0.0001 | 9q33 | 5.3 | 10.5 |
| miR-32 | 1891.71 | 1143.94 | 1.65 | 0.00010 | 0.0059 | 9q31 | 2.6 | 13.2 |
The miRNA array includes multiple spots for some miRNAs therefore there are multiple probe readouts. Additional probes for miRNAs are numbered in parenthesis after the probe name.
Mean values for miRNA expression are shown for AFP high and normal cases and were used to calculate fold change.
High AFP, n=40
Low AFP, n=74
Percent gain and loss were estimated by copy-number variation (CNV) data from 36 HCC tumor tissue samples. In cases where the miRNA is not in a gene, the genes upstream and downstream of the miRNA were used to estimate CNV.
We found that there is an inverse correlation between miR-29 and AFP where miR-29a decreased significantly as serum AFP levels increased (Figure 1C, top panel; miR-29b/c data in Suppl. Figure 1A). This coincides with an association between low levels of miR-29a and poor overall survival (Figure 1D, top panel; miR-29b/c data in Suppl. Figure 1B). Since miR-29 targets DNMT3A (21), we also examined DNMT3A expression in all 237 HCC cases included in the gene expression microarray. We found that DNMT3A is negatively correlated with the miR-29 family (r= −0.41; −0.36, −0.35 respectively; p<0.0001; Suppl. Figure 2A–C). DNMT3A showed the opposite trend of miR-29 as it increased significantly with serum AFP and high levels are associated with poor overall survival (bottom panels of Figure 1C and 1D). As a control, we compared AFP gene expression levels to AFP serum levels in patients and found a positive correlation (r= 0.66, p<0.0001, Suppl. Figure 2D). In addition, 10% of patients in the miRNA expression dataset were randomly selected to validate their miR-29a expression by quantitative RT-PCR. Suppl. Figure 2E shows a positive correlation between miR-29a expression in the microarray data and the expression detected by RT-PCR (r= 0.75, p<0.001).
Since DNMT3A expression is positively correlated with AFP, and associated with poor overall survival, we hypothesized that the global methylation profile of HCC patients with high AFP and DNMT3A levels would differ from patients with low AFP and DNMT3A. To test our hypothesis, we analyzed a subset of HCC cases (n=48) with Illumina 27k DNA methylation arrays. Unsupervised hierarchical clustering of AFP high, DNMT3A high (HH) cases (n=20) and AFP low, DNMT3A low (LL) cases (n=28) reveals that tumors with high AFP/DNMT3A expression (HH) are enriched in cluster #1 and have similar methylation profiles (Figure 1E). We performed a Fisher’s exact test and found that HH cases were significantly enriched in cluster #1 (p=0.012) compared to clusters #2 and #3. Consistent with AFP and DNMT3A data (Figure 1C and 1D, bottom panels), the patients in cluster #1 with high AFP/DNMT3A expression show significant survival differences when compared to patients in clusters #2 and #3 with predominantly low AFP/DNMT3A expression (Mantel-Cox p<0.05; Figure 1F). Here, we questioned whether AFP mRNA levels were associated with aberrant methylation. To test this we analyzed CpG island methylation on the AFP promoter in all 48 HCC patients in the methylation array (Suppl. Table 3). Using a quartile cut-off of tumor-specific AFP methylation, we compared low and high AFP methylation status to AFP mRNA expression in the same HCC cases and found no significant difference (Suppl. Figure 2F).
The inverse correlation between AFP and the miR-29 family provoked our curiosity as to whether the trend in cancer existed in normal physiological conditions. As an oncofetal protein, AFP is known to be highly expressed during development then decrease to <20ng/mL in adult serum. The same trend was observed when fetal tissue was compared to adult normal liver tissue (Suppl. Figure 3A). Conversely, the miR-29 family was hardly detectable in the fetal liver but highly expressed in adult normal liver tissue (Suppl. Figure 3B) whereas DNMT3A expression showed an increase in the fetal liver (Suppl. Figure 3C). These trends were also apparent in mouse fetal and adult normal liver tissue over time (Suppl. Figure 3D). In the mouse liver, the miR-29 family was minimally expressed until after birth when it began to rise significantly. Concurrent with the rise in miR-29, AFP begins to decrease and a switch point is observed at 1 week. It is this switch point in expression that led us to explore AFP’s role in functionally regulating miR-29 expression in HCC.
AFP expression transcriptionally regulates miR-29a
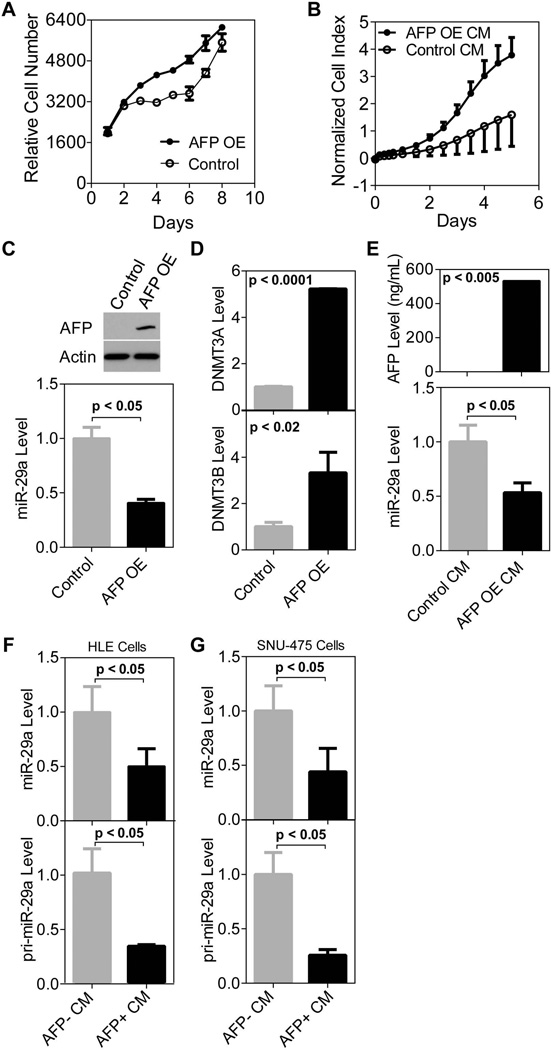
AFP has previously been shown to have an effect on HCC cell growth but the findings have been contradictory (4). We show that AFP can promote cell proliferation when over expressed in AFP− HLE cells (Figure 2A). In addition, HLE cells proliferate faster in the presence of conditioned media (CM) taken from HLE cells over expressing AFP compared to cells growing in CM taken from HLE cells transfected with an empty vector (Figure 2B, additional AFP negative cell line included in Suppl. Figure 4A).
Figure 2.
AFP transcriptionally regulates miR-29. A) Over expression of AFP significantly increases cell proliferation compared to control HLE cells after a 48hr transient transfection (p<0.0005 days 3–6 and p<0.005 days 7 and 8). Cell growth was monitored using a CalceinAM assay with 4 replicates per time point and error bars represent standard deviation. B) HLE cells in the presence of conditioned media taken from AFP over expressing HLE cells (AFP OE CM) proliferate faster than those in AFP− CM (taken from HLE cells transfected with an empty vector, Control CM). Cells were seeded on day 0, CM was applied on day 1 and cell growth was monitored by xCELLigence technology over a period of five days. There are four replicates per time point and error bars represent standard deviation. The growth rate of HLE cells growing in the presence of AFP is significantly faster on days 1–5 (p<0.05). C) Over expression of AFP in HLE cells leads to decreased mature miR-29a expression compared to control HLE cells (48 hour transient transfection). Top panel shows AFP protein expression by western blot. miR-29a expression was quantified using qRT-PCR in the bottom panel. D) DNMT3A and DNMT3B levels significantly increased after transient AFP over expression (48 hours) in HLE cells. E) Conditioned media taken from transfected cells was applied to HLE cells. AFP OE CM led to a significant decrease in mature miR-29a measured by qRT-PCR as compared to CM from control cells. ELISA data in the top panel shows that media taken from cells over expressing AFP has >500ng/ml of AFP present. F) AFP+ CM taken from HUH-7 cells led to a decrease in both the mature (top panel) and primary transcript (bottom panel) of miR-29a in HLE cells as measured by qRT-PCR. G) AFP+ CM from HUH-7 cells also lead to a decrease in miR-29a mature (top panel) and primary transcript (bottom panel) expression in SNU-475 cells.
Next, to determine if AFP functionally modifies miR-29a expression we transiently over expressed AFP in HLE cells (Figure 2C, top panel). A fifty percent reduction in mature miR-29a expression was observed in HCC cells over expressing AFP (Figure 2C, bottom panel; additional AFP negative cell line included in Suppl. Figure 4B). We also analyzed DNMT3A and DNMT3B expression in these cells and the levels of both DNMTs significantly increased after transient AFP over expression compared to control cells (Figure 2D and Suppl. Figure 4C). Next, we silenced AFP expression in HUH-7 cells using a lentiviral-shRNA construct. After a 96 hour transient infection with lentivirus, AFP protein expression decreased and we observed an induction of miR-29a expression (Suppl. Figure 4D). However, when we selected for cells that incorporated the control or shAFP lentiviral construct and compared their cell proliferation rate, we found no significant difference (Suppl. Figure 4E). Though the AFP protein level was reduced in the shAFP infected cells, abundant AFP remained in the media and was detected by ELISA as long as one week after antibiotic selection (Suppl. Figure 4F, media changed every three days). We reasoned that the AFP in the media may still be functional. The lack of AFP depletion in cultured media upon knockdown prompted us to only focus on adding AFP protein to AFP− cells either by over expression or AFP+ conditioned media. First, conditioned media was collected from AFP− cells over expressing AFP (referred to as AFP OE CM) and placed on freshly seeded HLE cells. Compared to HLE cells in the presence of AFP− conditioned media (referred to as Control CM), taken from HLE cells transfected with an empty vector, the AFP OE CM led to a significant decrease in mature miR-29a expression (Figure 2E, bottom panel). In addition to AFP OE CM, we applied conditioned media from HUH-7 cells, which highly express AFP (Suppl. Figure 4G), to two AFP negative HCC cell lines, HLE and SNU-475. The mature miR-29a level decreased at least fifty percent in each cell line (Figure 2F and 2G, top panel).
Then we questioned whether the regulation of miR-29 was transcriptional or through the processing of the miRNA. Though the OSU-CCC microarray includes over 1,700 probes for miRNAs, only 18 were significantly differentially expressed with a fold change greater than 20% in our class comparison analysis. Using a Taqman assay specific to a region on the miR-29a transcript, we tested whether or not expression of miR-29a decreased at the transcriptional level when AFP was present. Indeed, when the primary transcript of miR-29a was tested, a decrease was apparent in the presence of AFP suggesting a transcriptional method of regulation (Figure 2F and 2G, bottom panel; additional AFP negative cell line included in Suppl. Figure 4H).
c-MYC mediates the transcriptional down regulation of miR-29a
The above results reveal that AFP plays a functional role in down regulating miR-29a expression. Since AFP is a membrane-secreted protein and not abundant in the cell nucleus, we hypothesized that it acts through a transcription factor of miR-29a. To determine which transcription factors were activated in the presence of AFP we went back to the mRNA profiles of HCC patients and ran a class comparison analysis between 44 patients with high AFP, high DNMT3A, and low miR-29a (our phenotype of interest) versus 44 patients with low AFP, low DNMT3A, and high miR-29a expression using median cut-off. A total of 379 genes were differentially expressed with a fold change greater than 2 after 10,000 permutations were computed with an FDR of 1% and a p-value cut-off of 0.001. The gene list was imported into Ingenuity Pathway Analysis (IPA) to find transcription factors significantly activated or deactivated based on altered gene expression. The activation state of transcription factors is determined by comparing gene expression patterns to expression of transcriptional regulators in the input dataset and IPA then assigns each transcriptional regulator a z-score (more detailed information in Supplemental Experimental Procedures). In the gene list derived from our class comparison analysis, a total of eight transcription factors were assigned activated (Table 3) while twelve were inhibited with statistical significance (not shown). Among those activated were MYC and GLI1, both shown previously to bind the promoter of miR-29a/b-1 and inhibit its expression (22, 23). The miR-29a promoter region has been mapped to roughly 36kb upstream of the miR-29a/b-1 cluster (22, 23). In addition to MYC and GLI1, the UCSC Genome Browser displays chromatin immunoprecipitation (ChIP)-Sequencing data showing that FOXM1, E2F, and SP1 also have binding sites on the miR-29a/b-1 locus (Table 3). In our analysis, c-MYC seemed a likely transcription factor involved as it ranked second most activated by z-score and had been previously shown to inhibit miR-29a promoter expression experimentally.
Table 3.
Eight transcription factors are activated in patients with high AFP, high DNMT3A and low miR-29a expression.
| Transcription Factora | Ingenuity Regulation z-score |
Predicted Activation State |
p-value of Overlap |
UCSC ChIP- Seq binding sites? (#) |
|---|---|---|---|---|
| FOXM1 | 3.437 | Activated | 1.42E-07 | Yes (3) |
| MYC | 3.022 | Activated | 3.67E-11 | Yes (5) |
| TBX2 | 2.624 | Activated | 2.22E-05 | n.s |
| SREBF1 | 2.466 | Activated | 1.40E-06 | n.s |
| E2F | 2.398 | Activated | 1.53E-07 | Yes (1) |
| FOXO1 | 2.301 | Activated | 1.08E-13 | n.s. |
| SP1 | 2.165 | Activated | 2.24E-04 | Yes (6) |
| GLI1 | 2.138 | Activated | 1.18E-04 | n.s. |
Transcription factors were predicted in Ingenuity Pathway Analysis from 379 differentially expressed genes between HCC cases with high AFP/DNMT3A and low miR-29a expression versus cases with the opposite expression pattern.
n.s. = none shown
To determine if c-MYC regulated the transcript of miR-29a, we silenced c-MYC protein expression using siRNA in HLE cells (Figure 3A). HLE cells endogenously express c-MYC but are negative for AFP. Indeed, the absence of c-MYC induced expression of the miR-29a transcript compared to cells transfected only with an empty vector (Figure 3B). As a control we over expressed AFP and found that pri-miR-29a decreased as expected (Figure 3B). Interestingly, in the absence of c-MYC protein, AFP was unable to decrease miR-29a transcript expression (Figure 3B). To further test if AFP inhibits miR-29a through c-MYC, we designed primers around predicted c-MYC binding sites on the miR-29a/b-1 transcript (Figure 3C). A ChIP assay was used with anti-c-MYC to pull down genomic regions bound by c-MYC protein. Following ChIP, P-1 and P-2 primer sets were employed to quantify the amount of c-MYC bound to the specified regions on the miR-29a/b-1 transcript. Figure 3D shows that c-MYC was abundantly bound to the miR-29a/b-1 promoter and to the intron in the presence of AFP, but did not bind without AFP. Though the presence of AFP leads to increased binding of c-MYC to the miR-29a transcript, an increased steady state level of c-MYC protein during AFP overexpression was minimal (Figure 3A). To better understanding the possible mechanism by which AFP facilitates MYC binding to the miR-29a transcript, we conducted a set of experiments with cycloheximide (CHX). CHX interferes with the translation step during protein synthesis and effectively stops translational elongation. By treating cells first with AFP+ conditioned media (eight hours) then with CHX for 15 minutes, we found an accumulation of c-MYC protein (Suppl. Figure 5A), suggesting that AFP regulates the half-life of c-MYC.
Figure 3.
AFP mediates the down regulation of miR-29a through c-MYC. A) AFP is over expressed and c-MYC silenced 72 hours post-transfection in HLE cells. B) miR-29a expression is only silenced when AFP is present and c-MYC is functionally active. Upon c-MYC silencing, miR-29a expression is induced even in the presence of AFP. C) The miR-29a/b-1 transcript is shown with two sets of primers designed around c-MYC binding sites. P-1 is in the promoter region and P-2 located in the intron of the transcript. Transcript schematic is modified from that created by Chang et al (22). D) Chromatin Immunoprecipitation was performed to pull down c-MYC bound to genomic regions in HLE cells after AFP+ or AFP− CM was applied (rabbit IgG was used as a control). Both P-1 and P-2 primer sets were used to quantify the amount of c-MYC bound to two specified regions on the miR-29a transcript using qRT-PCR. Results show that c-MYC bound to both regions of miR-29a/b-1 only in the presence of AFP.
AFP expression promotes tumor growth in vivo
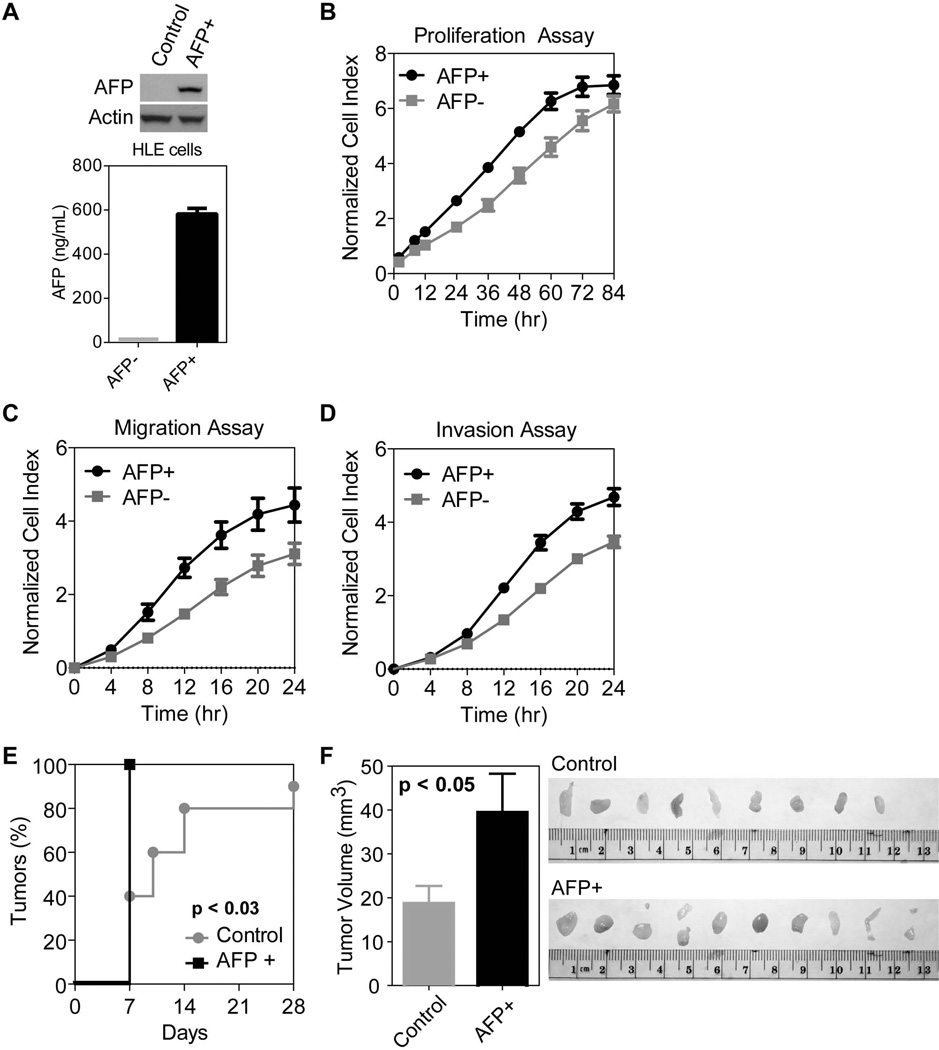
A stable AFP+ HLE cell line was created for a xenograft study in nude (Aythymic Nu/Nu) mice. Control and AFP+ HLE cells were infected with a lentiviral construct and selected using an antibiotic. Stable AFP+ HLE cells express AFP endogenously as well as in the media (Figure 4A). Consistently, stable AFP+ HLE cells proliferated much faster than control cells (Figure 4B). These cells also had faster migratory and invasive capacities than control cells (Figure 4C–D). Upon subcutaneous injection in nude mice, the group with subcutaneous injection of 0.5×106 AFP+ cells showed faster tumor incidence (Figure 4E) and enhanced tumor growth (Figure 4F). Similar results were obtained with 1×106 cells (data not shown).
Figure 4.
AFP expression promotes cell growth, cell migration and invasion in vitro and tumorigenesis in vivo. A) AFP expression detected by western (top) and ELISA (bottom) in stable AFP+ versus control HLE cells. AFP is only expressed and secreted in cells infected with a lentivirus incorporating the AFP gene. B) Stable AFP+ cells show more rapid proliferation compared to control cells. The growth rate of AFP+ cells is significantly faster on days 1–3 (p<0.01). There are three replicates per time point and error bars represent standard deviation. C) AFP expression induces increased migratory capacity, significantly different from control cells starting at 8 hours (p<0.01). There are three replicates per times point and error bars represent standard deviation. D) AFP expression also leads to increased invasion significantly different from control cells starting at 12 hours (p<0.01). There are three replicates per time point and error bars represent standard deviation. E) Tumor incidence is significantly faster in nude mice injected with 0.5×106 AFP+ cells AFP+ cells compared to 0.5×106 AFP− control HLE cells (p<0.05). After 1 week, all mice in the AFP+ group carried a tumor compared to less than half of the control group. F) Quantification of tumor volume (mm3) shows a significant difference between the size of control versus AFP+ tumors (p<0.05). Error bars represent mean plus standard error. Image of tumors extracted from the control and AFP+ groups after 4 weeks.
Discussion
AFP was first detected as a fetal-associated protein in 1957 and found to be tumor-associated six years later (24, 25). Almost a decade past before the protein was isolated and purified and it took yet another decade before biological studies were initiated (4).Currently, more than fifty years after its discovery, AFP is thought to exist mainly as a transport protein and biomarker for HCC. Though it has been shown to have growth regulatory properties and to bind and transport estrogen, its molecular mechanism in HCC is not well understood (4).
Recently, an interest in AFP and methylation has sparked. Zhang et. al examined the promoter methylation status of nine genes in 50 paired tumor and non-tumor HCC tissue samples (26). They defined a CpG island methylator phenotype (CIMP+) if five of the nine genes were concordantly methylated. Not only did this group observe a higher frequency of CIMP+ in tumor than in nontumor tissue, they found that CIMP+ was most frequent in HCC with elevated AFP (>30ng/ml) (26). In addition to this study, another group, Wu et. al, found similar results (27). They analyzed promoter methylation of seven genes in 65 HCC cases (CIMP+ if 3 or more genes were methylated) and found that CIMP+ was more prevalent in patients with >400ng/ml serum AFP. Furthermore, they observed that patients with CIMP+ often had multiple tumors and worse recurrence-free survival compared to CIMP-patients (27).
In our study, we find an association between AFP and methylation. Not only is AFP associated with increased expression of DNA methyltransferases, enzymes that catalyze the methylation process, AFP is also associated with increased methylation of many gene promoters. When we analyzed CpG island methylation of the AFP promoter itself, we found no association with mRNA expression in 48 HCC cases suggesting the gene is not epigenetically regulated in our HCC cohort and a feedback loop between the DNMT enzymes and AFP is not evident. Global methylation profiling of the same 48 HCC cases showed that a significant number of patients with high AFP and DNMT3A expression cluster together, suggesting that their poor outcome is driven by a common mechanism. Moreover, AFP was observed to induce cell growth in vitro and in vivo, a trait ubiquitous in cancer, and to increase the migratory and invasive properties of HCC cells indicating the oncofetal protein has a functional role in addition to its role as a biomarker. We have found AFP works by transcriptionally inhibiting miR-29a expression, which leads to the induction of DNMT3A, and we propose that AFP drives these epigenetic changes to shape the microenvironment in a way that promotes tumorigenesis. Based on this evidence, AFP, as an extracellular protein circulating in blood, changes the cell fate and tumorigenic capacity of HCC cells making it an ideal candidate to target therapeutically using pharmacological interventions.
DNA methylation is a key epigenetic component that regulates gene expression. DNA methyltransferases (DNMTs), enzymes that methylate DNA by binding to CpG dinucleotides on gene promoters, are associated with transcriptional silencing and may lead to aberrant methylation of genes when upregulated (28, 29). Though not much is known about the relationship between DNMTs and miRNA, a study by Croce and colleagues showed that miR-29 specifically targets both DNMT3A and DNMT3B in lung cancer (21). The regulation of DNMTs by miR-29 may contribute to the transcriptional silencing of tumor suppressors leading to poor prognosis of cancer patients. Additionally, the down regulation of miR-29 has been shown in HCC and lung cancer suggesting tumor suppressor properties (30, 31). In our study, we find that AFP induced miR-29a suppression leads to increased expression of both DNMTs in HCC.
It is interesting that c-MYC acts as the mediator between AFP and the miR-29a/b-1 transcript. In the early 1990’s the association between c-MYC and HCC was first described (32). Peng et. al found that the c-MYC gene was amplified (>1.5 fold) in nearly 40% of their HCC cases and showed that those patients not only had elevated serum AFP (>320ng/ml) but were more likely to have hepatitis B infection (32). They concluded that amplification of c-MYC was not uncommon in HCC and may be related to its biological behavior. Furthermore, HBx, a hepatitis B viral protein that transforms hepatocytes and has been implicated in HBV-driven HCC, has also been shown to activate c-MYC (33–35). Though we don’t see an amplification of the c-MYC gene when comparing AFP low to AFP high patients in our cohort (data not shown), that might be due to the fact that all patients have hepatitis B infection.
In our molecular studies we observe that c-MYC binds to the miR-29a/b-1 transcript in the presence of AFP. It is known that AFP does not localize to the nucleus, therefore the mechanism by which AFP promotes c-MYC binding to the nuclear transcript is unclear, but there are several possibilities. For example, AFP may induce the nuclear localization of c-MYC by transporting a number of factors into tumor cells or may even bind to and transport c-MYC itself. There is also evidence of a non-secreted form of AFP, which may have the ability to interact with transcription factors, co-activators, or regulators of cell cycle (36–38). In addition, the half-life of c-MYC is short and fluctuates substantially in response to many cellular activities (39). In our study we found that treatment with AFP+ conditioned media followed by a short treatment with cycloheximide led to an accumulation of c-MYC protein in HLE cells. It is possible that AFP extends the half-life of the c-MYC protein by increasing its stability, for even a relatively short extension of c-MYC expression could greatly change the cellular microenvironment in favor of tumor growth. Undoubtedly further functional analysis at the molecular level must be done to elucidate the function of AFP in regards to c-MYC signaling and epigenetic alterations that drive poor outcome in HCC.
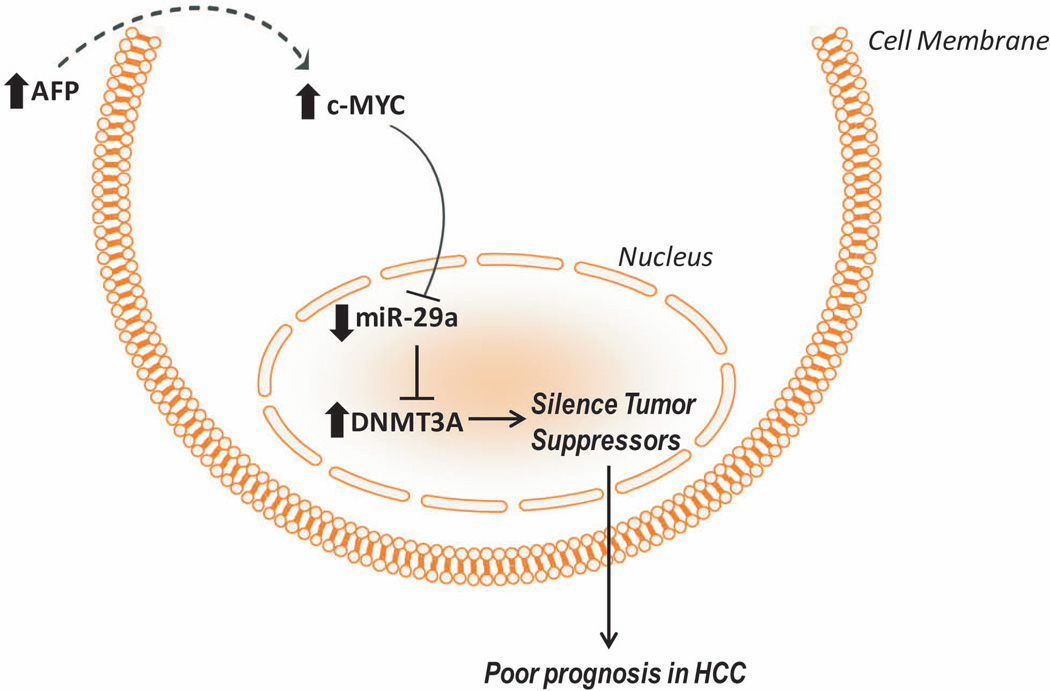
We propose that AFP may regulate miR-29a expression, which in turn alters DNA methyltransferase expression and modulates the epigenome in HCC (Figure 5). The increased methylation we observe may inhibit tumor suppressor gene expression provoking aggressive HCC and poor outcome. For the first time, striking differences at the molecular level are shown between AFP+ and AFP− HCC patients. In light of these findings, it is our hope that further work is done to determine the molecular functions of AFP in regards to c-MYC signaling and the epigenome. The more insight we gain at the molecular level, the better able we are to subgroup HCC patients and appropriately treat the heterogeneous disease.
Figure 5.
Schematic illustrating the mechanism by which AFP transcriptionally down regulates miR-29a and drives poor prognosis in HCC.
Supplementary Material
Acknowledgements
We appreciate the mouse-specific AFP primers given to us by Agnus Holczbauer in Snorri Thorgeirrson’s Laboratory of Experimental Carcinogenesis, NCI. We also acknowledge the SAIC-Protein Expression Lab for virus production.
Financial Support:
This work was supported by the grant (Z01 BC 010313) from the Intramural Research Program of the Center for Cancer Research, National Cancer Institute.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 3.Johnson PJ. Role of alpha-fetoprotein in the diagnosis and management of hepatocellular carcinoma. J Gastroenterol. Hepatol. 1999;14(Suppl):S32–S36. doi: 10.1046/j.1440-1746.1999.01873.x. [DOI] [PubMed] [Google Scholar]
- 4.Mizejewski GJ. Biological roles of alpha-fetoprotein during pregnancy and perinatal development. Exp. Biol. Med. (Maywood.) 2004;229:439–463. doi: 10.1177/153537020422900602. [DOI] [PubMed] [Google Scholar]
- 5.Terentiev AA, Moldogazieva NT. Structural and functional mapping of alpha-fetoprotein. Biochemistry (Mosc.) 2006;71:120–132. doi: 10.1134/s0006297906020027. [DOI] [PubMed] [Google Scholar]
- 6.Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis. 2001;5:145–159. doi: 10.1016/s1089-3261(05)70158-6. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez JM, Elahi A, Clark CW, Wang J, Humphries LA, Centeno B, et al. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann Surg Oncol. 2013;3:S625–S635. doi: 10.1245/s10434-013-3106-3. [DOI] [PubMed] [Google Scholar]
- 8.Saffroy R, Pham P, Reffas M, Takka M, Lemoine A, Debuire B. New perspectives and strategy research biomarkers for hepatocellular carcinoma. Clin Chem. Lab Med. 2007;45:1169–1179. doi: 10.1515/CCLM.2007.262. [DOI] [PubMed] [Google Scholar]
- 9.Pang RW, Joh JW, Johnson PJ, Monden M, Pawlik TM, Poon RT. Biology of hepatocellular carcinoma. Ann. Surg. Oncol. 2008;15:962–971. doi: 10.1245/s10434-007-9730-z. [DOI] [PubMed] [Google Scholar]
- 10.Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv. Genet. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 14.Ji J, Wang XW. New kids on the block: Diagnostic and prognostic microRNAs in hepatocellular carcinoma. Cancer Biol Ther. 2009;8:1686–1693. doi: 10.4161/cbt.8.18.8898. [DOI] [PubMed] [Google Scholar]
- 15.Budhu A, Ji J, Wang XW. The clinical potential of microRNAs. J. Hematol. Oncol. 2010;3:37. doi: 10.1186/1756-8722-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Research. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 19.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J. Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstrand CG CG, Czar B. Paper electrophoretic study of human fetal serum proteins with demonstration of a new protein fraction. Scand. J. Clin Lab Invest. 1957;9:277–286. doi: 10.3109/00365515709079971. [DOI] [PubMed] [Google Scholar]
- 25.Abelev GI, Perova SD, Khramkova NI, Postnikova ZA, Irlin IS. Production of embryonal alpha-globulin by transplantable mouse hepatomas. Transplantation. 1963;1:174–180. doi: 10.1097/00007890-196301020-00004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Li Z, Cheng Y, Jia F, Li R, Wu M, Li K, et al. CpG island methylator phenotype association with elevated serum alpha-fetoprotein level in hepatocellular carcinoma. Clin Cancer Res. 2007;13:944–952. doi: 10.1158/1078-0432.CCR-06-2268. [DOI] [PubMed] [Google Scholar]
- 27.Wu LM, Zhang F, Zhou L, Yang Z, Xie HY, Zheng SS. Predictive value of CpG island methylator phenotype for tumor recurrence in hepatitis B virus-associated hepatocellular carcinoma following liver transplantation. BMC. Cancer. 2010;10:399. doi: 10.1186/1471-2407-10-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulis M, Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 30.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 31.Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1:224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng SY, Lai PL, Hsu HC. Amplification of the c-myc gene in human hepatocellular carcinoma: biologic significance. J. Formos. Med. Assoc. 1993;92:866–870. [PubMed] [Google Scholar]
- 33.Balsano C, Avantaggiati ML, Natoli G, De Marzio E, Will H, Perricaudet M, Levrero M. Full-length and truncated versions of the hepatitis B virus (HBV) X protein (pX) transactivate the cmyc protooncogene at the transcriptional level. Biochem. Biophys. Res Commun. 1991;176:985–992. doi: 10.1016/0006-291x(91)90379-l. [DOI] [PubMed] [Google Scholar]
- 34.Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, Han YM, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J. Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 35.Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, Buendia MA. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 36.Sarcione EJ, Zloty M, Delluomo DS, Mizejewski G, Jacobson H. Detection and measurement of alpha-fetoprotein in human breast cancer cytosol after treatment with 0.4 M potassium chloride. Cancer Res. 1983;43:3739–3741. [PubMed] [Google Scholar]
- 37.Sarcione EJ, Hart D. Biosynthesis of alpha fetoprotein by MCF-7 human breast cancer cells. Int. J. Cancer. 1985;35:315–318. doi: 10.1002/ijc.2910350306. [DOI] [PubMed] [Google Scholar]
- 38.Seol W, Hanstein B, Brown M, Moore DD. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner) Mol. Endocrinol. 1998;12:1551–1557. doi: 10.1210/mend.12.10.0184. [DOI] [PubMed] [Google Scholar]
- 39.Lin CP, Liu CR, Lee CN, Chan TS, Liu HE. Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J. Hepatol. 2010;2:16–20. doi: 10.4254/wjh.v2.i1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.