Abstract
Background
Neural tube closure defects (NTDs) are among the most common congenital malformation in human, typically presenting in liveborns as spina bifida. At least 240 gene mutations in mouse are known to increase the risk of NTD. There is a growing appreciation that environmental factors significantly contribute to NTD expression, and that NTDs likely arise from complex gene-environment interactions. Because maternal folic acid supplementation reduces human NTD risk in some populations by 60–70%, it is likely that NTD predisposition is often associated with a defect in folate-dependent one-carbon metabolism. A comprehensive, untargeted metabolic survey of NTD-associated changes in embryo metabolism would provide a valuable test of this assumption. We sought to establish a metabolic profiling platform that is capable of broadly assessing metabolic aberrations associated with NTD-promoting gene mutations in early-stage mouse embryos.
Methods
An LC/MS-based untargeted metabolite profiling platform was employed to broadly identify significant differences in small molecule levels (50 – 1,000 Da) in NTD-affected E9.5 mouse embryos (Lrp6−/−) vs. unaffected (Lrp6+/+) control embryos.
Conclusion
Results provide proof-of-principal feasibility for the broad survey of the metabolome of individual E9.5 mouse embryos and identification of metabolic changes associated with NTDs and gene mutations. Levels of 30 different metabolites were altered in association with Lrp6 gene deletion. Some metabolites link to folate-dependent one-carbon transfer reactions, as anticipated, while others await structure elucidation and pathway integration. Whole-Embryo Metabolomics offers the potential to identify metabolic changes in genetically determined NTD-prone embryos.
INTRODUCTION
Spina bifida and other neural tube defects (NTDs) are common and severe congenital malformations that arise during early embryonic development (Blom and others, 2006; Greene and Copp, 2005; Wallingford and others, 2013). In humans, NTDs are the second most common structural birth defect, following congenital heart defects, and have an overall worldwide incidence of 0.5–2.0 per thousand live births (Detrait and others, 2005; Li and others, 2006; Mitchell, 2005). Notably, NTDs arise when proper formation of the neural tube is disrupted during embryogenesis by either gene mutations or environmental factors. Mutation of more than 240 different genes in mice have been shown to elicit an increased risk of NTDs (Harris and Juriloff, 2010; Juriloff and Harris, 2000; Ross, 2010; Wallingford and others, 2013). In accord with a long-standing appreciation that maternal folic acid (FA) supplementation can profoundly reduce the incidence of NTDs in humans, it is notable that at least 23 murine models of NTD have so far been tested and shown to be afforded protection by FA supplementation (Carter and others, 1999; Gray and others, 2010; Gray and Ross, 2009; Wallingford and others, 2013). Importantly, the genetic setting is a key determinant of whether folic acid supplementation will be protective against NTDs, and a number of mouse mutants have now been shown to suffer more severe embryopathy when provided additional folic acid (Gray and others, 2010; Marean and others, 2011).
This Lrp6 gene encodes a Wnt co-receptor and loss-of-function (LOF) as well as gain-of function (GOF) Lrp6 mutations result in an increased prevalence of NTDs, which manifest as exencephaly and spina bifida (Gray and others, 2010; Pinson and others, 2000). Notably, maternal FA-supplementation diminishes the prevalence of these NTDs in Lrp6 GOF mouse embryos, while increasing prevalence in Lrp6 LOF embryos (Gray and others, 2010). Studies of murine models of folate-responsive NTDs can provide fundamental insights into metabolic and environmental contributors to NTD risk in humans. Additionally, such studies may shed light on the relationship between Wnt signaling and folate metabolism, a connection that is presently ill-defined.
We previously used metabolomics as a powerful “-omic” approach for discovering differences in the expression of small molecule metabolites in complex biological matrices (Abbott and others, 2014; Chen and others, 2012; Witherspoon and others, 2013). Indeed, untargeted metabolomics provides a means to conduct comprehensive unbiased screening of the effects of genetic and environmental perturbations on the metabolism of developing embryos. By revealing an instantaneous snapshot of all metabolic processes occurring in a system of interest, this approach offers the potential to provide comprehensive information regarding the underlying molecular basis for developmental perturbations that lead to NTDs. Untargeted metabolite profiling may be the most effective strategy for discovering the molecular basis for NTDs that arise in association with embryonic-lethal gene mutations, i.e., a setting in which phenotypic analysis of liveborn offspring is not a possibility. In this case, whole-embryo metabolite profiling can be performed on NTD-affected embryos at a stage just prior to their incipient mortality.
The present investigation was performed as a proof-of-principal study to evaluate the potential of untargeted metabolite profiling to survey the small molecule content of individual E9.5 mouse embryos and identify metabolic aberrations that are associated with NTD in Lrp6 deletion mutants.
MATERIALS AND METHODS
Mice breeding and embryo harvest
All procedures involving animals were carried out in accordance with the guidelines established by the NIH and IACUC of Weill Cornell Medical College. Mice were housed in Weill Cornell’s animal facility on a 12 hr light-dark cycle. Mating pairs of Lrp6+/− mice were set in the late afternoon by introducing males to females (Carter and others, 2005; Carter and others, 1999; Gray and others, 2010). The following morning (embryonic day E0.5), males and females were separated and potential pregnancies were determined by the presence of a plug. Female mice were weighed immediately and again at day 7 to assess weight gain, indicative of pregnancy. Dams were sacrificed if a plug was observed following mating and if the dam gained over 2.5 g of weight during the 9.5 day pregnancy window. Potentially pregnant dams were sacrificed by cervical dislocation at E9.5. The uterus was removed and placed into ice-cold phosphate-buffered saline (PBS) for dissection. Individual implantations were dissected free from the uterus and visually inspected for the presence or absence of NTDs. Dissection and NTD scoring were conducted with the aide of a Leica dissection microscope and images were saved to hard disk via camera. Embryos were then washed briefly in ddH20 to removes excess PBS and snap frozen on dry ice. The yolk sac was removed from each embryo and saved for genotyping.
Lrp6 embryo genotyping
Yolk sacs removed from the Lrp6 embryos were genotyped by PCR followed by DNA gel electrophoresis. A GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA) was used for the PCR reaction and the products were resolved on a 2% agarose gel. Lrp6-specific primers were designed as follows: Forward 5′-CAGGCATGTAGCCCTTGGAG-3′, Reverse 5′-ACTACAAGCCCTGCACTGCC-3′, Neoreverse- 5′-GTAGAGTTCCCAGGAAGGAGCC-3′. Thermocycler parameters included an annealing step at 60°C and an extension step at 72°C, with 30 cycles in total. Lrp6+/+ embryos displayed a single 350bp PCR product band, Lrp6+/− embryos displayed both a 350bp and a 400bp bands and Lrp6−/− mice displayed a single band at 400bp.
Metabolite extraction from E 9.5 embryos
Methanol/ddH2O (80:20 at −70°C) was used to rapidly quench metabolism and extract small molecule metabolites from NTD-affected and non-affected E9.5 mouse embryos of various Lrp6 genotypes. Individual embryos were homogenized using a stainless steel bead beater (TissueLyser II; Qiagen, Valencia, CA) in a −20°C solution of 80% methanol. The homogenate was then pelleted by centrifugation (14,000 RPM, 4°C) and the supernatant was collected. Three successive 400 μL volumes of 80% methanol were added to each embryo pellet, followed by homogenization and centrifugation after each volume. The methanolic supernatants were pooled and vacuum centrifuged until dry using a vacuum concentrator (Eppendorf Vacufuge, Eppendorf, Germany), and stored at −80°C until the day of LC/MS metabolomic analysis.
Normalization of metabolites to total protein
To account for differences in embryo size, samples were normalized based on total protein in the tissue pellet from each embryo that remained after methanol extraction. 100 μL of 0.2 M NaOH was added to each tissue pellet and these samples were heated for 20 min at 95°C with frequent vortexing. The dissolved proteinaceous solution was spun down at 10,000 RPM for 5 min to remove debris. A DC protein assay (Bio-Rad, Hercules, CA) was conducted on the supernatant. Total protein (μg) was calculated and recorded for each sample so that the metabolite extracts could later be resuspended to equivalent concentrations.
Sample preparation for metabolomic analysis
Metabolite extract films were resuspended in 70% acetonitrile (ACN) containing 0.2% acetic acid to their appropriate concentrations based on protein content from normalization steps. These resuspensions were spun at 15,000 RPM for 10 min to remove insoluble debris. The metabolite-containing supernatant from these samples was then transferred to 250 μL conical polypropylene vials, capped, and loaded into the LC autosampler for subsequent LC/MS analysis. ACN-diluted samples, diluted to the desired protein concentration, were injected into the LC/MS system for chromatographic separation and MS detection.
LC/MS data acquisition and analysis
Samples were analyzed using on an Agilent model 1200/6220 Liquid Chromatography/Accurate-Mass Time-Of-Flight mass spectrometry (LC/MS) system (Agilent Technologies, Santa Clara, CA) equipped with dual electrospray ionization source, employing positive ion-monitoring and aqueous normal phase (ANP) chromatographic separation, as described previously (Chen and others, 2012). Data were saved in centroid mode using Agilent Masshunter workstation data acquisition software (B05.00). Acquired raw data files were processed with Agilent MassHunter Qualitative Analysis Software (B05.00; Agilent Technologies). Downstream comparative data analysis was performed using MassProfiler Professional (Agilent, B12.01) and MassHunter Profinder (B06.00). The algorithm used for MFE treats all of the mass spectral data from the experiment as a large, three-dimensional array of values (i.e., retention time, m/z, and ion abundance). It then searches for features (ions) with a common elution profile (e.g. identical m/z values and retention times), within the specified window of mass accuracy and RT. These aligned features are further grouped into one or more “compounds” based on their natural abundance isotope patterns as well as their detection as dimers, common adducts, including those with salts, and neutral loss of H2O. MFE was restricted to the m/z range of 50–1000 Da and completed within the time-span of the LC gradient. Singly charged ions with two or more ion species (including an M+1 isotope peak or adduct ion) and a minimum peak height of 250 ion counts were selected for MFE. The allowed adduct ions included H+, H−, Na+. The continuously-injected reference ions at m/z 121.0509 and 922.0093 were excluded from analysis.
Compounds/features with a minimum absolute peak height of 1000 counts were further filtered for statistical analysis. Recursive analysis was also conducted to re-mine the raw data for missed features in order to eliminate false-positives and false-negatives. MassProfiler Professional (Agilent, B12.01) was used to perform multivariate analysis on data sets. This chemometric software package employs an algorithm for data alignment across multiple groups and treats all imported files as a single dataset. Each aligned mass is associated with neutral mass, retention time, and ion intensity. Alignment of feature retention time (RT) and mass measurements within and between groups of replicates was performed at this point as well. All data were preprocessed by limiting the species considered to only singly-charge ions that were observed at 80% frequency in at least one treatment group. Following feature alignment and log transformation, the unsupervised pattern recognition algorithms principal component analysis (PCA) and hierarchical clustering analysis (HCA) were used to examine data sets for similarly changing features, expected and unexpected clusters, and the presence of outlying samples. Unpaired t-tests were also conducted to determine what metabolites were significantly different between experimental groups (p < 0.05). A Benjamini Hochberg correction was applied to adjust for false-positive discovery, arising from multiple hypothesis testing. MassHunter Profinder (B06.00) was used to examine raw data chromatograms for accuracy and confirm trends seen in statistical analysis by MassProfiler Professional.
Metabolite identification
As a first step in the structural identification of these unknown metabolites, features that were identified as being differentially expressed based on statistical criteria and PCA results were assigned tentative formulae by utilizing a molecular formula generator (MFG) algorithm. The MFG algorithm was used to generate and score molecular formula by calculating goodness-of-fit to monoisotopic mass accuracy, isotope abundance ratios, and spacing between predicted isotope peaks. A putative compound ID was tentatively assigned based on the MFG result and comparison to databases of known metabolites. These IDs can be further confirmed by comparison to pure chemical standards. Validation of the tentatively assigned compound ID was examined by attempting to match observed chromatographic retention times and/or fragmentation patterns with pure molecule standards in an in-house metabolite database.
RESULTS
Untargeted metabolite profiling of E9.5 embryos and technical reproducibility
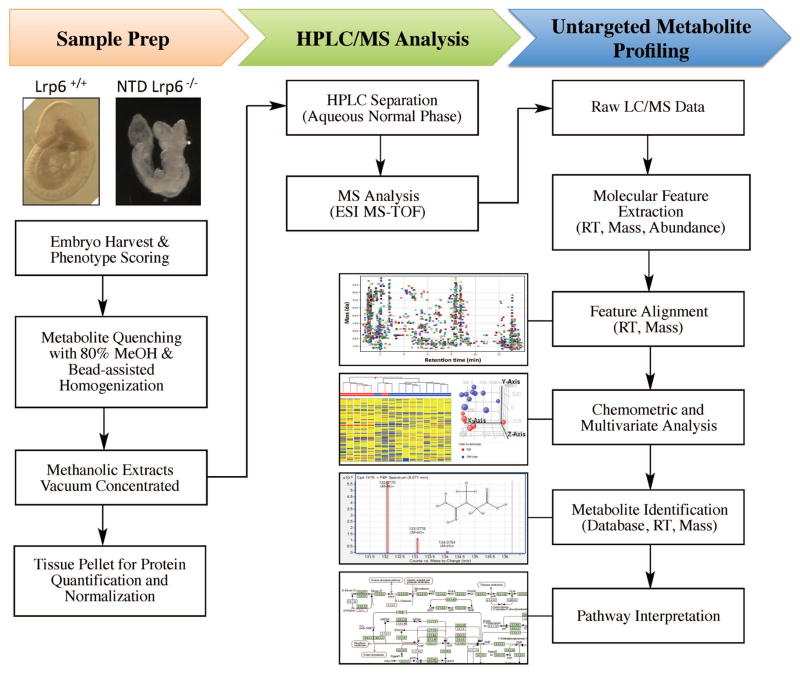
An LC/MS-based untargeted metabolite profiling platform was utilized in attempt to broadly survey small molecule changes (50–1,000 Da) that occur in Lrp6−/− embryo extracts in association with NTDs. Metabolite profiling was performed essentially as previously described (Abbott and others, 2014; Chen and others, 2012; Witherspoon and others, 2013), employing the workflow summarized in Fig. 1.
Figure 1. Whole-embryo metabolite profiling workflow.
E9.5 embryos from Lrp +/+ and Lrp6−/− gene-deleted mice were isolated from pregnant dams and analyzed for genotype, NTD phenotype and metabotype. Upon isolation, metabolism was rapidly quenched and metabolites were extracted by immersion in −70°C methanol (80%), followed by efficient bead-assisted homogenization. Metabolite extracts were vacuum concentrated and stored at −80°C until LC/MS analysis. Residual tissue pellets were retained for protein assay and results were used to normalize LC/MS injected embryo extracts. Dried down extracts were resuspended in 70% acetonitrile + 0.2% acetic acid and an equivalent of 1.0 μg protein was injected for metabolite profiling and chemometric analysis, as described in Methods.
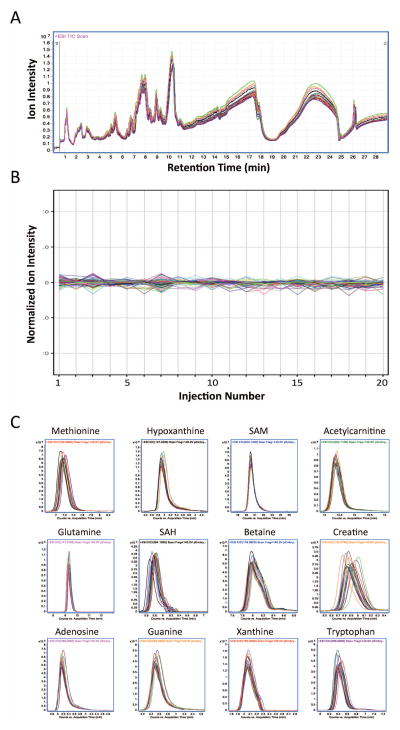
To assess the technical reproducibility of untargeted metabolite profiling of E9.5 whole embryos, a pooled quality control (QC) sample was prepared from 20 embryo extracts and an aliquot (5 μl) of this sample was sequentially analyzed to determine the consistency of profiling results. An overlay of total ion counts measured in each of 20 repeat measurements revealed a high degree of consistency (Fig. 2A). Over the mass and chromatographic span of the acquired LC/MS data (50 – 1000 Da and 29 min, respectively) we could quantify levels of 1,916 molecules, mostly undefined in terms of molecular structure (Fig. 2B). To precisely quantify the degree of technical reproducibility, levels of all 1,916 metabolites were determined for each of the 20 repeated analyses, and normalized to express the their variance in ion intensity over time. Considering all 1,916 metabolites, we observed a mean coefficient of variation (CV) of 1.38%, median CV of 0.98%, and a maximum/minimum CV of 9.59% and 0.13%, respectively (Figure 2B). Extracted ion chromatograms (EICs) for several common metabolites across these analyses further demonstrate the technical reproducibility of this approach (Figure 2C). This observed high degree of technical reproducibility would predictably allow for the confident discovery of bona fide biological changes in E9.5 embryo metabolite expression that are from differences in embryo genotype and/or phenotype.
Figure 2. Technical reproducibility of whole-embryo metabolite profiling.
Panel A: Overlay of 20 repeated LC/MS analyses using Aqueous Normal Phase (ANP) chromatography and positive ionization MS for a quality control (QC) sample isolated from a pool of E9.5 mouse embryos. Panel B: Profile plot overlay of normalized ion intensity for 1,916 distinct metabolite features, quantified as a function of injection number for 20 repeated analyses. Panel C: Extracted ion chromatograms (EICs) peak overlay of some common metabolites observed in each of 20 repeated analyses, demonstrating reproducibility of relative ion abundances and retention times for these molecules. EIC overlays were generated using MassHunter Qualitative Analysis B.05.00; normalization and profile plot visualization were performed using MassProfiler Professional B12.0 (Agilent Technologies).
Comparative metabolite profiling of Lrp6−/− vs. Lrp6+/+ embryos
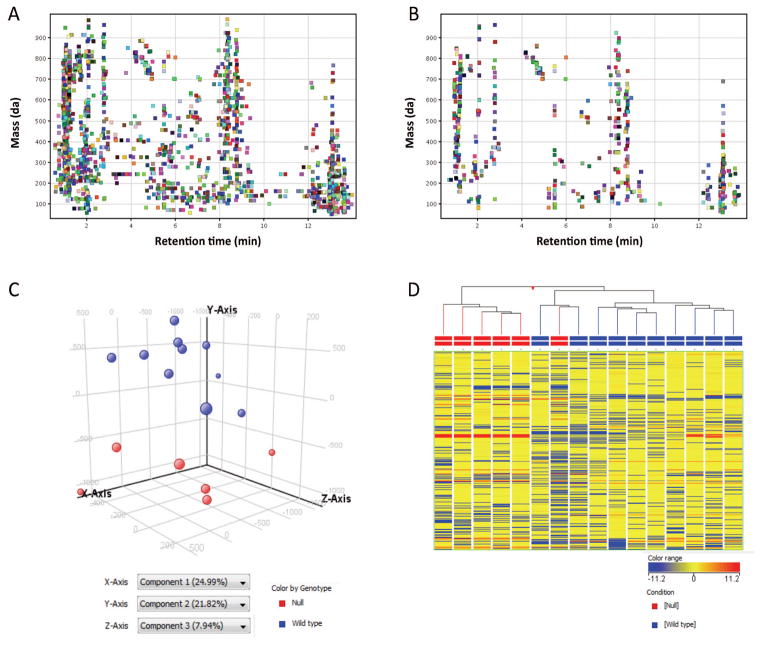
All Lrp6−/− embryos considered in analysis presented severe NTD in the form of open cranial neural folds, while all wild type embryos had completed normal neural tube closure at E9.5. The relative abundances of 2,596 molecular features across all Lrp6 embryo genotypes were determined following LC/MS analysis, raw data mining, and recursive processing of these data. Fig. 3A displays the distribution of these molecules according to observed neutral masses and retention times. Considering only those features that appear in at least 80% of samples from one or both experimental groups limits the likelihood of artifactual molecules that are not biologically relevant, but arise instead from extraneous sources and LC solvent background. Notably, of the 2,596 total features, 656 features were quantified in at least 80% of Lrp6+/+ or Lrp6−/− embryos (Fig. 3B). These molecular species were further considered in chemometric analyses using Agilent MassHunter Profinder in combination with MassProfiler Professional (Agilent Technologies, Santa Clara, CA),
Figure 3. Untargeted metabolite profiling comparison of Lrp6−/− vs Lrp6+/+ embryos.
LC/MS data were acquired using ANP chromatography and positive ion MS detection from 11 Lrp6+/+ and 6 Lrp6−/− embryos. Panel A: Plot of retention time vs. m/z for 2,596 aligned features detected by untargeted molecular feature extraction, quantified across all embryos. Panel B: Plot of retention time vs. m/z depicting 656 distinct features that were observed in at least 80% of embryos from at least one group. Panel C: PCA plot showing the 3D visualization of similarities and differences in the metabolite composition of each sample. Each data point corresponds with a single sample and is colored by genotype, representing the projection of 656 features considered in the first three principal components (PC). Data points are sized differently to give depth to the 3D visualization. PC1 accounted for 24.99% of the variance and PC2 accounted for 21.82%. As depicted by HCA and PCA, Lrp6+/+ embryos clustered independently of Lrp6−/− embryos. Notably, 30 features were found to be differentially-expressed between Lrp6+/+ and Lrp6 −/− embryos by unpaired t-testing (p<0.05, Table 1). This chemometric analysis was conducted using Mass Profiler Professional B12.0 (Agilent Technologies). Panel D: Unsupervised HCA, with samples color-coded by genotype and NTD-phenotype, displays the expression pattern and clustering of 656 metabolites quantified in individual Lrp6+/+ and Lrp6−/− embryos. Each column represents a single embryo sample with horizontal hash marks representing metabolite features, color-coded by normalized expression values. HCA was clustered by conditions, according to a Euclidean distance metric and Ward’s linkage rule.
Untargeted metabolomic datasets can offer a rather comprehensive snapshot of the expression of structurally diverse molecules; however, these analyses often result in very large datasets. Data analysis is facilitated by interpretations that utilize multiple statistical as well as visualization tools. The 656 features that met the above criteria were considered for unsupervised pattern recognition analysis. Without pre-specifying which experimental group each sample was a member of, principal component analysis (PCA; Fig. 3C) and hierarchical clustering analysis (HCA; Fig. 3D) were conducted to determine how the samples resembled one another in terms of metabolite profiles. Unsupervised PCA and HCA revealed that Lrp6−/− embryos clustered distinct from Lrp6+/+ embryos. PCA is a mathematical procedure that detects major trends in data by the linear combination of dimensions. This analysis transforms a number of potentially correlated variables (i.e. metabolite features) into a smaller number of uncorrelated linear variables called “principal components” that account for variability between samples. Each principal component represents the weighted linear combination of the untargeted LC/MS data. Principal component 1 (X-axis) accounted for over 24.99% of the variability between embryo metabolite samples and principal component 2 (y-axis) accounted for 21.82% (Fig. 3C). The hierarchical clustered dendrogram also highlighted differences in metabolite profiles, comparing Lrp6+/+ vs. Lrp6−/− embryos. Figure 3D displays the HCA dendrogram representing the relative abundances of the most confidently quantified 656 metabolites.
Differentially-expressed metabolites in Lrp6−/− embryos
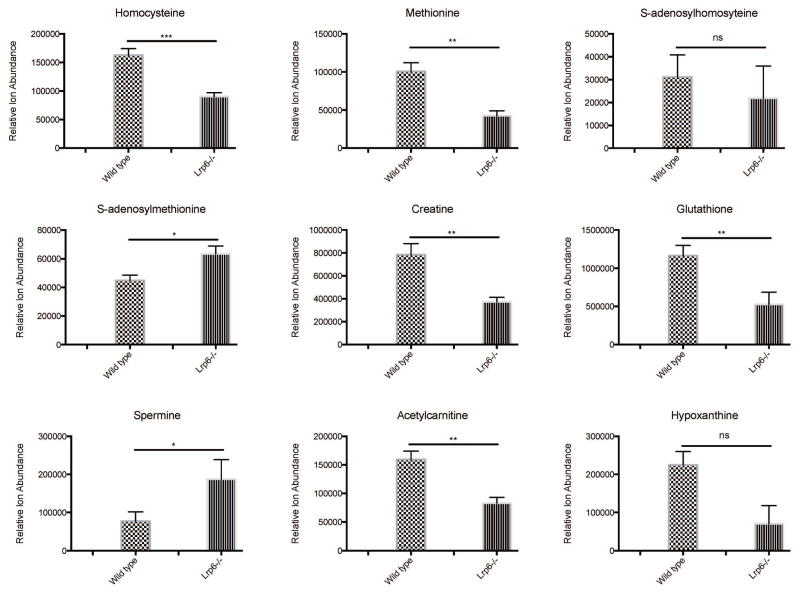
Unpaired t-tests identified 30 molecular features (i.e., metabolites) with significantly altered expression levels in Lrp6+/+ vs. Lrp6−/− embryos (p < 0.05; see Table 1). Of these, 19 could be structurally identified based on molecular formulae and retention time matches, or MS/MS fragmentation, relative to existing metabolite databases. These findings revealed Lrp6 deletion associated decreases in embryonic levels of threonine, homocysteine, methionine, glutathione, acetylcarnitine, creatine, creatinine, leucine/isoleucine, lysine, histidine, and dopamine. In contrast, Lrp6−/− embryos exhibited significant increases in the abundances of SAM, spermine and phosphatidylethanolamine (Table 1, Figure 4). In addition to these observed changes in structurally-identified metabolites, an additional 19 metabolites that await structure assignment were found to be significantly altered in embryonic expression levels in Lrp6−/− embryos. Pathway interrelationships involving these known and unknown molecular entities are currently under investigation.
Table 1.
| Mass | Retention Time | Compound | Regulation in Lrp6 −/− | p | p (Corr) | Fold Change |
|---|---|---|---|---|---|---|
| 85.09 | 5.19 | Piperidine | down | 1.17E-04 | 8.62E-04 | −4.23 |
| 113.06 | 8.04 | Creatinine | down | 2.02E-04 | 1.02E-03 | −1.75 |
| 119.057 | 6.32 | Threonine | down | 8.26E-05 | 7.43E-04 | −1.72 |
| 131.07 | 8.03 | Creatine | down | 5.54E-03 | 1.40E-02 | −1.99 |
| 131.095 | 5.19 | Leucine/Isoleucine | down | 7.44E-06 | 2.01E-04 | −3.85 |
| 132.055 | 7.34 | 132.055@7.34 | down | 1.93E-02 | 4.11E-02 | −1.98 |
| 133.039 | 9.37 | Aspartic Acid | down | 1.39E-04 | 8.65E-04 | −1.34 |
| 135.038 | 8.04 | Homocysteine | down | 3.66E-05 | 5.93E-04 | −1.79 |
| 136.039 | 2.05 | Hypoxanthine | down | 1.84E-03 | 6.23E-03 | −4.94 |
| 137.045 | 7.45 | p-Aminobenzoic acid | down | 3.07E-04 | 1.38E-03 | −1.83 |
| 146.106 | 12.38 | Lysine | down | 1.23E-02 | 2.93E-02 | −1.68 |
| 149.052 | 5.22 | Methionine | down | 8.43E-04 | 3.10E-03 | −2.34 |
| 153.051 | 8.03 | 153.051@8.03 | down | 1.32E-02 | 3.05E-02 | −1.54 |
| 155.074 | 12.20 | Histidine | down | 2.15E-03 | 6.95E-03 | −3.22 |
| 161.043 | 5.86 | 161.043@5.86 | down | 1.04E-04 | 8.44E-04 | −3.76 |
| 168.087 | 12.37 | 168.087@12.37 | down | 1.98E-02 | 4.11E-02 | −1.76 |
| 175.059 | 5.19 | Dopamine [M+Na]+ | down | 3.20E-06 | 1.61E-04 | −4.11 |
| 202.215 | 14.13 | Spermine | up | 1.93E-02 | 4.11E-02 | 2.62 |
| 203.116 | 8.48 | Acetyicarnitine | down | 7.53E-04 | 2.91E-03 | −1.90 |
| 241.093 | 1.38 | 241.093@1.38 | down | 2.40E-03 | 7.47E-03 | −2.37 |
| 241.277 | 1.90 | 241.277@1.90 | up | 4.43E-03 | 1.24E-02 | 1.97 |
| 256.14 | 9.38 | 256.14@9.38 | down | 1.90E-04 | 1.02E-03 | −2.21 |
| 297.017 | 6.71 | 297.017@6.71 | down | 1.56E-03 | 5.49E-03 | −2.14 |
| 307.085 | 5.79 | Glutathione | down | 2.68E-03 | 8.05E-03 | −2.83 |
| 315.914 | 8.76 | 315.914@8.76 | down | 7.36E-05 | 7.43E-04 | −3.56 |
| 398.139 | 13.10 | S-adenosylmethionine | up | 1.39E-02 | 3.14E-02 | 1.42 |
| 597.894 | 8.77 | 597.894@8.77 | down | 4.88E-03 | 1.27E-02 | −1.88 |
| 695.873 | 8.76 | 695.873@8.76 | down | 3.52E-04 | 1.50E-03 | −2.62 |
| 721.512 | 1.21 | PE(20:5/15:0) | up | 3.45E-03 | 9.99E-03 | 1.98 |
| 723.482 | 2.06 | PE(18:4/18:1) | up | 9.36E-03 | 2.30E-02 | 3.19 |
Figure 4. Statistically significant differences in metabolite expression in Lrp6−/− vs. Lrp6+/+ mouse embryos.
Unpaired t-test results are depicted for some metabolites from Table 1 that were observed statistically different between Lrp6+/+ and Lrp6−/− embryos. Raw data values were exported to Prism 6 (GraphPad Software Inc.) for repeated statistical testing (*= p<0.05, **= p<0.01, ***= p<0.001). Bars represent mean ion counts +/− standard error of the means.
DISCUSSION
To our knowledge, this is the first report to describe the application of untargeted metabolite profiling for the analysis of individual early-stage mouse embryo metabotypes. Accordingly, these studies establish a first proof-of-principal for effective application of LC/MS-based “whole-embryo metabolomics” to define the metabolic consequences of gene deletion in early development. The LC/MS platform employed in these analyses allowed broad metabolite coverage and provided ample sensitivity for profiling of single mouse embryos at the early developmental stage of E9.5. We further demonstrate a high level of technical reproducibility, allowing for the confident discovery of biologically-relevant alterations in metabolite levels that are changed by 20% or greater.
Whole-embryo metabolomics revealed 30 significant metabolite expression changes in NTD-affected Lrp6−/− embryos, relative to unaffected Lrp+/+ embryos (Table 1). In mammals, folate-activated one-carbon units serve as essential cofactors in many biosynthetic events. These include: the synthesis and catabolism of purines, the interconversion of serine and glycine, the synthesis of thymidylate from uridine monophosphate, as well as the remethylation of homocysteine to form methionine in the methionine cycle (Appling, 1991; Fox and Stover, 2008; Girgis and others, 1997; Herbig and others, 2002; Schirch and Strong, 1989; Shane, 1989; Wagner, 1995). The metabolic importance of folate extends to DNA- and histone-methylation reactions, owing to the essential requirement of reduced folate to enable regeneration of the universal methyl-group donor, S-adenosylmethionine (SAM). SAM is also essential for polyamine biosynthesis and hence, reduced folates are required to maintain cellular levels of polyamines (Avila and others, 2004; Bistulfi and others, 2009). Given the well-appreciated protection against NTDs afforded by maternal FA supplementation, alterations in the expression of metabolic intermediates in folate-associated biosynthetic pathways may be a common metabolic phenotype associated with NTD. Accordingly, it is notable that Lrp6 −/− embryos displayed significant alterations in the expression of several metabolites that contribute to one-carbon metabolism, methionine regeneration, purine metabolism, and polyamine biosynthesis (Table 1, Figure 4). These metabolites include SAM itself, along with methionine, homocysteine, creatine, creatinine, hypoxanthine, and spermine. In general, our metabolite profiling data are substantiated by gene expression analyses which showed that Lrp6 gene deletion is also associated with significant perturbations in the level of gene transcripts involved in methionine metabolism, purine metabolism, antioxidant defenses, and polyamine biosynthesis (Gray and others, 2010). We are currently unable to distinguish whether these changes in metabolite levels arise from difference between embryo genotype, NTD phenotype, or a combination of the two.
Further, we observed that glutathione was markedly diminished in NTD-affected Lrp6−/− mouse embryos – this is indicative of a decreased redox potential and likely increased oxidative stress, in comparison with Lrp6+/+ embryos that exhibit correct neural tube closure (Table 1 & Figure 4). It is likely that there is a molecular link between oxidative stress and one-carbon metabolism defects (Hoffman, 2011; Widner and others, 2002). The major methylation cycle in mammals relies on methionine synthetase and as an essential cofactor, utilizes 5-methylTHF as a methyl donor and vitamin B12 transferring a one-carbon unit to homocysteine and regenerating methionine. It has been demonstrated that methionine synthetase can be inactivated by oxidation of its cofactors (Christensen and Ueland, 1993). In Lrp6−/− embryos, an inability to mitigate damage from oxidative stress could therefore be a driver of aberrant neurulation by perturbing the methionine regeneration cycle and SAM-related pathways.
In sum, untargeted metabolite profiling revealed aberrant one-carbon metabolism, methionine regeneration, and antioxidant activity in NTD-affected Lrp6−/− embryos. These untargeted metabolite profiling data provide the first evidence of a metabolic link between a genetic lesion in Wnt signaling (Lrp6 receptor) and folate-related biosynthesis pathways. Further MS-based studies will be important to elucidate the structures of currently unidentified molecules listed in Table 1 that similarly exhibit expression level aberrations in Lrp6 gene-deleted mice. It will also be important to learn the extent to which similar metabolic perturbations are observed in NTD-affected mouse embryos that arise in association with other of the more than 240 known NTD-promoting gene mutations (Harris and Juriloff, 2010; Juriloff and Harris, 2000; Ross, 2010; Wallingford and others, 2013). Such future studies may provide fundamental insights and aid in the identification of common biochemical pathways that, when perturbed, drive NTDs in response to a multiplicity of gene mutations and environmental factors. This knowledge may lead to the identification of maternal supplements, beyond FA, that further diminish the risk of human birth defects.
Acknowledgments
This research was supported by NIH training grant T32 GM073546 (to AH), as well as research grants PO1 HD067244 (SSG, MER and RHF) and R37 HL87062 (SSG).
LITERATURE CITED
- Abbott GW, Tai KK, Neverisky DL, Hansler A, Hu Z, Roepke TK, Lerner DJ, Chen Q, Liu L, Zupan B, Toth M, Haynes R, Huang X, Demirbas D, Buccafusca R, Gross SS, Kanda VA, Berry GT. KCNQ1, KCNE2, and Na+-Coupled Solute Transporters Form Reciprocally Regulating Complexes That Affect Neuronal Excitability. Science signaling. 2014;7(315):ra22. doi: 10.1126/scisignal.2005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1991;5(12):2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- Avila MA, Garcia-Trevijano ER, Lu SC, Corrales FJ, Mato JM. Methylthioadenosine. The international journal of biochemistry & cell biology. 2004;36(11):2125–2130. doi: 10.1016/j.biocel.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Bistulfi G, Diegelman P, Foster BA, Kramer DL, Porter CW, Smiraglia DJ. Polyamine biosynthesis impacts cellular folate requirements necessary to maintain S-adenosylmethionine and nucleotide pools. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23(9):2888–2897. doi: 10.1096/fj.09-130708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom HJ, Shaw GM, den Heijer M, Finnell RH. Neural tube defects and folate: case far from closed. Nat Rev Neurosci. 2006;7(9):724–731. doi: 10.1038/nrn1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M, Chen X, Slowinska B, Minnerath S, Glickstein S, Shi L, Campagne F, Weinstein H, Ross ME. Crooked tail (Cd) model of human folate-responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor-related protein 6. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(36):12843–12848. doi: 10.1073/pnas.0501963102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M, Ulrich S, Oofuji Y, Williams DA, Ross ME. Crooked tail (Cd) models human folate-responsive neural tube defects. Human molecular genetics. 1999;8(12):2199–2204. doi: 10.1093/hmg/8.12.2199. [DOI] [PubMed] [Google Scholar]
- Chen Q, Park HC, Goligorsky MS, Chander P, Fischer SM, Gross SS. Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PloS one. 2012;7(6):e37149. doi: 10.1371/journal.pone.0037149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B, Ueland PM. Methionine synthase inactivation by nitrous oxide during methionine loading of normal human fibroblasts. Homocysteine remethylation as determinant of enzyme inactivation and homocysteine export. J Pharmacol Exp Ther. 1993;267(3):1298–1303. [PubMed] [Google Scholar]
- Detrait ER, George TM, Etchevers HC, Gilbert JR, Vekemans M, Speer MC. Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicology and teratology. 2005;27(3):515–524. doi: 10.1016/j.ntt.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitamins and hormones. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- Girgis S, Suh JR, Jolivet J, Stover PJ. 5-Formyltetrahydrofolate regulates homocysteine remethylation in human neuroblastoma. The Journal of biological chemistry. 1997;272(8):4729–4734. doi: 10.1074/jbc.272.8.4729. [DOI] [PubMed] [Google Scholar]
- Gray JD, Nakouzi G, Slowinska-Castaldo B, Dazard JE, Rao JS, Nadeau JH, Ross ME. Functional interactions between the LRP6 WNT co-receptor and folate supplementation. Human molecular genetics. 2010;19(23):4560–4572. doi: 10.1093/hmg/ddq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JD, Ross ME. Mechanistic insights into folate supplementation from Crooked tail and other NTD-prone mutant mice. Birth defects research Part A, Clinical and molecular teratology. 2009;85(4):314–321. doi: 10.1002/bdra.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene ND, Copp AJ. Mouse models of neural tube defects: investigating preventive mechanisms. American journal of medical genetics Part C, Seminars in medical genetics. 2005;135C(1):31–41. doi: 10.1002/ajmg.c.30051. [DOI] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Research Part A: Clinical and Molecular Teratology. 2010;88(8):653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. The Journal of biological chemistry. 2002;277(41):38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- Hoffman M. Hypothesis: hyperhomocysteinemia is an indicator of oxidant stress. Medical hypotheses. 2011;77(6):1088–1093. doi: 10.1016/j.mehy.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ. Mouse models for neural tube closure defects. Human molecular genetics. 2000;9(6):993–1000. doi: 10.1093/hmg/9.6.993. [DOI] [PubMed] [Google Scholar]
- Li Z, Ren A, Zhang L, Ye R, Li S, Zheng J, Hong S, Wang T, Li Z. Extremely high prevalence of neural tube defects in a 4-county area in Shanxi Province, China. Birth defects research Part A, Clinical and molecular teratology. 2006;76(4):237–240. doi: 10.1002/bdra.20248. [DOI] [PubMed] [Google Scholar]
- Marean A, Graf A, Zhang Y, Niswander L. Folic acid supplementation can adversely affect murine neural tube closure and embryonic survival. Human molecular genetics. 2011;20(18):3678–3683. doi: 10.1093/hmg/ddr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE. Epidemiology of neural tube defects. American journal of medical genetics Part C, Seminars in medical genetics. 2005;135C(1):88–94. doi: 10.1002/ajmg.c.30057. [DOI] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407(6803):535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Ross ME. Gene-environment interactions, folate metabolism and the embryonic nervous system. Wiley interdisciplinary reviews Systems biology and medicine. 2010;2(4):471–480. doi: 10.1002/wsbm.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirch V, Strong WB. Interaction of folylpolyglutamates with enzymes in one-carbon metabolism. Archives of biochemistry and biophysics. 1989;269(2):371–380. doi: 10.1016/0003-9861(89)90120-3. [DOI] [PubMed] [Google Scholar]
- Shane B. Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitamins and hormones. 1989;45:263–335. doi: 10.1016/s0083-6729(08)60397-0. [DOI] [PubMed] [Google Scholar]
- Wagner C. Biochemical role of folate in cellular metabolism. Folate in health and disease. 1995;1995:23–42. [Google Scholar]
- Wallingford JB, Niswander LA, Shaw GM, Finnell RH. The continuing challenge of understanding, preventing, and treating neural tube defects. Science. 2013;339(6123):1222002. doi: 10.1126/science.1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widner B, Enzinger C, Laich A, Wirleitner B, Fuchs D. Hyperhomocysteinemia, pteridines and oxidative stress. Current drug metabolism. 2002;3(2):225–232. doi: 10.2174/1389200024605091. [DOI] [PubMed] [Google Scholar]
- Witherspoon M, Chen Q, Kopelovich L, Gross SS, Lipkin SM. Unbiased metabolite profiling indicates that a diminished thymidine pool is the underlying mechanism of colon cancer chemoprevention by alpha-difluoromethylornithine. Cancer discovery. 2013;3(9):1072–1081. doi: 10.1158/2159-8290.CD-12-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]