SUMMARY
The first lineage choice in mammalian embryogenesis is that between the trophectoderm, which gives rise to the trophoblast of the placenta, and the inner cell mass, from which derive the embryo proper and the yolk sac. The establishment of these lineages is preceded by the inside-versus-outside positioning of cells in the early embryo and stochastic expression of key transcription factors, which is then resolved into lineage-restricted expression. The regulatory inputs that drive this restriction and how they relate to cell position are largely unknown. Here we show an unsuspected role of Notch signaling in regulating trophectoderm-specific expression of Cdx2 in cooperation with TEAD4. Notch activity is restricted to outer cells and is able to influence positional allocation of blastomeres, mediating preferential localization to the trophectoderm. Our results show that multiple signaling inputs at preimplantation stages specify the first embryonic lineages.
INTRODUCTION
Upon fertilization, the unicellular mammalian zygote undergoes a series of equal cell divisions that in four days produces a 60–100 cell blastocyst in which the first embryonic lineages the—trophectoderm (TE) and the inner cell mass (ICM)—have been established (Stephenson et al., 2012). How this initial lineage choice occurs, and what genetic components constitute the system that controls this process, has been the subject of active study for the past 20 years. Key transcriptional regulators of these early lineages identified in the mammalian preimplantation embryo include OCT4, NANOG and SOX2 for the ICM and CDX2 for the TE (Cockburn and Rossant, 2010). These factors determine blastocyst lineages, but the onset of their expression is stochastic (Dietrich and Hiiragi, 2007), and later restriction to a particular cell type occurs as a downstream effect of earlier events related to the position of blastomeres in the embryo (Rossant and Tam, 2009).
This process is believed to involve differences in polarity and adhesion between inner and outer cells that are related to differential activation of the Hippo signaling pathway (Hirate et al., 2013; Nishioka et al., 2009), which results in sustained and restricted expression of genes such as Cdx2 in the outer cells of the future trophectoderm (Cockburn and Rossant, 2010). Hippo signaling is switched off in outer cells, leading to nuclear localization of the transcriptional coactivator YAP, which can then activate downstream target genes through interaction with the transcription factor TEAD4. Embryos lacking Tead4 fail to develop the TE, and Cdx2 expression is not maintained (Nishioka et al., 2008; Yagi et al., 2007). Correspondingly, overexpression of LATS2 kinase (an activator of Hippo) reduces expression of CDX2 in outer cells, while embryos lacking Lats1 and Lats2 express CDX2 in inner cells (Lorthongpanich et al., 2013; Nishioka et al., 2009). Similar results are obtained when other components of the pathway, such as Nf2 and Amot, are disrupted (Cockburn et al., 2013; Hirate et al., 2013). How these components are integrated to fully define lineage restriction in the blastocyst is largely unknown, and additional inputs remain to be identified (Wennekamp et al., 2013). Reconstruction of this process will require detailed understanding of the transcriptional control of the key lineage regulators acting in the preimplantation embryo.
With this aim in mind, we have searched for cis-regulatory elements responsible for trophectoderm-restricted expression of Cdx2, and through the analysis of one such enhancer uncovered a role of the Notch signaling pathway. We find that Notch is active specifically in outer cells of the blastocyst. Analysis of double mutants for Tead4 and the Notch effector Rbpj shows that the Notch and Hippo pathways converge on Cdx2 to activate its expression in the trophectoderm, uncovering an unexpected role for Notch signaling during preimplantation development (Souilhol et al., 2006). Furthermore, we show that forced activation of Notch directs blastomeres to the trophectoderm.
RESULTS
Characterization of a trophectoderm specific enhancer from Cdx2
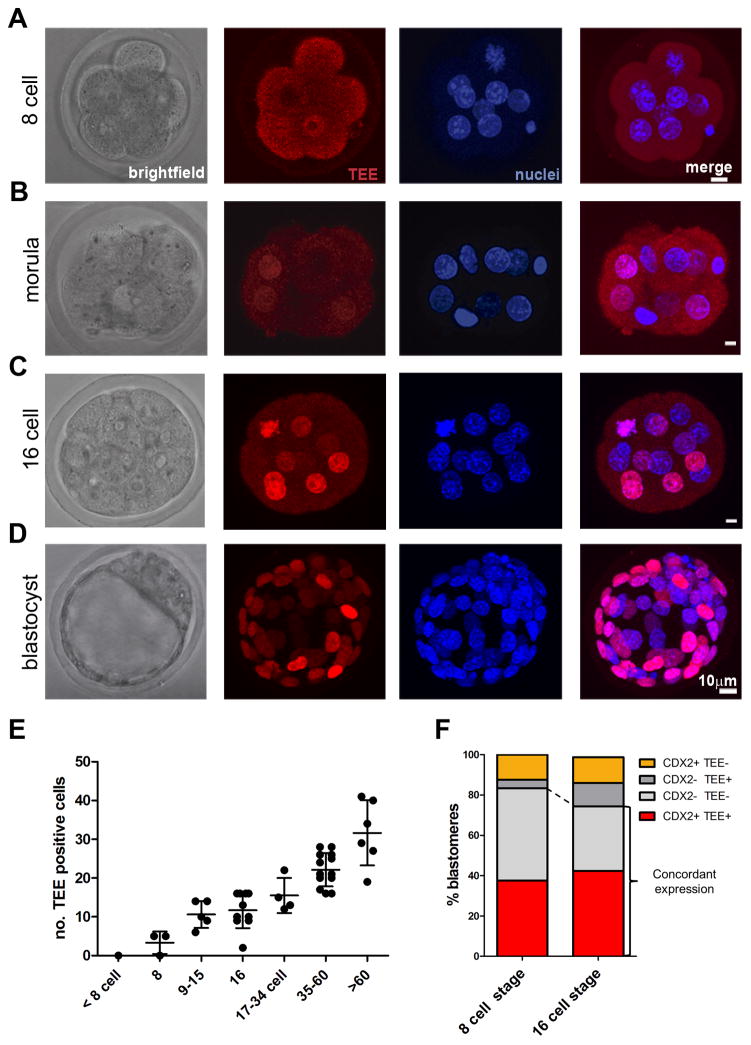
By means of transient transgenic analysis in mouse preimplantation embryos, we identified a cis-regulatory element located 5′ of Cdx2 that drives reporter expression in the trophectoderm (TE) of the blastocyst (Fig. S1A, B). Using fragment #3 of this TE enhancer (TEE) (Fig. S1A, B) linked to a H2B-mRFP reporter gene, we then generated several transgenic lines for further study. The reporter is only occasionally active in a few cells at the 8 cell stage (Fig. 1A), and upon compaction starts to be present in the outer cells of the morula (Fig. 1B), attaining strong activity at the 16 cell stage (Fig. 1C). At the blastocyst stage, TEE-driven reporter activity is localized throughout the TE and excluded from the ICM (Fig. 1D). We also generated TEE transgenic lines using a lacZ reporter, which behaved identically to the H2B-mRFP reporter lines (Fig. S1C). Tracking blastomeres in the mRFP reporter lines up to the 3.5 dpc blastocyst revealed a steady temporal increase in the number expressing mRFP (Fig. 1E). Reporter activity in either the mRFP or lacZ TEE lines was found throughout the TE, although occasionally we detected cells with lower levels of expression. Immunohistochemical analysis of the association between reporter activity and endogenous CDX2 showed concordant expression in >75% of blastomeres (CDX2+/TEE+, CDX−/TEE−; Fig. 1F), confirming that TEE activity closely matches the expression of CDX2 at preimplantation stages (Dietrich and Hiiragi, 2007; Strumpf et al., 2005).
Figure 1. A cis-regulatory element upstream of Cdx2 drives restricted expression in the trophectoderm.
(A–D) H2BmRFP reporter expression driven by the Cdx2 trophectoderm enhancer (TEE) at (A) the non-compacted 8-cell stage, (B) compacted 8-cell morula stage, (C) 16-cell stage, and (D) blastocyst stage. H2BmRFP was detected by immunohistochemistry and nuclei were stained with DAPI. Scale bars, 10 μm. (E) Quantification of the number of TEE-positive cells (y-axis) per embryo, staged by total cell number (x-axis). Each dot represents an individual embryo, and the bar and whiskers indicate means and standard deviations. (F) Correlation of TEE activity with endogenous CDX2 protein expression at the 8-cell (n=24, 3 embryos) and 16-cell stages (n=80, 5 embryos). Individual blastomeres were scored for H2BmRFP and CDX2 immunostaining. See also Figure S1.
The TE-enhancer is active in Cdx2 and Tead4 mutant blastocysts
We next aimed to determine the upstream regulatory factors acting on the TEE to restrict Cdx2 expression. To date, the only transcription factors proposed to regulate Cdx2 in the trophectoderm are CDX2 itself, through an auto-regulatory loop (Niwa et al., 2005), and TEAD4 acting together with YAP downstream of the Hippo pathway (Nishioka et al., 2009). To test whether these factors are required for TEE activity in the blastocyst, we first examined homozygous Cdx2 knockout (KO) mice (Strumpf et al., 2005) containing the mRFP reporter, finding that TEE reporter activity was still restricted to the TE (Fig. S2A, B) and that the number of positive cells was the same as in wild type littermates (Fig. S2C, D).
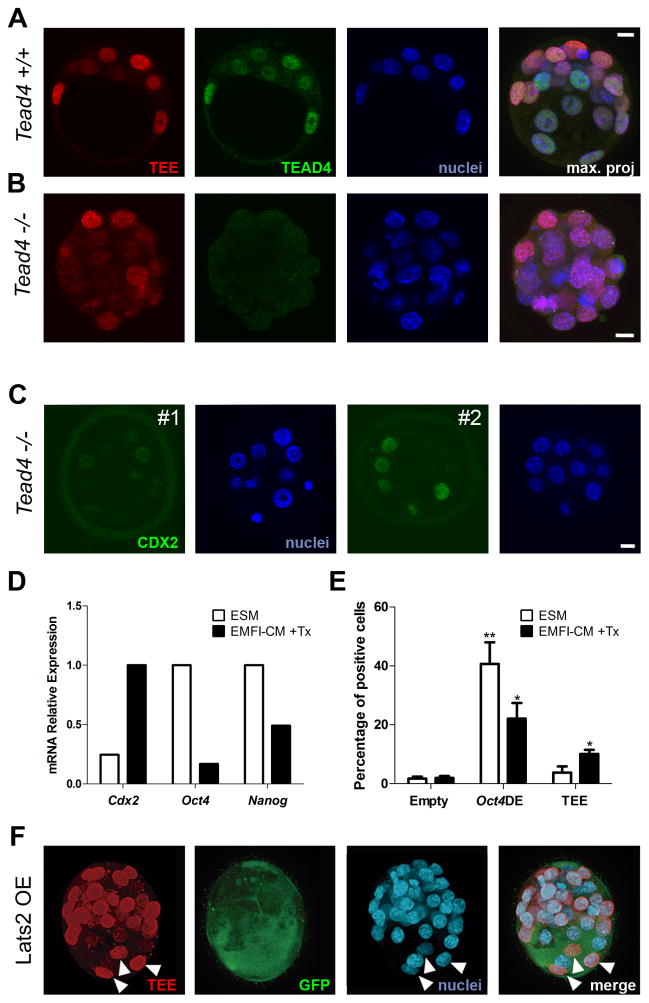
We next analyzed the regulation of the TEE by the Hippo pathway. To check to what extent the TEE was a YAP/TEAD response element, we bred the mRFP reporter line into the Tead4 KO background (Nishioka et al., 2008). The TEE was fully active in homozygous Tead4−/− 3.5 dpc embryos (Fig. 2A, B), and even conserved preferential activity in outer blastomeres (Fig. 2B). Consistent with previous reports (Kaneko and Depamphilis, 2013; Nishioka et al., 2008), we also found that some Tead4−/− embryos retained CDX2 expression (Fig. 2C), suggesting that additional inputs are necessary for the early expression of Cdx2. Furthermore, we did not detect significant differences in total cell number, or mRFP+ blastomeres per embryo between wild type, heterozygous and homozygous embryos (Fig. S2E, F), precluding an effect of the loss of Tead4 on the total number of cells in which the TEE is active.
Figure 2. The Hippo pathway is not sufficient to regulate the Cdx2 TEE.
(A–B) TEE activity (red) and TEAD4 immunodetection (green) in (A) wild type (wt) and (B) Tead4 knockout embryos (Tead−/−). Nuclei were stained with DAPI (blue). Maximal projections (max. proj) of merged images are shown in the right panels. (C) Immunohistochemistry for CDX2 (green) in two Tead4 mutant embryos from the same litter. Nuclei were stained with DAPI (blue). Scale bars, 10 μm. (D) Relative expression of Cdx2, Oct4 and Nanog in the 5TVER7 cell line in response to tamoxifen (Tx) induction in EMFI-CM medium of TEAD4-mediated transdifferentiation of ES to TS cells. (E) Percentage of 5TVER7 cells that activate the TEE in response to tamoxifen (Tx) induction in EMFI-CM medium of TEAD4-mediated transdifferentiation of ES to TS cells. Data are means ± s.e.m. n=3. **p<0.01, *p<0.05 by Student’s t-test. (F) TEE activity (red) and GFP expression (green) in blastocysts injected with Lats2-GFP mRNA. White arrowheads mark TEE+ nuclei in which Lats2-GFP is expressed. Nuclei were stained with DAPI (blue). A merged image is shown in the right panel. See also Figure S2.
To investigate this further, we took advantage of the 5TVER7 embryonic stem (ES) cell line, which stably expresses a tamoxifen-inducible active TEAD4 protein (Tead4VP16ER) (Nishioka et al., 2009). Upon tamoxifen induction, these cells activate Cdx2 expression and acquire a trophoblast stem (TS) cell phenotype. We transfected 5TVER7 cells with the TEE, or with the Oct4 distal enhancer (Oct4DE) (Yeom et al., 1996) as a control, and treated them with tamoxifen in TS culture medium (Tanaka et al., 1998). After 48 hours in these conditions, qPCR analysis showed robust activation of Cdx2 and down-regulation of Oct4, accompanied by a strong decrease in Oct4DE activity (Fig. 2D, E). Despite the forced expression of the active TEAD4 form, we did not observe a strong effect on TEE activity compared with culture in ES cell culture medium without tamoxifen, although treatment did increase its activity slightly but significantly compared with control empty vector (Fig. 2E). These experiments provide additional evidence that TEAD4 alone is not sufficient to fully activate the TEE. We next tested if forced activation of the Hippo pathway in outer cells would silence the TEE. Embryos of the mRFP-TEE line were injected with Lats2 RNA, which turns the Hippo pathway on and results in decreased expression of CDX2 in outer cells (Nishioka et al., 2009). This did not influence the expression of the reporter in the TE (Fig. 2F), confirming that the TEE does not respond exclusively to the activation state of the Hippo pathway in the blastocyst.
The Notch signaling pathway is active in the trophectoderm of the blastocyst
To identify other transcriptional inputs acting on the TEE, we searched the minimal 1.3 kb sequence active in the TE (fragment #6, Fig. S1A, B) for putative transcription factor binding sites; this search detected four high-confidence sites for RBPJ (Tun et al., 1994), the transcriptional effector of the Notch pathway. RBPJ acts together with the intracellular domain of Notch, product of the proteolytic cleavage of the receptor upon its activation that is then translocated to the nucleus (Kopan and Ilagan, 2009). Additionally, we detected two putative TEAD binding sites (Anbanandam et al., 2006) (Fig. S3A).
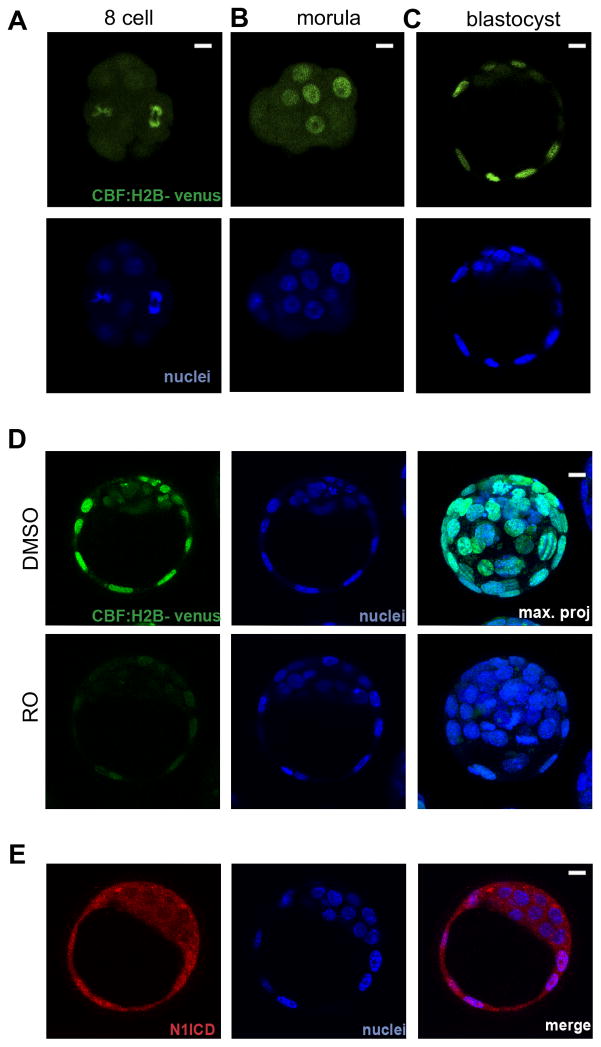
This observations suggested that Notch could have a role in development of the blastocyst, but to date there is scant evidence available for the expression of Notch pathway components at this stage (Cormier et al., 2004; Wang et al., 2004), and no studies have addressed their differential localization. To examine the activation status of the Notch pathway during pre-implantation stages we used a mouse line containing a CBF:H2B-Venus reporter, which contains multiple RBPJ binding sites driving the expression of a nuclear-targeted Venus fluorescent protein and that faithfully reports the activation state of the pathway (Nowotschin et al., 2013). The reporter was active in a few cells at the 8 cell stage, and activity became progressively restricted to outer cells by the 3.5 dpc blastocyst stage (Fig. 3A–C). Treatment of embryos from this line with the gamma-secretase inhibitor RO4929097 (RO), a well characterized and widely used tool that interferes with Notch signaling by blocking the processing of the receptor (Munch et al., 2013), resulted in a strong down-regulation of the reporter (Fig. 3D). We next examined the distribution by immunofluorescence of the endogenous NOTCH1 intra-cellular domain (N1ICD), finding that it is localized to the nucleus only in the outer cells of the blastocyst (Figs. 3E, S3B). Together, these data show that the Notch signaling pathway is active specifically in the developing TE.
Figure 3. The Notch signaling pathway is active in the trophectoderm.
(A–C) CBF:H2B-Venus reporter expression (green) at (A) the non-compacted 8-cell stage, (B) morula stage, and (C) blastocyst stage. (D) Treatment of embryos form the CBF:H2B-Venus line with the γ-secretase inhibitor RO4929097 (RO) downregulates reporter activity. (E) Immunodetection of Notch1 intracellular domain (N1ICD, red). Nuclei were stained with DAPI. Scale bars, 10 μm. See also Figure S3.
The TE-enhancer contains functional RBPJ and TEAD binding sites
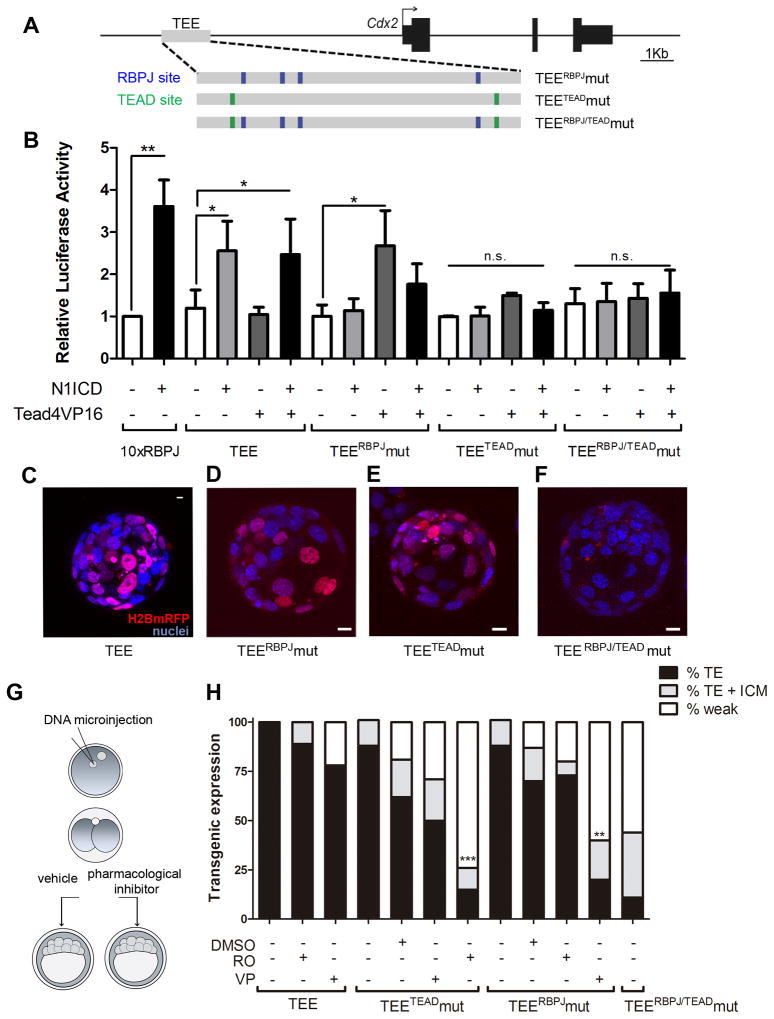
To investigate the function of the sites identified in the TEE, we transfected HEK293 cells with the wild type minimal TEE and versions mutated in the RBPJ sites alone (TEERBPJmut), the TEAD sites alone (TEETEADmut) or in the RBPJ and the TEAD sites (TEERBPJ/TEADmut) (Fig. 4A). We then tested their response to constitutively active forms of N1ICD (Kopan et al., 1994) and TEAD4 (Tead4VP16) (Nishioka et al., 2009), expressed either alone or in combination. Whereas the wild type TEE responded to N1ICD, the TEERBPJmut version did not (Fig. 4B), demonstrating that this regulatory element is a transcriptional target of the Notch pathway. Tead4VP16 alone did not affect the wild type TEE, consistent with the results from 5TVER7 ES cells (Fig. 2E); however, it was sufficient to transactivate the TEERBPJmut version (Fig. 4B). This suggests that TEAD4 acts together with N1ICD/RBPJ to activate the TEE, but cannot activate it when RBPJ is bound in the absence of N1ICD (and thus acts as a repressor (Lai, 2002)). Mutation of the RBPJ sites lifts this restriction, allowing transactivation of the TEE by TEAD4. The TEETEADmut or TEERBPJ/TEADmut versions did not respond to either factor (Fig. 4B). Surprisingly, transgenic embryos carrying the TEERBPJmut or the TEETEADmut version retained TE lineage-restricted activity (Fig. 4C–E), while TEE activity was abolished when the RBPJ and TEAD sites were both mutated (Fig. 4F). The lack of response of the TEETEADmut to N1ICD in vitro while retaining activity in vivo, suggests that TEAD sites are critical for enhancer activity in HEK cells but not in the blastocyst.
Figure 4. RBPJ and TEAD binding sites are necessary for the activity of the Cdx2 TEE.
(A) Diagram of the Cdx2 locus showing the TEE fragment with the putative sites for RBPJ (blue) and for TEAD (green) and the corresponding mutated versions. (B) Luciferase activity of wild type and mutated versions of the TEE (TEERBPJmut, TEETEADmut and TEERBPJ/TEADmut) in response to overexpression of active N1ICD and/or TEAD4 (TeadVP16) in tissue culture assays. A construct carrying ten copies of a consensus RBPJ binding site (10xRBPJ) was used as a positive control. Data are means ± s.e.m. n=8 10xRBPJ), 7 (TEE), 5 (TEERBPJmut), 4 (TEETEADmut) and 4 (TEERBPJ/TEADmut). **p<0.01, *p<0.05 by Bonferroni post test; n.s., not significant. (C) Activity of wild type TEE, (D) TEERBPJmut, (E) TEETEADmut and (F) TEERBPJ/TEADmut in transient transgenic embryos. A representative transgenic embryo is shown for each construct (red, mRFP; blue, DAPI). Scale bars, 10 μm. (G) Diagram of the experimental setup of inhibitor versus DMSO treatment of embryos from the same batch of microinjected embryos. (H) Percentage of transgenic expression (TE, TE + ICM, or weak) in embryos microinjected with the wildtype (n=30 transgenic/150 total), TEETEADmut (n=91/359), TEERBPJmut (n=66/296) or TEERBPJ/TEADmut (n= 9/79) versions of the TEE and treated with the indicated inhibitor (RO, RO49229097; VP, Verteporfin). **p<0.01, ***p<0.001 by Chi squared test. See also Figure S4.
To confirm the function of the identified binding sites, we combined microinjection of the mutated TEE versions for binding sites of one pathway with pharmacological disruption of the other (Fig. 4G). To disrupt the Notch pathway, we treated microinjected embryos with RO, that as shown above blocks Notch activity in the TE (Fig. 3D). As for the Hippo pathway, we used Verteporfin (VP), a small molecule that inhibits TEAD–YAP association (Liu-Chittenden et al., 2012). To confirm that VP disrupts Hippo signaling as expected, we treated embryos from the mRFP reporter line and examined reporter activity together with endogenous CDX2 expression (Fig. S4A). A high percentage of VP treated embryos phenocopied Tead4−/− embryos (Fig. 2B), with strong reduction of CDX2 but TEE activity still present (Fig. S4A).
As expected, treatment of TEERBPJmut microinjected embryos with RO, or TEETEADmut microinjected embryos with VP had no significant effect on TE specificity of reporter activity (Fig. 4H; S4B, C). RO treatment of TEETEADmut microinjected embryos strongly reduced the number of transgenic embryos showing TE specific expression compared to controls microinjected with TEETEADmut and incubated with DMSO, or with wild type TEE treated with RO (Figs. 4H, S4B). This result confirms that attenuation of Notch signaling diminishes activity of a TEE that lacks functional TEAD sites.
In the converse experiment, treatment with VP of embryos microinjected with the TEERBPJmut significantly reduced TE specificity, as compared to controls incubated with DMSO or with wild type TEE treated with VP (Figs. 4H, S4C). This shows that interfering with the transcriptional activity of YAP and TEAD4 when the TEE RBPJ sites are missing decreases enhancer activity.
Taken together, these results show that transcriptional inputs from both pathways are responsible for full enhancer activity through the specific binding sites identified, acting in a redundant fashion in vivo.
Notch and Hippo pathways converge on Cdx2 regulation
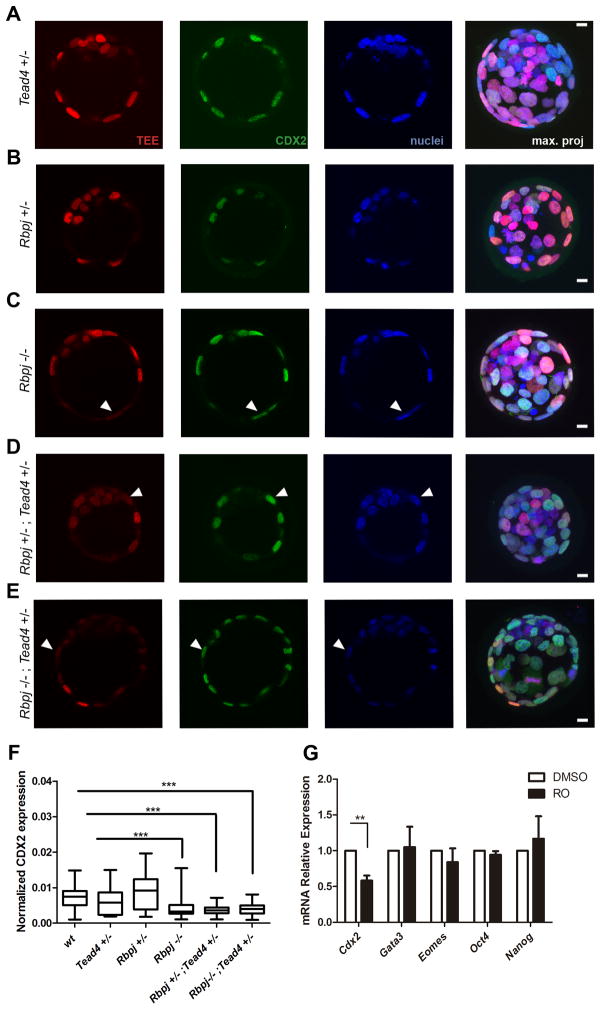
To further explore the regulation of the TEE by Notch, we tested its activity in the absence of Notch signaling by breeding the mRFP-TEE line in the Rbpj KO background (Oka et al., 1995). Consistent with the activity of TEERBPJmut in the TE (Fig. 4D), Rbpj+/− and Rbpj−/− embryos showed normal TEE-driven expression, as is the case for Tead4+/− embryos (Fig. 5A–C). These embryos also had normal numbers of cells and of TEE+ blastomeres, although Rbpj−/− embryos showed a possible but statistically non-significant tendency to have fewer TEE+ cells than wt embryos (Fig. S5A). To test for interaction between the Notch and Hippo on the TEE we generated embryos in the mRFP-TEE line containing different combinations of Tead4 and Rbpj mutant alleles, (Fig. 5D, E). Double heterozygote embryos (Rbpj+/−;Tead4+/−) contained significantly fewer than normal TEE+ cells (Figs 5D, S5A), and this effect was more marked in Rbpj−/−;Tead4+/− embryos, where only a few mRFP cells were detected in the TE (Figs 5E, S5A). This was not due to disruption of overall cell number or the inside-outside distribution of blastomeres in these embryos, as these parameters showed no significant difference compared with wild type, Tead4+/−, Rbpj+/− or Rbpj−/− blastocysts (Fig. S5B). Furthermore, we observed no difference in the total number of CDX2+ cells per embryo in the different genotypes (Fig. S5B), detecting outer blastomeres that express CDX2 but not the reporter (Fig. 5C–E, arrowheads). Unexpectedly, we did not recover any double homozygote knock-out embryos at 3.5 dpc (Fig. S5C), suggesting that the combined lack of these two factors causes the death of embryos at an earlier stage.
Figure 5. TEE activity and Cdx2 expression require transcriptional inputs from Notch and TEAD4.
(A–E) TEE activity (red) and CDX2 immunodetection (green) in (A) Tead4+/−, (B) Rbpj+/−, (C) Rbpj−/−, (D) Rbpj+/−;Tead4+/−, and (E) Rbpj−/−;Tead4+/− mutant embryos. Nuclei were stained with DAPI. Arrowheads in C–E indicate TEE-;Cdx2+ outer blastomeres. Maximal projections of merged images are shown in the right panels. Scale bars, 10 μm. (F) Quantified CDX2 expression in outer cells of wild type blastocysts (n=89, 3 embryos) and in Tead4+/− (n=140, 3 embryos), Rbpj+/− (n=128, 4 embryos), Rbpj−/− (n=150, 4 embryos), Rbpj+/−;Tead4+/− (n=115, 4 embryos) and Rbpj−/−;Tead4+/− allelic combinations (n=159, 3 embryos). Boxes span the 25th to the 75th percentile, internal horizontal lines indicate median values, and whiskers show minima and maxima. ***p<0.001 by Bonferroni post test. (G) Relative expression of Cdx2, Gata3, Eomes, Oct4 and Nanog in pools of 25 embryos (n=6) treated from 2-cell until blastocyst stage with DMSO or RO. Data are means ± s.e.m. **p<0.01 by Student’s t-test. See also Figure S5.
We next investigated whether Notch was required for endogenous levels of Cdx2 expression. Quantification of CDX2 protein per blastomere revealed significantly below-normal expression in Rbpj−/−, Rbpj+/−;Tead4+/− and Rbpj−/−;Tead4+/− embryos, but not in Rbpj or Tead4 single heterozygotes (Fig. 5F). The effect of Notch on Cdx2 expression was confirmed by treating embryos from the 2-cell to blastocyst stages with the γ-secretase inhibitor RO, which significantly decreased mRNA expression of Cdx2, but not Oct4 or Nanog. We also examined changes in expression of other TE-expressed genes, finding that neither Gata3 nor Eomes changed significantly (Fig. 5G), suggesting a specific requirement of Notch for Cdx2 expression. Notch on its own does not impact events downstream or parallel to Cdx2 function, what could be explained by the earlier requirement of Cdx2 as compared to Gata3 and Eomes (Ralston et al., 2010; Strumpf et al., 2005), and is consistent with Notch signaling not being strictly required for trophectoderm development (Souilhol et al., 2006). In line with the genetic analysis, combined treatment of embryos from the mRFP-TEE transgenic line with the pharmacological disruptors of both the Notch and Hippo pathways previously used (RO+VP) strongly reduced TEE activity and endogenous CDX2 expression (Fig. S5D).
Notch signaling and RBPJ are thus necessary for proper expression of endogenous Cdx2 in the embryo, in cooperation with TEAD4. Our results thus show that the Notch and Hippo pathways act together in the pre-implantation embryo, affecting TEE activity, CDX2 expression and embryo viability.
Notch activation increases CDX2 levels and drives blastomeres to the trophectoderm
To further explore the role of Notch in trophectoderm development, we overexpressed the active form of NOTCH1 in the blastocyst by crossing a R26-stop-N1ICD-ires-EGFP line (Murtaugh et al., 2003) with a line carrying a maternal Sox2-Cre allele (Hayashi et al., 2003). Most blastocysts obtained were viable (80%, n=58), as shown by the presence of a blastocoele and proper CDX2 expression in outer cells (Fig. 6A, Fig. S6A–C). Although this strategy predicts uniform recombination in all cells of the blastocyst (Fig. S6G), we found mosaic expression of the reporter at 3.5–4.0 dpc (23 % EGFP-blastomeres; Fig. 6A, E). The degree of mosaicism in R26-stop-N1ICD-ires-EGFP embryos ranged from zero recombined cells to fully recombined blastocysts (Fig. S6A–C). This was confirmed by PCR detection of the non-recombined allele in blastocysts obtained from this cross (Fig. S6E), and we were also able to detect mosaic recombination using a R26-stop-YFP reporter and the maternal Sox2-Cre allele (Srinivas et al., 2001) (Fig. S6F, H).
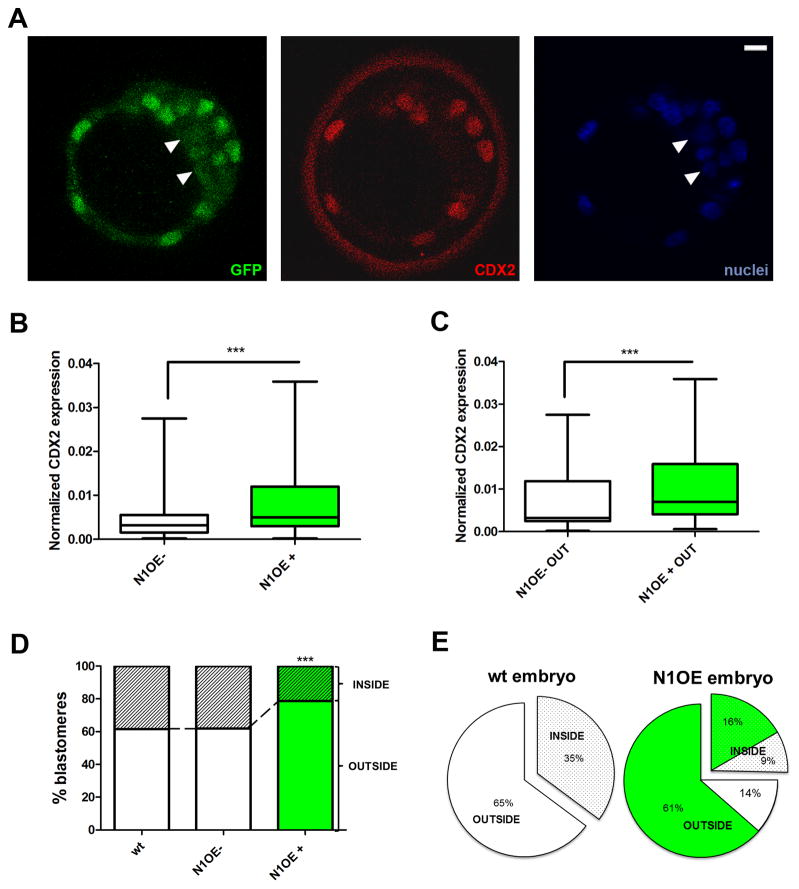
Figure 6. Notch regulates Cdx2 expression and instructs cells to adopt an outer position in the blastocyst.
(A) Expression of the reporter from the R26-stop-N1ICD-ires-EGFP line (green) and CDX2 (red) when recombined by maternal Sox2-Cre. White arrowheads mark non-recombined cells. Nuclei were stained with DAPI. Scale bars, 10 μm. (B) Quantified CDX2 expression in all N1ICD-overexpressing (N1OE+; green) and non-overexpressing (N1OE−) blastomeres in embryos generated from the ♂R26-stop-N1ICD-ires-EGFP X ♀Sox2-Cre cross (n=313 N1OE+; 93 N1OE− blastomeres/10 embryos). (C) Quantified CDX2 expression in outer N1OE+ (n=247) and N1OE− (n=59) blastomeres. In (B and C), boxes span the 25th to the 75th percentile, internal horizontal lines indicate median values, and whiskers show minima and maxima. ***p<0.001 by Student’s t-test. (D) Inside/outside distribution of all N1OE+ and N1OE− blastomeres compared with the distribution in wild type (wt) blastomeres (n=304, 6 embryos). ***p<0.001 by Chi squared test. (E) Distribution per embryo of inside (grey) and outside cells (white) in wild type (wt; n=6) and N1ICD over-expressing embryos (N1OE; n=10). For N1OE embryos, the contribution of EGFP-positive cells (green) to each population is also shown. See also Figure S6.
We took advantage of this mosaicism to investigate whether activation of the Notch pathway could upregulate endogenous Cdx2. Quantification of CDX2 staining at the blastocyst stage showed that blastomeres overexpressing N1ICD have higher levels of CDX2 protein (Fig. 6B). The same pattern was seen when we analyzed only outer cells (those that would normally express Cdx2) (Fig. 6C). These results complement the earlier observation of reduced CDX2 in Rbpj mutant embryos (Fig. 5F), confirming that the Notch pathway directly regulates Cdx2 expression.
In this set of experiments, we also noticed that EGFP+ blastomeres were more often localized to the TE (Fig. 6A), so we analyzed the spatial distribution of blastomeres overexpressing N1ICD in more detail. Whereas EGFP− blastomeres from mosaic embryos showed an inside-outside distribution similar to that of blastomeres from wild type blastocysts, EGFP+ blastomeres were significantly more abundant in outside positions (Fig. 6D). Analysis of individual blastocysts revealed that those overexpressing N1ICD had significantly more outside cells (75% vs 65% in wild type embryos) and that EGFP+ cells were overrepresented in this population compared with inside cells (81.3% vs 64%; Fig. 6E). These results indicate that blastomeres in which the Notch pathway is active localize preferentially to the outer TE population, and that cells can be directed to this population by forced overexpression of the active form of Notch1.
DISCUSSION
In this study, we have dissected the transcriptional regulation of Cdx2 in the preimplantation mouse embryo, and by characterizing an early TE-specific enhancer we have identified Notch signaling as a component of the process. The role of Notch in cell fate decisions during development is well established (Koch et al., 2013), but previously no function had been described in the initial lineage choices in the early embryo, due to the fact that no loss of function mutant of the Notch pathway has a phenotypic effect on preimplantation development (Souilhol et al., 2006). We find that Notch directly regulates Cdx2 expression in cooperation with Tead4, and can also influence the fate of blastomeres by driving them to outer positions in the embryo. This cooperation is also supported by the genetic interaction of the two pathways, as no double homozygous mutants for Tead4 and Rbpj are obtained at preimplantation stages. While other instances of interaction between Notch and Hippo have been described (Barry and Camargo, 2013; Camargo et al., 2007; Chen et al., 2011; Graves et al., 2012; Tschaharganeh et al., 2013), the results of this study show that the Hippo and Notch pathways act in parallel and converge during the first lineage specification in the blastocyst. In this scenario, outside cells more prone to a TE fate would have the Notch pathway active and the Hippo pathway inactive. It is interesting to note that in Drosophila wing epithelial cells, inactivation of Hippo signaling results in apical accumulation of the Notch receptor (Genevet et al., 2009).
Compound mutants for Rbpj and Tead4 show a reduction in the total number of TEE-positive cells but not of CDX2-positive cells. However, they have below-normal expression levels per blastomeres, suggesting that additional regulatory inputs act on Cdx2 in the blastocyst. A possible mechanism is auto-regulation of Cdx2 (Barros et al., 2011; Cockburn and Rossant, 2010; Niwa et al., 2005), which would maintain and stabilize Cdx2 expression levels after initial inputs from Notch and Tead4.
Sustained activity of the Notch pathway in outer cells would ensure that they maintain a TE fate, allowing for correct specification of blastomeres that reposition during cell division. Therefore the forced activation of the pathway we achieve by overexpressing N1ICD drives cells to relocate to the outside and adopt a TE fate. It could be argued that Notch simply elevates Cdx2 levels and that this is the trigger for cell relocation (Jedrusik et al., 2008). However, the fact that blastomeres mutant for Cdx2 can adopt an outer position argues against this possibility (Ralston and Rossant, 2008), and suggests that Notch must target other mechanisms operating in outer cells that results in early lineage segregation.
The Notch signaling pathway’s role in generating heterogeneity from an otherwise homogenous group of cells has been extensively studied (Artavanis-Tsakonas et al., 1999). On the other hand, it has also been shown how Notch can produce local homogeneity in a tissue by signaling between adjacent cells (de la Pompa and Epstein, 2012; Lewis, 1998; Neves et al., 2011). It would be too early to speculate which of these mechanisms, lateral inhibition or lateral induction, is involved in lineage determination in the blastocyst. Future work on how pathway components are differentially localized during preimplantation development and on the precise timing of expression of ligands and receptors will be key to resolve the role of Notch in TE specification.
The early mouse embryo has been proposed to be a self-organizing system (Wennekamp et al., 2013), such that individual cells choose their fate according to a combination of intrinsic and extrinsic cues. In this scenario, the robustness for TE specification would be conferred though combinatorial inputs that would initiate and refine the expression of TE factors such as Cdx2. The partially redundant activities of these inputs would be integrated via regulatory elements such as the Cdx2 TEE, and could underlie the regulatory capacities of the mammalian embryo and the initial stochastic lineage-specific transcriptional programs.
The structure of the Cdx2 TE-enhancer described here can be used to screen for cis-regulatory elements that integrate signals from both pathways, with a similar organization of RBPJ and TEAD sites and thereby identify other components of the TE specification gene regulatory network. Future work will provide information about how modulation of Notch together with other signaling pathways leads to the differentiation of the totipotent cell, and how symmetry is first broken during mammalian development.
EXPERIMENTAL PROCEDURES
Construct generation for microinjection
Cdx2 genomic regions were amplified by PCR using BAC RP245I065 as template. This BAC covers the whole intergenic region containing mouse Cdx2 and was obtained from the BACPAC Resources Center (http://bacpac.chori.org/). The restriction enzyme strategy or primers used for PCR, together with the lengths of corresponding amplified fragments, were as follows: Fragment #1, GTTTGGAGAGAAGAAAGGAG and GGGTGAAGTGAAGAAGATCAG (5428 bp product size); Fragment #2, 1856 bp ApaI digestion of Fragment #1; Fragment #3, 3572 bp ApaI digestion of Fragment #1; Fragment #4, TGCTAACACAAGCTCCCTCA and AAAGCAGGGAAGAGCACTTTA (766 bp); Fragment #5, GACTGGCTGCCTTACCAGAG and TCTTCCAAAGACGCTGGAGT (1487 bp); Fragment #6, CACACGGATGAATTGTCTGG and AACAGGGACAGGTGAGATGG (1329 bp); Fragment #7, GCCTAGGATGCTGACTGAGG and CCCAAGTTGGAAAGGTTTGA (809 bp); Fragment #8, ATCTCACCTGTCCCTGTTGG and CCCTGGGTGAAGTGAAGAAG (1107 bp).
As a positive control, we used the Pou5f1 distal enhancer element (Oct4DE) (Pernaute et al., 2010; Yeom et al., 1996). Each fragment was subcloned in pGEM-T Easy Vector and then excised and cloned into a modified pBluescript vector (Yee and Rigby, 1993) containing either a lacZ reporter gene or H2BmRFP1 reporter gene under the control of the human beta-globin minimal promoter and including an SV40 polyadenylation signal. Constructs were linearized and plasmid sequences removed before microinjection.
Transient transgenic analysis
For the generation of transient transgenics, F1 (C57Bl/6xCBA) females were superovulated to obtain fertilized oocytes as described (Nagy et al., 2003). Each construct was microinjected into the pronucleus of fertilized oocytes at E0.5 at a concentration range of 3–6 ng/μl. Microinjected oocytes were cultured in microdrops of M16 medium (Sigma) coveredwith mineral oil (Sigma) at 37°C, 5% CO2 until the blastocyst stage.
A minimum of 50 blastocysts were used to calculate the percentage of lacZ or H2BmRFP positive embryos per construct. When using the empty vector containing only the minimal promoter and the lacZ reporter as a negative control, we routinely obtain low-level punctuated lacZ expression or weak H2BmRFP expression in approximately 10% of blastocysts. For lacZ staining, blastocysts were fixed in buffer containing 1% formaldehyde, 0.2% glutaraldehyde, 2 mM MgCl2, 5 mM ethyleneglycoltetraacetic acid (EGTA), and 0.02% Igepal for 5 min at room temperature. After washes in phosphate buffered saline (PBS), blastocysts were transferred to X-Gal staining solution for 24 hours at room temperature in the dark. For H2BmRFP detection, embryos were fixed in 4% paraformaldehyde for 10 min at room temperature and either analyzed for endogenous fluorescence or immunostaining. To visualize nuclei, embryos were incubated in DAPI at 1 μg/ml (Vector Laboratories).
Generation of TEE mouse lines
Three independent transgenic mouse lines where obtained for the fragment #3 TEE constructs linked to each lacZ or H2BmRFP. All lines reproduced the TE-restricted expression pattern in early pre-implantation stages. Genotyping was performed by PCR with primers SV40 (TCACTGCATTCTAGTTGTGG) and 5′-rev comp (CTGATCTTCTTCACTTCACCCAG), which generate a 150 bp product for the TEE-lacZ lines, and 5′Cdx2 (GCAGGTGGCTGATCTTCTTC) and mRFP (GAGCCGTACTGGAACTGAGG), which generate a 900 bp product for the TEE-H2BmRFP lines.
PCR conditions were 95°C for 5min, 35 cycles of 94 °C for 1min, 60°C for 1min and 72°C for 1 min, followed by 72 °C for 10 min. Embryos were collected from crosses of TEE-lacZ or TEE-H2BmRFP males with outbred superovulated ICR females.
Mouse breeding
The different mouse lines used are described in the main text. Adults were genotyped by PCR of tail-tip DNA using primers and conditions previously described for each line. For preimplantation embryos, genotyping was performed directly on individually isolated embryos, after observation in culture and X-gal or antibody staining.
Animal procedures were approved, as appropriate, by the CNIC Animal Experimentation Ethics Committee in accordance with Spanish and European regulations or by the Toronto Centre for Phenogenomics Animal Care Committee in accordance with guidelines from the Canadian Council for Animal Care.
Embryo collection and culture
For TEE mouse line characterization, E0.5 embryos were collected from swollen ampulas, treated with hyaluronidase (Sigma) to remove cumulus cells and cultured until the blastocyst stage at 37.5°C in 5% CO2 in air, in M16 medium (Sigma) covered with mineral oil (Sigma). For experiments using Tead4, Cdx2 and Rbpj mutant mouse strains, E2.5 embryos were collected by flushing the oviduct through the infundibulum, and were cultured up to blastocyst stage. For experiments with R26YFP and R26N1OE transgenics, E3.5 embryos were recovered by flushing uteri with M2 medium (Sigma).
Lats2 mRNA injection
The Lats2-GFP construct was transcribed and microinjected into 2-cell embryos of the TEE-H2B-mRFP mouse line as previously described (Cockburn et al., 2013). After injection, embryos were cultured up to the blastocyst stage.
Immunohistochemistry of preimplantation embryos
Immunohistochemistry was performed as previously described (Dietrich and Hiiragi, 2007). The following antibodies and dilutions were used: monoclonal mouse anti-CDX2 (MU392-UC, BioGenex) 1:200, rabbit polyclonal living colors DsRed (632496 Clontech) 1:500, rabbit polyclonal living colors GFP (632460 Clontech) 1:200, mouse anti-TEAD4 (ab58310 Abcam) 1:100, and rabbit anti-Cleaved NOTCH1 (Val1744) (2421, Cell Signaling Technology) 1:100. Cleaved NOTCH1 was also immunodetected with amplification of the signal with a tyramide amplification kit (TSA) coupled to Cy3 (1:100, Perkin Elmer). Nuclei were visualized by incubating embryos in DAPI at 1 μg/ml.
Pharmacological inhibitor treatments
Two or four-cell embryos were cultured in drops of M16 medium (Sigma) covered with mineral oil (Sigma) at 37°C, 5% CO2, containing the corresponding pharmacological inhibitor or control (DMSO) until the blastocyst stage. The following inhibitors and concentrations were used: 10 μM of the γ-secretase inhibitor RO4929097 (S1575, Selleckchem) (Munch et al., 2013) and 2.5 μM of the TEAD/YAP inhibitor Verteporfin (Sigma) (Liu-Chittenden et al., 2012).
Cell culture
5TVER7-ES cells were cultured on gelatin-coated dishes with 1000 U/μl LIF (ESGRO-LIF; Millipore) as previously described (Nishioka et al., 2009). Tead4VP16ER was induced by incubation of cells for 48 h with 4-hydroxytamoxifen (Sigma) at 0.1 μg/ml in EMFI-CM medium (Tanaka et al., 1998) in the presence of FGF4 (1:1000 dilution; R&D Systems) and 1 μg/ml heparin (Sigma). Cell transfections were performed with Lipofectamine 2000 (Invitrogen) and 400 ng DNA (eGFP, empty vector, Oct4DE or TEE) followed by growth for 48 hours.
Quantitative-PCR
RNA was isolated from 5TVER7 cells with the RNeasy Mini Kit (Qiagen) and then reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For γ-secretase inhibitor experiments, RNA from pools of 25 embryos was isolated using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems) and reverse transcribed using the Quantitect Kit (Qiagen). cDNA was used for quantitative-PCR (qPCR) with Power SYBR® Green (Applied Biosystems) in a 7900HT Fast Real-Time PCR System (Applied Biosystems). Expression of each gene was normalized to the expression of the housekeeping genes Actin and Ywhaz (Vandesompele et al., 2002). The following primers were used: Actin, CAGAAGGAGATTACTGCTCTGGCT (forward) and TACTCCTGCTTGCTGATCCACAT (reverse); Cdx2, TCAACCTCGCCACAACCTTCCC (forward) and TGGCTCAGCCTGGGATTGCT (reverse); Eomes, TTCACCTTCTCAGAGACACAGTTCAT (forward) and GAGTTAACCTGTCATTTTCTGAAGCC (reverse); Gata3, GGGTTCGGATGTAAGTCGAG (forward) and CCACAGTGGGGTAGAGGTTG (reverse); Nanog, CTTACAAGGGTCTGCTACTGAGATGC (forward) and TGCTTCCTGGCAAGGACCTT (reverse); Oct4, ATCAGCTTGGGCTAGAGAAGGATG (forward) and AAAGGTGTCCCTGTAGCCTCATAC (reverse); Ywhaz, CGTTGTAGGAGCCCGTAGGTCAT (forward) and TCTGGTTGCGAAGCATTGGG (reverse).
Mutagenesis
Mutated versions of fragment #6 (TEERBPJmut, TEETEADmut and TEERBPJ/TEADmut) were generated by site-directed mutagenesis (Mutagenex Inc.). RBPJ binding sites were located according to the consensus motif and changes that abolish binding were made as described (Tun et al., 1994). TEAD binding sites were based on the MCAT consensus motif (5′-CATTCCA/T-3′) (Anbanandam et al., 2006). The following changes were introduced: RBPJ binding sites, TTCCCACCG to TTCggACCG, TGTGGGAAA to TGTccGAAA, TTCCCAGGT to TTCggAGGT, and TTCCCACTT to TTaggACTT; TEAD binding sites, AATTCCTA to AcggaaTA, and ATTCCAG to cggaaAG. Lower case indicates the altered residues.
Luciferase assays
To generate the pGL3prom-TEE, pGL3prom-TEERBPJmut, pGL3prom-TEETEADmut and pGL3prom-TEERBPJ/TEADmut constructs, the wild type and mutated versions of the 1.3 Kb TEE sequence were subcloned into the SacI/SmaI sites of the pGL3-Promoter Vector (Promega).
HEK 293 cells were seeded at a density of 1.5 × 105 cells per well on 24-well plates 24 hours before transfection. They were then transfected using Lipofectamine2000 (Invitrogen) with the appropriate DNA mix containing pGL3prom-TEE (280 ng), pGL3prom-TEERBPJmut (280 ng), pGL3prom-TEETEADmut (280 ng), pGL3prom-TEERBPJ/TEADmut (280 ng), pGL3-10xRBPJ (McKenzie et al., 2005) (280 ng), N1ICD (100 ng) or Tead4VP16 (Nishioka et al., 2009) (100 ng); pGL3-Renilla (20 ng) was included in all cases. Twenty-four hours after transfection firefly and Renilla luciferase activities were measured using the Dual Luciferase Assay System (Promega). Data are expressed as ratios of firefly to Renilla luciferase activities. Experiments were performed in quadruplicate. Data were normalized to the basal activity of pGL3-10xRBPJ.
Imaging and quantification
Confocal images of microinjected or antibody-stained embryos were acquired with a Leica SP5 confocal microscope. Images were acquired with a 63x objective and 2x zoom every 2.5μm. Images of lacZ-stained embryos were obtained with a Leica DMIRE2 inverted microscope. Images were prepared for figures using Adobe Photoshop CS5.
For quantification, unmodified images were analyzed as previously described using IMARIS imaging software version 7.6.3 (Bitplane AG) (Dietrich and Hiiragi, 2007) with some modifications. Nuclei were segmented in 3D reconstructions based on DAPI staining with an 8 μm isosurface. After computer segmentation, segments were visually inspected and corrected when necessary. The number of nuclei staining positive for TEE, CDX2 or GFP was evaluated visually (8-cell stage embryos) or by segmentation (IMARIS software; E2.5 to E4.5). CDX2 protein level was estimated from unmodified mean fluorescence intensities within segmented nuclei. Mean DAPI fluorescence intensity was used to minimize error caused by staining and confocal imaging variability. CDX2 intensity values for each blastomere were normalized to the mean DAPI fluorescence intensity for each nucleus, and these ratios were normalized to the average mean DAPI intensity per whole embryo.
Statistics
Statistical analyses were performed with GraphPad Prism 5. Data are presented as means ± s.e.m or ± s.d. as indicated in the figures. Differences were considered statistically significant at p < 0.05. p values were calculated by t-test for comparisons of two groups, and ANOVA with Bonferroni post-test for multiple pair wise comparisons. For the data presented in Figs 4H, 6D and E, a Chi square test was performed. All data used for quantitative analysis are supplied in Table S1.
Supplementary Material
Highlights.
A Cdx2 enhancer region drives reporter expression in outer cells of the blastocyst
Notch and Hippo pathways converge on this enhancer to regulate Cdx2
The Notch pathway is activated specifically in the trophectoderm of the blastocyst
Forced expression of Notch drives cells to outer positions and a trophectoderm fate
Acknowledgments
We thank Tristan Rodriguez and Miguel Torres for comments and suggestions, Isabel Rollan, Inmaculada Ors and members of the Manzanares and de la Pompa labs for technical help and support, Simon Bartlett (CNIC) for English editing, and the CNIC Transgenics Unit for generation of mouse lines. This study was funded by grants from the Ministerio de Economia y Competitividad (grant BFU2011-23083 to MM, grant SAF2010-17555 to JLP, FPU Doctoral Fellowship to TR), Comunidad Autónoma de Madrid (grant CELLDD-CM to MM), Human Frontiers Sciences Program (KH), National Institutes of Health (grants RO1-HD052115 and RO1-DK084391 to KH), and Canadian Institutes of Health Research (Doctoral Research Award to KC, grant FRN13426 to JR). The CNIC is supported by the Spanish Ministerio de Economia y Competitividad and the Pro-CNIC Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci USA. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Barros R, da Costa LT, Pinto-de-Sousa J, Duluc I, Freund JN, David L, Almeida R. CDX2 autoregulation in human intestinal metaplasia of the stomach: impact on the stability of the phenotype. Gut. 2011;60:290–298. doi: 10.1136/gut.2010.222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Camargo FD. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol. 2013;25:247–253. doi: 10.1016/j.ceb.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cockburn K, Biechele S, Garner J, Rossant J. The Hippo Pathway Member Nf2 Is Required for Inner Cell Mass Specification. Curr Biol. 2013;23:1195–1201. doi: 10.1016/j.cub.2013.05.044. [DOI] [PubMed] [Google Scholar]
- Cockburn K, Rossant J. Making the blastocyst: lessons from the mouse. J Clin Invest. 2010;120:995–1003. doi: 10.1172/JCI41229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M. Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns. 2004;4:713–717. doi: 10.1016/j.modgep.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Wang CM, Wang TW, Liaw GJ, Hsu TH, Lin TH, Yu JY. The Hippo pathway controls polar cell fate through Notch signaling during Drosophila oogenesis. Dev Biol. 2011;357:370–379. doi: 10.1016/j.ydbio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, Tapon N. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves HK, Woodfield SE, Yang CC, Halder G, Bergmann A. Notch signaling activates Yorkie non-cell autonomously in Drosophila. PLoS One. 2012;7:e37615. doi: 10.1371/journal.pone.0037615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Tenzen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue KI, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al. Polarity-Dependent Distribution of Angiomotin Localizes Hippo Signaling in Preimplantation Embryos. Curr Biol. 2013;13:1181–1194. doi: 10.1016/j.cub.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko KJ, Depamphilis ML. TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development. 2013;140:3680–3690. doi: 10.1242/dev.093799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Messerschmidt DM, Chan SW, Hong W, Knowles BB, Solter D. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 2013;27:1441–1446. doi: 10.1101/gad.219618.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Stevenson P, Ward G, Papadia S, Bading H, Chawla S, Privalsky M, Hardingham GE. Nuclear Ca2+ and CaM kinase IV specify hormonal- and Notch-responsiveness. J Neurochem. 2005;93:171–185. doi: 10.1111/j.1471-4159.2005.03010.x. [DOI] [PubMed] [Google Scholar]
- Munch J, Gonzalez-Rajal A, de la Pompa JL. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development. 2013;140:1402–1411. doi: 10.1242/dev.087346. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsensten M, Vintersten K, Behringer R. Manipulating the mouse embryo: a laboratory manual. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Neves J, Parada C, Chamizo M, Giraldez F. Jagged 1 regulates the restriction of Sox2 expression in the developing chicken inner ear: a mechanism for sensory organ specification. Development. 2011;138:735–744. doi: 10.1242/dev.060657. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Xenopoulos P, Schrode N, Hadjantonakis AK. A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev Biol. 2013;13:15. doi: 10.1186/1471-213X-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Pernaute B, Canon S, Crespo M, Fernandez-Tresguerres B, Rayon T, Manzanares M. Comparison of extraembryonic expression of Eomes and Cdx2 in pregastrulation chick and mouse embryo unveils regulatory changes along evolution. Dev Dyn. 2010;239:620–629. doi: 10.1002/dvdy.22176. [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Souilhol C, Cormier S, Tanigaki K, Babinet C, Cohen-Tannoudji M. RBP-Jkappa-dependent notch signaling is dispensable for mouse early embryonic development. Mol Cell Biol. 2006;26:4769–4774. doi: 10.1128/MCB.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson RO, Rossant J, Tam PP. Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb Perspect Biol. 2012;4:a008235. doi: 10.1101/cshperspect.a008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530–1542. e12. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wennekamp S, Mesecke S, Nedelec F, Hiiragi T. A self-organization framework for symmetry breaking in the mammalian embryo. Nat Rev Mol Cell Biol. 2013;14:454–461. doi: 10.1038/nrm3602. [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.