Abstract
AIM: To investigate the protein profile of human hepatocarcinoma cell line SMMC-7721, to analyze the specific functions of abundant expressed proteins in the processes of hepatocarcinoma genesis, growth and metastasis, to identify the hepatocarcinoma-specific biomarkers for the early prediction in diagnosis, and to explore the new drug targets for liver cancer therapy.
METHODS: Total proteins from human hepatocarcinoma cell line SMMC-7721 were separated by two-dimensional electrophoresis (2DE). The silver-stained gel was analyzed by 2DE software Image Master 2D Elite. Interesting protein spots were identified by peptide mass fingerprinting based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) and database searching.
RESULTS: We obtained protein profile of human hepatocarcinoma cell line SMMC-7721. Among the twenty-one successfully identified proteins, mitofilin, endoplasmic reticulum protein ERp29, ubiquinol-cytochrome C reductase complex core protein I, peroxisomal enoyl CoA hydratase, peroxiredoxin-4 and probable 3-oxoacid CoA transferase 1 precursor were the six novel proteins identified in human hepatocarcinoma cells or tissues. Specific functions of the identified heat-shock proteins were analyzed in detail, and the results suggested that these proteins might promote tumorigenesis via inhibiting cell death induced by several cancer-related stresses or via inhibiting apoptosis at multiple points in the apoptotic signal pathway. Other identified chaperones and cancer-related proteins were also analyzed.
CONCLUSION: Based on the protein profile of SMMC-7721 cells, functional analysis suggests that the identified chaperones and cancer-related proteins have their own pathways to contribute to the tumorigenesis, tumor growth and metastasis of liver cancer. Furthermore, proteomic analysis is indicated to be feasible in the cancer study.
Keywords: Human hepatocarcinoma cell line SMMC-7721, Protein identification, Functional analysis, Heat-shock protein, Tumorigenesis
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the five most common cancers with the highest incident rate worldwide. Its pathogenesis is complex and long, in which multiple factors are involved. The analysis of its pathogenesis and the effects of regulators would give suggestions to the further diagnostic and therapeutic schemes.
The proteomic approach is used to analyze the complete complements of proteins. Proteomics includes not only the identification and quantification of proteins, but also the determination of their localization, modifications, interactions, activities, and, ultimately, their function. The proteomic analysis is one of the valuable tools in the cancer research[1]. The abundant proteomic information about proteins in the aimed tissues or cells of various tumors could help us find the key proteins related to the potential metabolic processes of cancer progression, signaling and development; furthermore, it is useful to discover the cancer-specific protein markers for developing the new schemes for early prediction and diagnosis, even to explore the new drug targets for cancer therapy. In the cancer proteomic analysis, comparative and comprehensive proteomics are two different key methodologies involved. In the comparative proteomics, the protein profiles in differential physiological or pathological status are compared and various displayed proteins are identified. It is useful to find the key proteins related to the changed status of tissues and cells. In the comprehensive proteomics, all the expressed proteins are discovered and counted. The comprehensive proteomics is not only the basis for acquiring the global protein profiles, but also is significant to the exploratory research of proteins interrelations and the related metabolic processes.
The genesis and growth of HCC is complexly and multi-factorially regulated, so it is regarded that several candidate proteins may be involved in this process. To find and identify these candidate proteins, a technology which can screen out the candidate proteins and then acquire their abundant information is needed. Associated with internet query from the related databases, proteomic methods, especially consisting of two-dimensional electrophoresis (2DE) and the subsequent mass spectrometry (MS), are available to analyze the candidate proteins related to HCC genesis and growth. These identified proteins can also become the novel biomarkers for the prediction, diagnosis and therapy of HCC.
Some studies on the comparative proteomic analysis of HCC focus on the novel biomarkers identification for the HCC pathogenesis, diagnosis and therapy. Persistent viral infections, mainly including hepatitis B viruses (HBV) or hepatitis C viruses (HCV) infection[1], are regarded as the main etiological factors of HCC. The chronic liver injuries, including inflammation, regeneration, fibrosis and cirrhosis which are resulted from hepatitis viral infections, are always preformed to HCC too. To get the candidate biomarkers related to the potential etiological factors of HCC, proteomic analyses were performed with the cirrhosis[2], HBV[3] and HCV-associated[4,5] HCC, and various differently expressed proteins related to the etiological factors were identified. Additionally, some of them were suggested to be candidate biomarkers for HCC diagnosis and therapy.
Other proteomic analyses of HCC pay attention to the comprehensive identification of proteins in the hepatocarcinoma cell lines. The comprehensive protein identification in hepatocarcinoma cell lines can give abundant information about the characteristics of expressed proteins in hepatocarcinoma cells, and then help to analyze the interrelations of the expressed proteins and the specific functions of the identified proteins in the cancer-related metabolic processes. Proteins expressed in various hepatocarcinoma cell lines, including BEL7404[6], HepG2[7,8] and HCC-M[9], have been identified. And a few proteins or protein families are pointed out to be cancer-related; meanwhile the associated metabolic processes are also suggested in these studies. Prohibitin, calreticulin and some members of heat-shock protein family, which are identified in the present study, are also introduced in these studies.
To our knowledge, there has not been the comprehensive proteomic analysis of SMMC-7721 cells so far. Only in the study on all-trans retinoic acid (ATRA)-inhibited cell proliferation and migration of SMMC-7721 cell line, the comparative proteomic analysis of SMMC-7721 cells was performed to find the different expressed proteins after the ATRA treatment[10]; however, the comprehensive proteomic analysis is not included. Hepatocarcinoma cell line SMMC-7721 is established from a Chinese HCC patient and is commonly used in the research of HCC in China. Anticancer effects and mechanisms of different anti-cancer drugs, including adriamycin (ADR)[11], paclitaxel (PTX)[12] and docetaxel (Taxotere®)[13], etc., have been experimented on SMMC-7721 cell line. The physical and chemical factors, such as oxidative stress[14], free radical, heat-therapy, ultraviolet radiation, and chemical reagents, could act on SMMC-7721 cells and change their growing status. These physical and chemical factors, combined with suitable clinical schemes, were then introduced to the new methods for HCC prevention and therapy. SMMC-7721 cells were also used to evaluate the targeted and binding efficiency of the targeted drug delivery system and the anti-human HCC monoclonal antibody[15].
In the present work, proteins from hepatocarcinoma cell line SMMC-7721 were well separated by 2DE and identified by peptide mass fingerprinting based on MALDI-TOF-MS. Among 21 successfully identified proteins, six were found to be the novel proteins in the cells or tissues from HCC. Specific functions of the identified proteins were analyzed and their potential contribution to the pathogenesis and development of HCC were explored. The results are useful to study on HCC pathogenesis, and to give support to establish the effective diagnostic and therapeutic schemes.
MATERIALS AND METHODS
Chemicals and materials
Immobilized pH gradient (IPG) strips (pH 3-10, nonlinear, 13 cm) and IPG buffer (pH 3-10, nonlinear) were purchased from Pharmacia, Amersham Biotech. Dithiothreitol (DTT), PMSF and CHAPS were purchased from Sigma Company (USA). All the buffers were made by using high purity MilliQ water. IPGphor isoelectric focusing equipment, Hoefer SE 600 vertical chambers, electrophoresis apparatus, and Image Master 2D Elite 4.01 software were purchased from Pharmacia, Amersham Biotech. Voyager-DE MALDI-TOF MS was product of Applied Biosystems (USA).
SMMC-7721 cell line culture and sample collection
The human hepatocarcinoma cell line SMMC-7721 was supplied by the Center of Laboratory Animals of the Forth Military Medical University, China. SMMC-7721 cells were cultured in RPMI 1640 culture medium, and were generated once in every two days. After five generations, every two flasks of cells were collected into one 1 mL-eppendorf tube after being centrifuged at 1000 × g for 5 min and then resuspended with cooled PBS. The suspended cells were washed with cooled PBS for 3 times and then were centrifuged at 1500 × g for 10 min to get the cell pellet. The supernatant was abandoned carefully and the pellet in eppendorf tube was stored in -80°C ultra low temperature freezer (LegaciTM Refregeration system, REVCO Technologies) for the subsequent 2D PAGE analysis. All the processes were performed in sterile condition.
Sample prepared for 2D-PAGE
Before cell lysis, cooled and distilled water was used to rinse the surface of cell pellets gently for two times to remove the remnant phosphate. For two bottles of cells, 100 μL of lysis solution containing 8 mol/L urea, 40 g/L CHAPS and 20 g/L Pharmalyte 3-10 was added into the eppendorf tube, and well-established freeze-thaw lysis method was used to extract the proteins from SMMC-7721 cells with added 0.1 mmol/L PMSF as protease inhibitor. Then the suspension was centrifuged at 40000 × g for 1 h at 4°C (Beckman TL100 Ultracentrifuge, TLA 100.3 rotor, Fullerton, CA, USA). All the process was carried through in ice bath or at 4°C. The precipitation was removed and the supernatant was retained for the subsequent 2D-PAGE analysis. Protein concentration was measured by the Bradford assay using bovine serum albumin as standard.
First dimension: Isoelectric focusing
Proteins were isoelectrofocused on the IPGphor isoelectric focusing equipment. Extracted protein was mixed with reswelling buffer (8 mol/L urea, 20 g/L CHAPS, 0.7 mg DTT, 1 μL IPG buffer 3-10 and 0.02 g/L bromophenol blue). The mixture was oscillated completely by Vortex Mixer and centrifuged at 2000 × g for 2 min at 4°C to remove the foam. Then 250 μL of supernatant (containing 200 μg proteins) was added in the ceramic strip holder and IPG strip was placed in it with Mineral oil (Pharmacia, Amersham Biotech) covered. The dried IPG strips were rehydrated for 12 h with the sample in reswelling buffer (250 μL) in the ceramic strip holder (1 h without current, followed by 5 h at 30 V and then 6 h at 60 V). After rehydration for 12 h, protein samples were focused at 200 V for 3 h and 500 V for 3 h to remove any ions contained, and then for 30 min each at 1000 V, 5000 V and 8000 V, respectively, followed by 8000 V, for a total of 35000 Vhs.
Second dimension: SDS-PAGE
IPG strips were removed into the first equilibrated buffer (50 mmol/L Tris-HCl, 6 mol/L urea, 20 g/L SDS, 300 mL/L glycerol, 30 mmol/L DTT, pH 6.8) and incubated for 15 min after isoelectric focusing, followed by incubation in the second equilibrated buffer (50 mmol/L Tris-HCl, 6 mol/L urea, 20 g/L SDS, 300 mL/L glycerol, 25 g/L iodoacetamide, pH 6.8) for 15 min. After two-steps equilibration, IPG strips were loaded on the 2D gels (14 cm × 15 cm, 1 mm thickness) which contained 120 g/L acrylamide and were sealed by 5 g/L agarose containing 0.02 g/L bromophenol blue. The second dimension separation was performed using Hoefer SE 600 standard vertical system with the current of 10 mA per strip for 20 min, followed by 20 mA per strip for 5 h. The 2D gels were stained using optimized classic silver staining method which is more suitable for mass spectrometry identification.
Analysis of 2D gel images
The 2D gel images were acquired from a Sharp JX-330 scanner and analyzed by the Image Master 2D Elite software. After intensity calibration, spot detection, background subtraction, normalization and one dimension calibration, the pI and molecular weight of each spot were calculated, followed by the identification based on MALDI-TOF-MS.
Identification of proteins using MALDI-TOF-MS
The selected protein spots were cut from the gel and tryptic digested in gel. The extracted enzymolyzed peptides were identified by peptide mass fingerprinting based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS, Voyager-DE), analyzed using Data explore (Applied Biosystems) and matched in ProFound-Peptide Mapping (http://prowl.rockefeller.edu/profound_bin/WebProFound.exe).
RESULTS
2D-PAGE analysis
The representative examples of cell proteins separated on a 2D-gel are shown in Figure 1, where 200 μg of total proteins extracted from SMMC-7721 cells were loaded. Nearly 1000 proteins spots were obtained in the range of Mr 14.4-94.0 kDa, pI 3-10. We successfully identified 21 proteins and marked in this map with their NCBI/SWISS-PROT number. The 2D map shows that there are various proteins abundantly expressed in the high molecular mass region at acidic end and this characteristic appears in all the 2D gels with different protein sample load. The different lysis solutions (7 mol/L urea, 2 mol/L thiourea, 40 g/L CHAPS, 60 mmol/L DTT, 20 g/L Pharmalyte pH 3-10 and 0.02 g/L bromophenol blue) led to a few changes of protein profile at basic end; however, no change in the high expression of proteins in high molecular mass region at acidic end.
Figure 1.
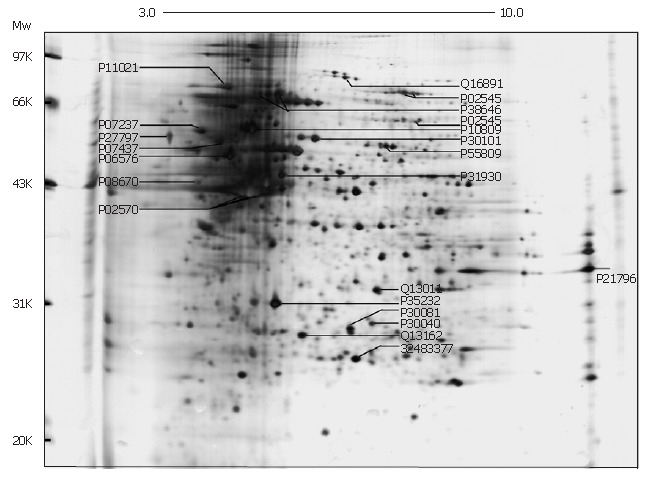
Two-dimensional electrophoresis map of human hepatocarcinoma cell line SMMC-7721.
Protein identification by MS
The information of proteins marked in Figure 1 are shown in Table 1, including the theoretical pI/Mr (kDa) value of identified proteins from Profound, experimental pI/Mr (kDa) value from one-dimensional calibration of 2D image in Image Master 2D Elite, Profound scores and normalization volume. Comparison between experimental and theoretical pI/Mr (kDa) of proteins was used to ensure the protein identification based on the MS results. The Profound score represented the possibility of correct identification in ProFound-Peptide mapping. score of 1.65 for a search means that the search was in the 95th percentile, higher than which is regarded as probably correct match. The volume of each spot was divided by the total volume of all the spots in the gel, and then multiplied by 100, which produced spot percentage volume. This final value is related to the expressing quantity of proteins in cells. "-" means the protein did not perform in normalization calculation. Among the identified proteins (Table 1), several proteins have been reported in the previous studies on other HCC cell lines, such as HSP70, chaperonin CH60, calreticulin, voltage-dependent anion channel protein 1, GRp78 (Bip), prohibitin, vimentin, lamin A/C, protein disulfide isomerase ER60, prolyl 4-hydroxylase beta subunit, ATP synthase beta chain, peroxiredoxin-3 isoform b and mitochondrial short-chain enoyl-coenzyme A hydratase 1. In addition, chaperonin CH60, calreticulin, protein disulfide isomerase ER60, actin and endoplasmic reticulum protein ERp29 were included in the identified proteins in human liver shown in SWISS-2DPAGE[7]. However, to our knowledge, mitofilin, endoplasmic reticulum protein ERp29, ubiquinol-cytochrome C reductase complex core protein I, peroxisomal enoyl CoA hydratase, peroxiredoxin-4 and probable 3-oxoacid CoA transferase 1 precursor are the novel proteins identified in hepatocarcinoma cell lines or tissues. Identified proteins may be potentially involved in the metabolic processes of HCC pathogenesis, development and metastasis, and some of them could become the novel candidate biomarkers in diagnosis and therapy of liver cancer. Five protein spots, although with the good spectra, were returned unmatched in the profound search. They may be the novel proteins and need to be identified further.
Table 1.
Information of the identified proteins in human hepatocarcinoma cell line SMMC-7721
| No. | NCBI/SWISS-PROT accession No. | Protein description | Theoretical pI/Mr (kDa) | Experimental pI/Mr (kDa) | Profound score | Normal volume |
| 159 | P38646 | HSP70, heat shock 70-ku protein 9B precursor | 5.9/73.7 | 5.4/71.0 | 2.37 | 1.228 |
| 149 | P38646 | HSP70, heat shock 70-ku protein 9B precursor | 5.9/73.7 | 5.4/71.8 | 2.30 | - |
| 259 | P10809 | Chaperonin CH60, | 5.7/61.2 | 5.3/58.7 | 2.33 | 0.271 |
| 60-ku heat-shock protein, mitochondrial precursor | ||||||
| 136 | P11021 | GRP78, Bip, 78-ku glucose-regulated protein precursor | 5.2/71.0 | 5.1/75.4 | 2.37 | - |
| 275 | P27797 | Calreticulin precursor | 4.3/48.2 | 4.4/56.0 | 2.35 | 0.383 |
| 757 | P30040 | Endoplasmic reticulum protein ERp29 precursor (ERp31) (ERp28) | 6.8/29.0 | 6.0/29.3 | 2.26 | 0.374 |
| 278 | P30101 | Protein disulfide isomerase ER60 precursor | 6.0/57.1 | 5.7/55.6 | 2.42 | 0.958 |
| 265 | P07237 | Protein disulfide isomerase precursor | 4.8/57.0 | 4.8/57.7 | 2.41 | 0.734 |
| (Prolyl 4-hydroxylase, beta subunit) | ||||||
| 789 | Q13162 | Peroxiredoxin-4 (Prx-IV) (Thioredoxin peroxidase AO372) (Thioredoxin-dependent peroxide reductase A0372) (antioxidant enzyme AOE372) | 5.9/30.7 | 5.6/28.5 | 2.38 | 0.785 |
| 849 | 32483377 | (NP_054817.2) Peroxiredoxin 3 isoform b; thioredoxin-dependent peroxide reductase precursor | 7.1/26.1 | 6.0/26.5 | 1.73 | 1.141 |
| 645 | P21796 | Voltage-dependent anion channel protein I (VDAC1) (plasmalemmalporin) (outer mitochondrial membrane protein porin) (PORIN 31NL) (porin 31HM) | 8.8/30.8 | 9.2/34.4 | 2.23 | 1.572 |
| 372 | P31930 | Ubiquinol-cytochrome C reductase complex core protein I | 5.9/53.3 | 5.5/45.5 | 2.29 | 0.121 |
| 699 | Q13011 | Peroxisomal enoyl-CoA hydratase | 6.6/36.1 | 6.0/32.3 | 2.14 | 0.927 |
| 319 | P06576 | ATP synthase beta chain, mitochondrial precursor | 5.3/56.6 | 5.1/51.1 | 2.39 | 0.222 |
| 774 | P30084 | Mitochondrial short-chain enoyl-coenzyme A hydratase 1 | 6.9/32.0 | 5.9/29.0 | 2.26 | 0.773 |
| 295 | P55809 | Probable 3-oxoacid CoA transferase 1 precursor | 7.2/56.6 | 6.0/53.5 | 1.65 | 0.449 |
| 108 | Q16891 | Mitochondrial inner membrane protein (Mitofilin) (p87/89) | 6.1/84.0 | 5.8/81.8 | 2.24 | 0.410 |
| 714 | P35232 | Prohibitin | 5.6/29.9 | 5.5/31.1 | 2.18 | 1.923 |
| 397 | P08670 | Vimentin | 4.8/41.7 | 4.8/43.2 | 2.19 | - |
| 283 | P07437 | Tubulin beta-2 chain | 4.7/50.2 | 5.0/55.0 | 2.37 | - |
| 153 | P02545 | Lamin A/C isoform 1 precursor | 6.6/74.0 | 6.2/71.6 | 2.30 | 0.297 |
| 150 | P02545 | Lamin A/C isoform 2 | 6.4/65.1 | 6.1/71.8 | 2.28 | 0.444 |
| 246 | P02545 | Lamin A | 6.4/57.7 | 6.2/60.4 | 1.80 | 0.16 |
| 415 | P02570 | Actin cytoplasmic 1 | 5.3/41.7 | 5.3/42.4 | 2.39 | - |
| 416 | P02570 | Actin cytoplasmic 1 | 5.3/41.7 | 5.4/42.6 | 2.23 | - |
| 417 | P02570 | Actin cytoplasmic 1 | 5.3/41.7 | 5.4/42.7 | 2.18 | - |
DISCUSSION
Chaperones are one main kind of proteins identified in SMMC-7721. They are always involved in the regulation of protein synthesis, folding, assembly, modification and isomerization. They are also commonly correlative to the process of transport across membrane and digesting of incorrect peptide folding. Chaperones can form the essential defense mechanism for the protection of cells from a variety of harmful conditions (e.g. temperature elevation or heat shock, decrease in pH, oxidative stress, hypersalinity, fever, inflammation, etc.), and so are suggested to be not only the potential regulator involved in tumor pathogenesis but also the candidate biomarkers for tumor diagnosis and therapy.
Heat-shock proteins (HSPs) belong to the chaperone family, in which HSP60, HSP70 and GRP78 are identified in SMMC-7721. Expression of HSPs could protect cells and tissues from harmful assaults, including lethal temperature and stress. Conversely, the stressful microenvironments in intra- or extra-tumor cells may also stimulate the high expression of HSPs. So it is suspected that the high expression of HSPs may be the response of cells to the stressful microenvironments and may promote tumorigenesis via inhibiting cell death induced by a wide range of stimuli, including several cancer-related stresses like hypoxia, inflammatory cytokines, irradiation, oxidative stress and anticancer drugs[16]. The ability of HSPs to protect cells from stressful stimuli also suggests that these proteins play a role in tumorigenesis, with the fact that cells or tissues from various tumors have been shown to express unusually high levels of one or more HSPs. Furthermore, as the key regulatory points in the control of apoptosis, HSPs function at multiple points in the apoptotic signal pathway and sometimes their expressing levels depend on the expression of other proteins involved in activation of apoptotic pathway[17]. So they could promote tumor cell growth by inhibiting apoptosis or stabilizing the lysosomal membranes[18]. Moreover, HSPs chaperone activity can also influence tumorigenesis by regulating the activity of proteins involved in the cell cycle machinery[19]. However, sometimes the expressing level of HSPs depends on the tumoral level of the neoplasm and is related to the origin of neoplasm tissue[20]. Hence, on the one hand, the high expression of a group of HSPs (i.e. HSP70 and HSP60) found in SMMC-7721 cells can reflect the stress response and self-protective effort of the cells during the genesis of cancer. On the other hand, their involvement in apoptotic process needs further proving. As for the GRP78, many endoplasmic reticulum (ER) proteins interact transiently with it, so it may play a key role in monitoring protein transport through the cell. High expression of GRP78 in SMMC-7721 cells in this study may indicate that the endoplasmic reticulum responds to the stresses in microenvironments of tumor cells at the cellular level and evokes specific signal transduction pathways to induce GRP78 as a result of accumulating malfolded proteins, glycosylation block or calcium ionophore, etc. in the ER. Moreover, GRP78 can reduce the sensitivity of tumor cell to be killed by cytotoxic T lymphocytes, increase its tumorigenesis and prevent its apoptosis[21]. So, GRP78 can be the potential candidate marker for hepatocarcinoma diagnosis and therapy, furthermore, destructing the synthesis of GRP78 may be useful as a new approach to cancer therapy.
Calreticulin (P27797, Figure 1) is pivotal molecule mediating ER-initiated apoptosis, which is involved in ER stress. Associated with other ER chaperones, such as Bip and PDI, calreticulin participates in the assembly and matureness of MHC class I molecule and then cooperates with MHC class I molecule to act on the correct folding and assembly of protein and the presentation of antigen[22], which shows key effect on tumor pathogenesis and development. Calreticulin, down-regulated in the highly metastatic hepatocarcinoma MHCC97-H cells, was involved in the inhibition of angiogenesis and suppression of tumor growth[23]. However, calreticulin is highly expressed in SMMC-7721 cells and human colorectal carcinoma, in which the mechanism is not clear. Thus, the specific cancer-related function of calreticulin needs to be further studied.
Considering the chaperone activity of protein disulfide isomerases (PDIs) involved in the proper folding and formation of proteins, it is reasonable that PDI expression is often up-regulated under stress conditions. This may be useful to explain the high expression of PDIs (i.e., ER60, proly 4-hydroxylase complex and ER29) in the stressful microenvironment in SMMC-7721 cells. For example, ER60 (P30101, Figure 1) with CH60 (P10809) and other enzymes participating in the catalysis of protein folding and assembly were reported to be differently expressed in differentiating Caco-2 cells, a colorectal carcinoma cell line, thereby suggesting that the process of the correct protein folding and assembly may be involved in Caco-2 differentiation[24]. Proly 4-hydroxylase β-subunit (P4HB) was reported to be highly expressed in HER-2/neu-positive breast cancer and may be associated with antioxidant and detoxification pathways which are involved in the microenvironment changes during the tumorigenesis process[25]. ER29 (P30040, Figure 1) is hypothesized to function as a molecular chaperone facilitating not only folding of the secretory proteins in ER[26], but also transporting of thyroglobulin and other secretory proteins to the cell exterior. However, whether ER29 is involved in the tumorigenesis needs to be proved further because its specific functions are not clear so far.
Although the molecular basis for high expression of chaperones and HSPs in tumors is not completely understood and there may be different etiologies involved, the suboptimal cellular environment in poorly vascularised hypoxic tumors or the stressful growth conditions within the solid tumors may contribute to the over-expression of chaperones and HSPs[27]. Moreover, the contribution of chaperones and HSPs to tumorigenesis may be attributed to their pleiotropic activities as molecular chaperones, which provide the cancer cell an opportunity to alter protein activities, in particular the components of the cell cycle machinery, kinases and other proteins implicated in tumor progression[19]. HSPs are useful biomarkers for carcinogenesis in some tissues and indicate the degree of differentiation and aggressiveness of some cancers. The circulating levels of HSPs and anti-HSP antibodies in cancer patients may be also useful in tumor diagnosis[27]. Moreover, compared with the low metastatic subpopulations of lung cancer, the protein profiles showed protein disulfide isomerase ER60, heat-shock protein CH60 and thioredoxin peroxidase (P30101, P10809, Q13162; Figure 1) were significantly down-regulated in high-metastasis subpopulation. These candidate proteins are evidenced to be somehow associated with various aspects of tumor metastasis, such as cell growth, motility, invasion, adhesion, apoptosis and tumor immunity.
Another cancer-related protein identified in SMMC-7721 cells was prohibitin (P35232, Figure 1), which was also identified in hepatocarcinoma HCC-M cell line[9]. Prohibitin is an anti-proliferative protein and may function as a tumor suppressor. Commonly, it is involved in blocking DNA synthesis and the negatively regulation of cell proliferation. However, prohibitin was highly expressed in SMMC-7721 cells and increased expression of prohibitin was also reported in gastric cancer[28], human epithelial ovarian cancer cell line[29] and papillary carcinomas[30]. The high expression of prohibitin may be a reciprocal indicator for the inhibition of cell proliferation or be related to other interaction of prohibitin with various cell-cycle regulatory proteins rather than inducing anti-proliferation.
The identified proteins in the present study may be involved in the tumorigenesis, tumor growth and metastasis of hepatocarcinoma via their own way and some of them may be the candidate biomarkers for hepatocarcinoma diagnosis and therapy. However, some proteins also exist in normal liver cells and may behave different functions during the carcinogenesis in tumor cells or tissues compared with in normal ones. Thus, the expression of these proteins in normal and tumor tissues need to be examined and compared further, and the cloning and functional studies need to be carried out to ensure whether these proteins are significantly involved in the process of carcinogenesis.
COMMENTS
Background
In the processes of hepatocarcinoma genesis, growth and metastasis, proteins are always the main participators involved. So, to get the protein profile of hepatocarcinoma cells or tissues would give us useful information to analyze the protein interactions happened, as well as how the physiological or pathological metabolic processes are activated and developed. Proteomic analysis, based on two-dimensional electrophoresis and MALDI-TOF mass spectrometry, is a useful tool to analyze protein functions in the tumor cells or tissues.
Research frontiers
The present study gives the protein profile of human hepatocarcinoma cell line SMMC-7721. Based on the functional information of proteins identified, their potential involvements in the process of hepatocarcinoma genesis, development and metastasis are presumably suggested.
Innovations and breakthroughs
Some previous studies gave the comprehensive proteomic analysis of other human hepatocarcinoma cell lines. The present study shows the protein profile of human hepatocarcinoma cell line SMMC-7721 and finds six novel proteins which have never been reported before in the liver cancer cells or tissues. The functions of identified proteins are also analyzed, which suggest their involvements in the tumor pathogenesis and development.
Applications
The present study would help better analysis of hepatocarcinoma development, which can promote new therapeutic schemes. Further comparison with normal liver cells or tissues can give more useful information.
Peer review
The current manuscript describes a comprehensive proteomics analysis of the hepatocarcinoma cell line SMMC-7721. Six novel proteins in the setting of hepatocellular carcinoma were identified. My main criticism with regard to the approach of the authors is that they did not use proteins of normal human hepatocytes as reference material. It is clearly significantly more informative to know to which extent a protein expression profile differs from normal cells.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30370403; the Key Project of Chinese Ministry of Education, No. 705046; the Doctoral Foundation of Xi’an Jiaotong University, grants No. DFXJTU2005-05
S- Editor Liu Y L- Editor Kumar M E- Editor Zhou T
References
- 1.Seow TK, Liang RC, Leow CK, Chung MC. Hepatocellular carcinoma: from bedside to proteomics. Proteomics. 2001;1:1249–1263. doi: 10.1002/1615-9861(200110)1:10<1249::AID-PROT1249>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 2.Paradis V, Degos F, Dargère D, Pham N, Belghiti J, Degott C, Janeau JL, Bezeaud A, Delforge D, Cubizolles M, et al. Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology. 2005;41:40–47. doi: 10.1002/hep.20505. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Tan YX, Zhou H, Ding SJ, Li SJ, Ma DJ, Man XB, Hong Y, Zhang L, Li L, et al. Proteomic analysis of hepatitis B virus-associated hepatocellular carcinoma: Identification of potential tumor markers. Proteomics. 2005;5:1125–1139. doi: 10.1002/pmic.200401141. [DOI] [PubMed] [Google Scholar]
- 4.Blanc JF, Lalanne C, Plomion C, Schmitter JM, Bathany K, Gion JM, Bioulac-Sage P, Balabaud C, Bonneu M, Rosenbaum J. Proteomic analysis of differentially expressed proteins in hepatocellular carcinoma developed in patients with chronic viral hepatitis C. Proteomics. 2005;5:3778–3789. doi: 10.1002/pmic.200401194. [DOI] [PubMed] [Google Scholar]
- 5.Takashima M, Kuramitsu Y, Yokoyama Y, Iizuka N, Fujimoto M, Nishisaka T, Okita K, Oka M, Nakamura K. Overexpression of alpha enolase in hepatitis C virus-related hepatocellular carcinoma: association with tumor progression as determined by proteomic analysis. Proteomics. 2005;5:1686–1692. doi: 10.1002/pmic.200401022. [DOI] [PubMed] [Google Scholar]
- 6.Yu LR, Zeng R, Shao XX, Wang N, Xu YH, Xia QC. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis. 2000;21:3058–3068. doi: 10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez JC, Appel RD, Golaz O, Pasquali C, Ravier F, Bairoch A, Hochstrasser DF. Inside SWISS-2DPAGE database. Electrophoresis. 1995;16:1131–1151. doi: 10.1002/elps.11501601190. [DOI] [PubMed] [Google Scholar]
- 8.Fujii K, Kondo T, Yokoo H, Okano T, Yamada M, Yamada T, Iwatsuki K, Hirohashi S. Database of two-dimensional polyacrylamide gel electrophoresis of proteins labeled with CyDye DIGE Fluor saturation dye. Proteomics. 2006;6:1640–1653. doi: 10.1002/pmic.200401346. [DOI] [PubMed] [Google Scholar]
- 9.Seow TK, Ong SE, Liang RC, Ren EC, Chan L, Ou K, Chung MC. Two-dimensional electrophoresis map of the human hepatocellular carcinoma cell line, HCC-M, and identification of the separated proteins by mass spectrometry. Electrophoresis. 2000;21:1787–1813. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1787::AID-ELPS1787>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 10.Wu N, Zhang W, Yang Y, Liang YL, Wang LY, Jin JW, Cai XM, Zha XL. Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics. 2006;6:6095–6106. doi: 10.1002/pmic.200500321. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Cai M, Wang Z, Liu X, Liang Z. Preparation and in vitro killing effect of adriamycin-loaded immunonanosphere against hepatoma led by F (ab')2 Fragment of monoclonal antibodies. Huaxi Yike Daxue Xuebao. 2002;33:8–11, 14. [PubMed] [Google Scholar]
- 12.Yuan JH, Wang XW, Luo D, Xie Y, Xie H. Anti-hepatoma activity of taxol in vitro. Acta Pharmacol Sin. 2000;21:450–454. [PubMed] [Google Scholar]
- 13.Geng CX, Zeng ZC, Wang JY. Docetaxel inhibits SMMC-7721 human hepatocellular carcinoma cells growth and induces apoptosis. World J Gastroenterol. 2003;9:696–700. doi: 10.3748/wjg.v9.i4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren JG, Zheng RL, Shi YM, Gong B, Li JF. Apoptosis, redifferentiation and arresting proliferation simultaneously triggered by oxidative stress in human hepatoma cells. Cell Biol Int. 1998;22:41–49. doi: 10.1006/cbir.1998.0226. [DOI] [PubMed] [Google Scholar]
- 15.Chan KT, Cheng SC, Xie H, Xie Y. A humanized monoclonal antibody constructed from intronless expression vectors targets human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2001;284:157–167. doi: 10.1006/bbrc.2001.4837. [DOI] [PubMed] [Google Scholar]
- 16.Garrido C, Gurbuxani S, Ravagnan L, Kroemer G. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem Biophys Res Commun. 2001;286:433–442. doi: 10.1006/bbrc.2001.5427. [DOI] [PubMed] [Google Scholar]
- 17.Faried A, Sohda M, Nakajima M, Miyazaki T, Kato H, Kuwano H. Expression of heat-shock protein Hsp60 correlated with the apoptotic index and patient prognosis in human oesophageal squamous cell carcinoma. Eur J Cancer. 2004;40:2804–2811. doi: 10.1016/j.ejca.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Nylandsted J, Gyrd-Hansen M, Danielewicz A, Fehrenbacher N, Lademann U, Høyer-Hansen M, Weber E, Multhoff G, Rohde M, Jäättelä M. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200:425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zylicz M, King FW, Wawrzynow A. Hsp70 interactions with the p53 tumour suppressor protein. EMBO J. 2001;20:4634–4638. doi: 10.1093/emboj/20.17.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappello F, Zummo G. HSP60 expression during carcinogenesis: a molecular "proteus" of carcinogenesis? Cell Stress Chaperones. 2005;10:263–264. doi: 10.1379/1466-1268(2005)10[263:HEDCAM]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang ZW, LeBourhis X, Hondermarck H. Progressing growth of tumor cell and synthesis of Bip/GRP78. Zhongguo Yaolixue Tongbao. 2002;18:79–83. [Google Scholar]
- 22.Diedrich G, Bangia N, Pan M, Cresswell P. A role for calnexin in the assembly of the MHC class I loading complex in the endoplasmic reticulum. J Immunol. 2001;166:1703–1709. doi: 10.4049/jimmunol.166.3.1703. [DOI] [PubMed] [Google Scholar]
- 23.Ding SJ, Li Y, Shao XX, Zhou H, Zeng R, Tang ZY, Xia QC. Proteome analysis of hepatocellular carcinoma cell strains, MHCC97-H and MHCC97-L, with different metastasis potentials. Proteomics. 2004;4:982–994. doi: 10.1002/pmic.200300653. [DOI] [PubMed] [Google Scholar]
- 24.Stierum R, Gaspari M, Dommels Y, Ouatas T, Pluk H, Jespersen S, Vogels J, Verhoeckx K, Groten J, van Ommen B. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim Biophys Acta. 2003;1650:73–91. doi: 10.1016/s1570-9639(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4:1686–1696. doi: 10.1074/mcp.M400221-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Hubbard MJ, McHugh NJ. Human ERp29: isolation, primary structural characterisation and two-dimensional gel mapping. Electrophoresis. 2000;21:3785–3796. doi: 10.1002/1522-2683(200011)21:17<3785::AID-ELPS3785>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He QY, Cheung YH, Leung SY, Yuen ST, Chu KM, Chiu JF. Diverse proteomic alterations in gastric adenocarcinoma. Proteomics. 2004;4:3276–3287. doi: 10.1002/pmic.200300916. [DOI] [PubMed] [Google Scholar]
- 29.Gagné JP, Gagné P, Hunter JM, Bonicalzi ME, Lemay JF, Kelly I, Le Page C, Provencher D, Mes-Masson AM, Droit A, et al. Proteome profiling of human epithelial ovarian cancer cell line TOV-112D. Mol Cell Biochem. 2005;275:25–55. doi: 10.1007/s11010-005-7556-1. [DOI] [PubMed] [Google Scholar]
- 30.Srisomsap C, Subhasitanont P, Otto A, Mueller EC, Punyarit P, Wittmann-Liebold B, Svasti J. Detection of cathepsin B up-regulation in neoplastic thyroid tissues by proteomic analysis. Proteomics. 2002;2:706–712. doi: 10.1002/1615-9861(200206)2:6<706::AID-PROT706>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]


