Abstract
AIM: To identify alterations in genes and molecular functional pathways in esophageal cancer in a high incidence region of India where there is a widespread use of tobacco and betel quid with fermented areca nuts.
METHODS: Total RNA was isolated from tumor and matched normal tissue of 16 patients with esophageal squamous cell carcinoma. Pooled tumor tissue RNA was labeled with Cy3-dUTP and pooled normal tissue RNA was labeled with Cy5-dUTP by direct labeling method. The labeled probes were hybridized with human 10K cDNA chip and expression profiles were analyzed by Genespring GX V 7.3 (Silicon Genetics).
RESULTS: Nine hundred twenty three genes were differentially expressed. Of these, 611 genes were upregulated and 312 genes were downregulated. Using stringent criteria (P ≤ 0.05 and ≥ 1.5 fold change), 127 differentially expressed genes (87 upregulated and 40 downregulated) were identified in tumor tissue. On the basis of Gene Ontology, four different molecular functional pathways (MAPK pathway, G-protein coupled receptor family, ion transport activity, and serine or threonine kinase activity) were most significantly upregulated and six different molecular functional pathways (structural constituent of ribosome, endopeptidase inhibitor activity, structural constituent of cytoskeleton, antioxidant activity, acyl group transferase activity, eukaryotic translation elongation factor activity) were most significantly downregulated.
CONCLUSION: Several genes that showed alterations in our study have also been reported from a high incidence area of esophageal cancer in China. This indicates that molecular profiles of esophageal cancer in these two different geographic locations are highly consistent.
Keywords: Esophageal cancer, Gene expression profile, Tobacco consumption, Betel quid chewing, North East India
INTRODUCTION
Environmental carcinogens have repeatedly been shown to affect the genetic material of host cells, leading to uncontrolled growth and ultimately malignant tumors[1]. The development of esophageal cancer is a leading example in which environmental carcinogens in addition to geographic and genetic factors appear to play major etiologic roles[2]. Esophageal cancer occurs at very high frequencies in certain parts of China, Iran, South Africa, Uruguay, France, Italy and in some regions of India[1]. The highest incidence of this cancer in India has been reported from Assam in the North-east region where it is the second leading cancer in men and third leading cancer in women[3].
Tobacco smoking, betel quid chewing, and alcohol consumption are the major known risk factors for esophageal cancer[4]. Tobacco smoke contains over 60 established carcinogens including polycyclic aromatic hydrocarbons and nitrosamines. Tobacco specific nitrosamines such as 4- (methyl nitrosamino)-1-butanone (NNK) and N’-nitrosonornicotine (NNN) that are carcinogens in smokeless tobacco, have been shown to enhance the risk of cancer development by forming adducts with DNA[5]. Betel quid chewing, a common habit in Southeast Asia, has been found to increase the risk of developing esophageal cancer by 4.7-13.3 fold, although other exogenous risk factors may also be involved[4]. Betel quid usually comprises a piece of areca nut, which contains many polyphenols and several alkaloids, Piper betle, and lime with or without Piper betle leaves[6]. Arecoline, a major component of areca nut can produce 3-methyl nitrosamine propionitrile (MNPN), a potent carcinogen and safrole-like DNA adducts that have been shown to be genotoxic and mutagenic. Furthermore, contamination of areca nuts by fungi has been reported to produce carcinogenic aflatoxins. This assumes importance since using fermented areca nut with any form of tobacco is a common habit of people in Assam and has been reported to be a potential risk factor of esophageal cancer in this region[3].
The molecular mechanisms that may lead to the development of esophageal cancer in betel quid chewers and tobacco users are unknown. Recent studies are focusing on mechanisms that can explain the carcinogenic effects of tobacco and areca nut on epithelial cell lines. Incubation of areca nut extract or arecoline with primary oral keratinocytes has been reported to promote cell survival and an inflammatory response by induction of prostaglandin E2, interleukin-6 (IL-6) and cyclooxygenase-2 (COX-2) production via activation of MEK1/ERK/c-Fos pathway[6]. Genotoxic stress as well as tissue inflammation and release of inflammatory mediators have been suggested to be key factors in carcinogenesis of gastrointestinal system. Genotoxic chemicals may induce the release of inflammatory mediators via mitogen activated protein kinase (MAPK) activation. Phosphorylated ERK1/2, JNK, p38 and ERK5 are reported to be significantly increased by exposure to tobacco smoke, indicating the activation of MAPK pathways[7]. NNK has recently been identified as a ligand of neuronal nicotinic acetylcholine receptors, which belong to G-protein-coupled receptors (GPCRs). GPCR induces proliferation through activation of members of the family of MAPKs[8,9].
The gene expression profile of esophageal cancer in a high incidence region of Assam where tobacco use and alcohol consumption are widespread and the users of these two substances are also betel quid chewers, has so far not been investigated. In the current study, cDNA microarray gene expression analysis was done to identify the genes differentially expressed in esophageal cancer associated with prevalent risk factors such as tobacco use and betel quid chewing in a high-risk Indian population.
MATERIALS AND METHODS
Collection of tumor samples
Endoscopic tissue biopsy specimens were taken from 16 patients at Dr. Bhubaneshwar Borooah Cancer Institute (BBCI), Guwahati, Assam. Routine histopathologic analysis was done to confirm the diagnosis. Tumor tissue and matched normal tissue distant to the tumor were collected during endoscopy in RNA later (Ambion, Austin, USA), snap-frozen in liquid nitrogen and stored at -70°C until processed. Informed consent was obtained from all patients. Data of clinicopathologic parameters were obtained from patients’ clinical records, operative notes and pathologic reports. Institutional Human Ethics Committee approved the study.
Sample preparation and chip hybridization
Total RNA isolation: Tissues were ground into powder in -196°C liquid nitrogen and homogenized using Trizol reagent (Invitrogen Life Technologies, CA) for extraction of total RNA following the instruction of the manufacturer. The integrity of total RNA was checked by 1.2% formaldehyde agarose gel electrophoresis (visual presence of 28S and 18S bands). Total RNA with OD260/OD280 > 1.8 was used for microarray experiments.
Experimental design: Total RNA was isolated from normal tissue of the esophagus from all the patients involved in this study and combined to make one common control. We pooled total RNA from biopsy samples of three to four patients in all five experiments on the basis of matching the histological grade to get sufficient amounts of total RNA for direct labeling. Pooled tumor RNAs were labeled with Cy-3 and hybridized against the Cy-5 labeled pooled samples of normal esophageal RNA, which generated a constant control to be used on chips analyzed. Figure 1 shows the study design. Sample pooling was done to rapidly identify tumor markers that were expressed by the majority of tumors in a population. Pooling of RNA samples isolated from tissue is a strategy that can be implemented in microarray experiments when the amount of sample RNA is limited or when variations across the samples must be reduced. The reason behind this approach is that the concentration of an mRNA molecule in a pooled sample is likely to be closer to the average concentration for the class than the concentration in a sample from a single individual. Pooled samples have been shown to accurately reflect gene expression in individual samples and yield reproducible data[10,11].
Figure 1.
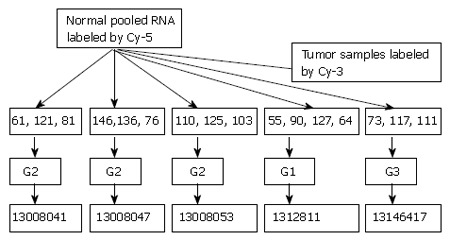
Experimental design: G1, G2 and G3 indicate well-differentiated, moderately and poorly differentiated squamous cell carcinoma respectively. 13008041-53, 1312811 and 13146417 indicated barcode of microarray chips.
Labeling and hybridization: Twenty μg of total RNA from the tumor and matched normal tissue were labeled with cyanine 3-dUTP and cyanine 5-dUTP by direct labeling method (Perkin Elmer Life Sciences, USA: Micromax Direct labeling kit). The labeled probes were denatured at 95°C for 5 min and hybridized with a human 10K cDNA chip (University Health Network, Microarray Centre, Toronto, Canada), which contains 9914 well-characterized human clones, in a hybridization chamber (Corning Life Sciences, USA) at 65°C water bath for 18 h. Before hybridization, slides were pre-hybridized in 5XSSC, 0.1% SDS and 1% BSA solution at 65°C for 45 min to prevent nonspecific hybridization. After hybridization, the slides were washed in 2XSSC with 0.1% SDS, 0.1X SSC with 0.05% SDS and 0.1XSSC sequentially for 20 min each and then spin-dried.
Microarray image analysis: Hybridized arrays were scanned at 5 μm resolution on a Gene Pix 4200A scanner (Axon Instruments Inc. Foster City, CA) at various PMT voltage settings to obtain maximal signal intensities with < 0.1% probe saturation. The Cy5-labelled cDNAs were scanned at 635 nm and the Cy3-labeled cDNA samples were scanned at 532 nm. The resulting TIFF images were analyzed by Gene Pix Pro 6.0.1.27 software (Axon Instrument). Both digital images were overlaid to form a pseudo colored image and a detection method was then used to determine the actual target region based on the information from both red (Cy5) and green (Cy3) pixel values. The ratios of the sample intensity to the reference intensity (green: red) for all of the targets were determined and ratio normalization was performed to normalize the center of the ratio distribution to 1.0. Image processing analysis was used for estimation of spot quality by assigning a quality score to each ratio measurement[12].
Data analysis
Quality assurance: The data sets were imported into Microsoft Excel spreadsheets. Four parameters were used to assess quality of spots with the following features excluded: diameter < 50 μm; > 50% saturated pixels in both channels; < 54% of the pixels with an intensity greater than the median background intensity plus one standard deviation in either channel; flagged by Gene Pix as “not found” or “absent” or manually flagged as “bad” due to high background, misshapen features, scratches or debris on the slide undetected by Gene Pix[13].
Normalization: The results were normalized for labeling and detection efficiencies of the two fluorescence dyes, prior to determining differential gene expression between tumor and normal tissue samples. Intensities of selected spots were transformed into log2 (Cy3/Cy5) and data were normalized by locally weighted linear regression (LOWESS) method. Genespring software version GX7.3.1 (Silicon Genetics, Redwood, CA) was used to normalize values for each gene for data analysis.
Ranking of genes: Data analysis was performed using Genespring Software GXV 7.3 (Silicon Genetics, Redwood City, CA). Differentially regulated genes were ranked on the basis of signal intensity, normalized ratio, flag value and variance across replicate experiments. Top ranked genes had a higher intensity, high-normalized ratio for up and low for down, unflagged, very low variance or standard deviation. Filtered genes identified to be differentially expressed by 1.5 fold or greater in three of five chips were analyzed for functional gene clusters using GeneSpring software GXV 7.3. GeneSpring used data found publicly in genomic databases to build gene ontology based on annotation information. t-test was performed at the 0.05% significant level to find genes that vary significantly across samples. P-Value or probability value is the chance of set of genes involved in a particular function to be present in any given gene list with reference to the number of genes known to be involved in the function.
Hierarchical clustering: Average linkage hierarchical clustering was done using the Cluster Software version 3.0 written by Michael Eisen[14]. The Euclidean distance metric was used as a measure of similarity between the gene expression patterns for each pair of samples based on log-transformed ratios across all genes. The results were analyzed and visualized with the Tree View Program Version 1.50 also written by Michael Eisen. Those genes showing progressive fold increases or decreases in gene expression relative to normal mucosa were shown proportionally in red and green, respectively.
Pathway prediction analysis: We obtained annotations of the bioprocesses, molecular function and cellular localization using the freely available Gene Ontology and Source database[15]. The significant gene clusters were queried with known components of the biological pathways on the freely available KEGG database[16]. We also used the Biointerpreter software (http://www.genotypic.co.in/biointerpreter) for gene ontology.
RESULTS
Sixteen esophageal biopsy samples were compared with normal pooled esophageal tissue. All patients were male and gave a history of tobacco consumption and betel nut chewing (Table 1). Gene expression was measured using microarrays to detect changes in tumor samples compared to normal. Nine hundred and twenty three genes were differentially expressed at least 1 fold in 3 out of 5 experiments. Of these, 611 genes were upregulated and 312 genes were down regulated. The scaled data generated from Gene Pix Pro 6.0.1.27 software were imported into GeneSpring for fold change analysis, filtering, and cluster analysis. Hierarchical Clustering analysis of 923 genes selected from the 10 000-gene sets was performed (Figure 2). To identify highly reproducible changes, data were filtered based on select criteria. Transcripts modulated by a minimum 1.5 fold in at least three of five chips were used for further analyses. However, genes that had a ≥ 1.5 fold cutoff value or had a P value of ≤ 0.05 were also included for analysis to seek for subtle changes in gene expression. Using stringent criteria (P ≤ 0.05 and ≥ 1.5 fold change), 127 differentially expressed genes (87 upregulated and 40 down regulated) were identified in tumor tissue. Using the Gene Ontology database, we categorized the 923 differentially expressed genes into known or probable functional categories. Genes involved in dimethylallyltransferase activity and farnesyltransferase activity (CTLA4), cation antiporter activity (SLC9A2) and cation transporter activity (KCNN2, SLC30A4, KCNJ15, CACNA2D3), G-protein coupled receptor (GPCR) activity (GPR87, NPY), MAPK signaling pathway (FGF12), and protein serine or threonine kinase activity (GRK4) were significantly upregulated (Table 2). Out of 87 upregulated genes, two genes were involved in GPCR group, five genes were involved in cation transporter activity, one gene each was involved in protein serine or threonine kinase activity and MAPK activity. Genes involved in anti-apoptosis activity (BIRC1), omega peptidase activity (UCHL1) and cellular proliferation (EGR2) were also significantly activated. Genes involved in structural constituent of the ribosome (RPL32, RPS4X), structural constituent of cytoskeleton (KRT17, KRT8, PLA2G1B), cysteine protease inhibitor activity (CSTB, CSTA), anti-oxidant activity (PRDX6), acyl groups transferase activity (TGM3), and translation elongation (EEF1A1) were significantly down-regulated (Table 3). Genes involved in humoral immune response (CD24) and base-excision repair (MPG) were significantly down regulated. Out of 40 down regulated genes, five genes were involved in structural constituent of the ribosome, four genes were involved in structural constituent of cytoskeleton, two genes were involved in cysteine protease inhibitor activity and one gene each was involved in anti-oxidant activity, acyl group transferase activity and translation elongation.
Table 1.
Demographic and clinical characteristics of esophageal squamous cell carcinoma cases
| Patient ID | Age | Sex | Tobacco/ smoking habit | Alcohol use | Betel nut chewing | Pathological grade |
| EC-61 | 55 | M | Yes | Yes | Yes | G2 |
| EC-64 | 45 | M | Yes | Yes | Yes | G1 |
| EC-55 | 52 | M | Yes | Yes | Yes | G1 |
| EC-103 | 59 | M | Yes | Yes | Yes | G2 |
| EC-111 | 55 | M | Yes | Yes | Yes | G3 |
| EC-90 | 50 | M | Yes | Yes | Yes | G1 |
| EC-117 | 60 | M | Yes | No | Yes | G3 |
| EC-73 | 50 | M | Yes | No | Yes | G3 |
| EC-125 | 71 | M | Yes | No | Yes | G2 |
| EC-110 | 70 | M | Yes | Yes | Yes | G2 |
| EC-146 | 45 | M | Yes | No | Yes | G2 |
| EC-136 | 48 | M | Yes | Yes | Yes | G2 |
| EC-127 | 58 | M | Yes | Yes | Yes | G1 |
| EC-121 | 55 | M | Yes | Yes | Yes | G2 |
| EC-76 | 50 | M | Yes | Yes | Yes | G2 |
| EC-81 | 55 | M | Yes | Yes | Yes | G1 |
G1 = Well differentiated squamous cell carcinoma; G2= Moderately differentiated squamous cell carcinoma; G3= Poorly differentiated squamous cell carcinoma.
Figure 2.
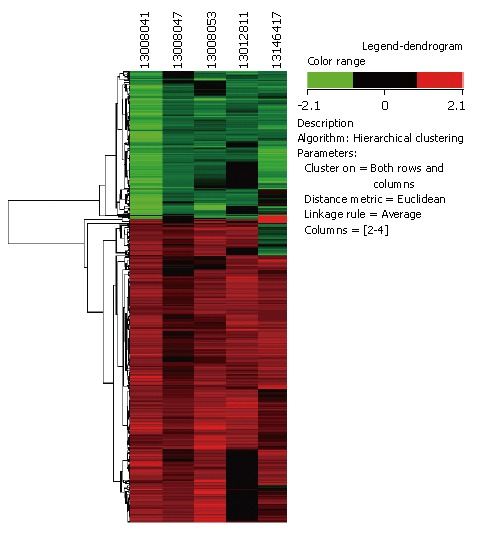
Hierarchical Clustering (Average Linkage Clustering) of the genes that were over or under expressed in tumor versus normal tissue in the 16 ESCC patients.
Table 2.
List of significantly upregulated genes in esophageal cancer patients
| Category | Genes in category | % Genes in category | Genes in list in category | % Genes in list category | P-value |
| GO: 4930: G-protein coupled receptor activity | 150 | 2.148 | 23 | 3.433 | 0.0156 |
| GO: 8324: cation transporter activity | 307 | 4.396 | 39 | 5.821 | 0.0404 |
| GO: 4674: protein serine/threonine kinase activity | 285 | 4.081 | 38 | 5.672 | 0.022 |
| GO: 4707: MAP kinase activity | 20 | 0.286 | 5 | 0.746 | 0.0368 |
| GO: 4161: dime- thylallyltransferase activity | 4 | 0.0573 | 3 | 0.448 | 0.00327 |
Table 3.
List of significantly down-regulated genes in esophageal cancer patients
| Category | Genes in category | % Genes in category | Genes in list in category | % Genes in list category | P-value |
| GO: 3735: structural constituent of ribosome | 115 | 1.647 | 32 | 4.992 | 5.54E-09 |
| GO: 4869: cysteine protease inhibitor activity | 27 | 0.387 | 11 | 1.716 | 1.19E-05 |
| GO: 5200: structural constituent of cytoskeleton | 63 | 0.902 | 12 | 1.872 | 0.011 |
| GO: 16746: transferase activity, transferring acyl groups | 90 | 1.289 | 19 | 2.964 | 0.000422 |
| GO: 16209: antioxidant activity | 25 | 0.358 | 9 | 1.404 | 0.000232 |
| GO: 3746: translation elongation factor activity | 14 | 0.2 | 5 | 0.78 | 0.00636 |
DISCUSSION
Several tobacco constituents, including nitrosamines, polycyclic aromatic hydrocarbons, aromatic amines, various aldehydes and phenols, may be causally related to esophageal cancer[1]. A previous report has shown that betel quid chewing with or without tobacco consumption is associated with the development of esophageal cancer in Assam[3]. However, there are very few studies of gene expression profiles of tumors that may be associated with betel quid chewing as well as tobacco consumption. The current study was aimed to analyze genes and pathways that may be involved in tobacco and betel quid chewing related esophageal malignancies in this high incidence region of India.
Gene expression levels showed that there were four different molecular functional pathways that were most significantly upregulated and six different molecular functional pathways that were most significantly down regulated. Some of the significantly overexpressed molecular functional pathways like MAPK signaling pathway, G-protein coupled receptor and cation transporter activity have earlier been reported in esophageal and other cancers. However, genes such as CTLA4 (involved in dimethylallyltransferase activity and farnesyltransferase activity) and UCHL1, NPY, FGF12, KCNN2, and KCNJ15 were found to be significantly upregulated in our study and have not been reported earlier. Genes involved in structural constituent of ribosome (RPL32, RPS4X, RPL7A) and anti-oxidant activity (PRDX6) that were found to be significantly down regulated in our study, have also not been reported earlier. Some of the significantly down regulated molecular functional pathways like structural constituent of cytoskeleton (KRT17, KRT8, KRT4), acyl groups transferase activity (TGM3) and cysteine protease inhibitor activity (CSTB, CSTA), have already been reported previously in esophageal carcinoma. The data may be used for selection of a limited number of markers that can be screened in large populations by RT-PCR. All the genes identified here are of interest because of their potential roles in the natural history of esophageal squamous cell carcinoma.
The mitogen activated signaling cascade showed significant up-regulation in our study. Activated MAPK pathway has been detected in many human tumors including carcinomas of the breast, colon, kidney, and lung suggesting the possibility that MAPK may play a major role in tumor progression and metastasis. MAPK induces proteolytic enzymes that degrade the basement membrane, enhance cell migration, initiate several pro-survival genes and maintain growth[17]. Oxidants in cigarette smoke have previously been reported to activate MAPK signaling cascades in lung epithelial cells in vitro and in vivo. These signaling pathways lead to the enhanced ability of Jun and Fos family members to activate transcription of a number of AP-1 dependent target genes involved in cell proliferation, differentiation, and inflammation[18]. Phosphorylated ERK 1/2, JNK, p38 and ERK5 genes were significantly increased upon exposure to tobacco smoke, indicating the activation of MAPK pathways[7]. Benzo (a) pyrene quinines, which is the non-volatile component of cigarette smoke, have also been reported to increase cell proliferation, generate reactive oxygen species, and transactivate the epidermal growth factor receptor in breast epithelial cells[19].
TNF- alpha was found to be upregulated in our study. Cigarette smoke is known to enhance the induction of TNF-alpha by differentiated macrophage that is regulated primarily via the ERK ½ pathway[20]. TNF receptor super family member 17 has been shown to specifically bind to the TNF (ligand) super family member 13b (TNFSF13B/TALL-1/BAFF) , which leads to activation of NF-kappaB and MAPK8/JNK.
Neuropeptide Y (NPY), which belongs to G-protein coupled receptor family, was found to be significantly upregulated in our study. Aberrant NPY expressions are early events in prostrate cancer development and are associated with a poor prognosis[21]. Activation of the Y1 receptor by NPY regulates the growth of prostate cancer cells. Estrogen upregulates NPY receptor expression in a human breast cancer cell line[22].
Ion channel regulating genes showed significant upregulation in our study. Several tumors such as prostrate, uterus, glial cells, stomach, pancreas, breast and colorectum are known to express Ca2+activated K+ channels. The complex process of carcinogenesis is triggered by oncogenic pathways via activation of K+ channels. For example, p21 ras and the Raf kinase are known to induce oncogenic transformation via activation of Ca2+ dependent K+ channels. Enhanced cell migration and tumor metastasis are associated with fluctuations in the activity of membrane transporters and ion channels that require K+ channel activity[23]. Genes involved in calcium regulation and calcium signaling also seemed to be important in the progression of esophageal squamous dysplasia[24].
The deleted in colorectal cancer (DCC) gene, which was found to be upregulated in our study, is a candidate tumor suppressor gene which may be associated with differentiation and proliferation of normal cells. DCC protein expression seems to be a significant prognostic factor in high-risk resected gastric cancer. This gene may play a role in the metastatic potential of these tumors[15].
Transglutaminase-3 (TGase-3) that showed significant down regulation in our study has been reported in earlier studies. TGase-3 has been implicated in the formation and assembly of the cornified cell envelope of the epidermis, hair follicle and perhaps other stratified squamous epithelia. Alterated TGase-3 expression is a common event in human esophageal cancer[25]. The lowest levels of TGases 3 and 7 have been reported in patients with metastatic disease[26]. Tissue transglutaminase (tTG) is a high level phenotypic biomarker down regulated in prostate cancer[27].
Intermediate filaments form the cytoskeleton of cells and maintain the integrity of cells. Keratins (KRT4 and KRT8) showed significant down regulation in our study. Overexpression of epithelial cell intermediate filaments and their isoforms (KRT8) have been reported earlier in colorectal polyp and cancer[28].
Reduced expression of cystatin B as found in our study, has earlier been reported to be associated with lymph node metastasis and may therefore prove to be a useful marker for predicting the biologic aggressiveness of esophageal cancer[29]. Overexpression of cystatin A has been shown to inhibit tumor cell growth, angiogenesis, invasion, and metastasis in esophageal cancer[30]. High levels of cystatin A and cystatin B have been reported to correlate with more favorable patient outcome in breast, lung and head and neck tumors.
A group of ribosomal proteins may function as cell cycle checkpoints and compose a new family of cell proliferation regulators. They play an important role in translational regulation and control of cellular transformation, tumor growth, aggressiveness and metastasis. Thus in addition to protein synthesis, they are involved in neoplastic transformation of cells. Ribosomal proteins were found to be significantly down regulated in our study. Expression of human ribosomal protein L13a has been shown to induce apoptosis, presumably by arresting cell growth in the G2/M phase of the cell cycle. In addition, a closely related ribosomal protein, L7, arrests cells in G1 and also induces apoptosis[15]. Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. MRPL41 is reported to be either expressed at reduced levels or absent in most tumor types and cell lines[31]. Human apurinic apyrimidinic endonuclease (RPLP0) and its N terminal truncated form (AN34) are involved in DNA fragmentation during apoptosis. Down regulation of RPLP0 expression is associated with the induction of apoptosis in differentiating myeloid leukemia cells[32]. Simultaneously, three ribosomal proteins (RPL10, RPL32, and RPS16) also showed up-regulation in C81 cells. Overexpression of several ribosomal proteins including RPS16 has been reported in colon, breast, liver, and pancreatic tumors[33].
This is the first study to provide gene expression profiles of esophageal cancer in a high-risk region of Assam in India. The most salient finding was identification of down regulated genes involved in structural constituents of ribosome and upregulated genes involved in cation transporter activity. In a similar study of gene expression profiles in a high-risk area of China, Taylor et al[12] and Liu et al[35,36] have reported down regulation of CSTB, CSTA, KRT4, and TGM3 and upregulation of G-coupled signaling, ion transport activity and MAPK activity[24]. In a recent study from a low risk area of India, deregulation of genes associated with zinc homeostasis in esophageal squamous cell carcinoma (ESCC) has been reported[34]. These data indicate the consistency of molecular profiles of esophageal cancer in two different geographic locations that have a high incidence of ESCC but different food habits and customs.
ACKNOWLEDGMENTS
The authors are thankful to Madavan Vasudevan, Dr. Sudha Rao and Dr. Raja Maugasimangalam of Genotypic Technology (P) Ltd. Bangalore, India for their valuable support in Microarray Data Analysis. The authors are thankful to Indian Council of Medical Research for their financial support. The authors are thankful to Jagannath Sharma of BBCI, Guwahati for histopathological analysis.
Footnotes
Supported by Non Communicable Disease Division, Indian Council of Medical Research
References
- 1.Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis. 2001;22:1737–1746. doi: 10.1093/carcin/22.11.1737. [DOI] [PubMed] [Google Scholar]
- 2.Murtaza I, Mushtaq D, Margoob MA, Dutt A, Wani NA, Ahmad I, Bhat ML. A study on p53 gene alterations in esophageal squamous cell carcinoma and their correlation to common dietary risk factors among population of the Kashmir valley. World J Gastroenterol. 2006;12:4033–4037. doi: 10.3748/wjg.v12.i25.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phukan RK, Ali MS, Chetia CK, Mahanta J. Betel nut and tobacco chewing; potential risk factors of cancer of oesophagus in Assam, India. Br J Cancer. 2001;85:661–667. doi: 10.1054/bjoc.2001.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CH, Lee JM, Wu DC, Hsu HK, Kao EL, Huang HL, Wang TN, Huang MC, Wu MT. Independent and combined effects of alcohol intake, tobacco smoking and betel quid chewing on the risk of esophageal cancer in Taiwan. Int J Cancer. 2005;113:475–482. doi: 10.1002/ijc.20619. [DOI] [PubMed] [Google Scholar]
- 5.Rohatgi N, Matta A, Kaur J, Srivastava A, Ralhan R. Novel molecular targets of smokeless tobacco (khaini) in cell culture from oral hyperplasia. Toxicology. 2006;224:1–13. doi: 10.1016/j.tox.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Chang MC, Wu HL, Lee JJ, Lee PH, Chang HH, Hahn LJ, Lin BR, Chen YJ, Jeng JH. The induction of prostaglandin E2 production, interleukin-6 production, cell cycle arrest, and cytotoxicity in primary oral keratinocytes and KB cancer cells by areca nut ingredients is differentially regulated by MEK/ERK activation. J Biol Chem. 2004;279:50676–50683. doi: 10.1074/jbc.M404465200. [DOI] [PubMed] [Google Scholar]
- 7.Zhong CY, Zhou YM, Douglas GC, Witschi H, Pinkerton KE. MAPK/AP-1 signal pathway in tobacco smoke-induced cell proliferation and squamous metaplasia in the lungs of rats. Carcinogenesis. 2005;26:2187–2195. doi: 10.1093/carcin/bgi189. [DOI] [PubMed] [Google Scholar]
- 8.Arredondo J, Chernyavsky AI, Jolkovsky DL, Pinkerton KE, Grando SA. Receptor-mediated tobacco toxicity: cooperation of the Ras/Raf-1/MEK1/ERK and JAK-2/STAT-3 pathways downstream of alpha7 nicotinic receptor in oral keratinocytes. FASEB J. 2006;20:2093–2101. doi: 10.1096/fj.06-6191com. [DOI] [PubMed] [Google Scholar]
- 9.Sud N, Sharma R, Ray R, Chattopadhyay TK, Ralhan R. Differential expression of G-protein coupled receptor 56 in human esophageal squamous cell carcinoma. Cancer Lett. 2006;233:265–270. doi: 10.1016/j.canlet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Helm J, Enkemann SA, Coppola D, Barthel JS, Kelley ST, Yeatman TJ. Dedifferentiation precedes invasion in the progression from Barrett's metaplasia to esophageal adenocarcinoma. Clin Cancer Res. 2005;11:2478–2485. doi: 10.1158/1078-0432.CCR-04-1280. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal D, Chen T, Irby R, Quackenbush J, Chambers AF, Szabo M, Cantor A, Coppola D, Yeatman TJ. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. J Natl Cancer Inst. 2002;94:513–521. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- 12.Su H, Hu N, Shih J, Hu Y, Wang QH, Chuang EY, Roth MJ, Wang C, Goldstein AM, Ding T, et al. Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Cancer Res. 2003;63:3872–3876. [PubMed] [Google Scholar]
- 13.Samimi G, Manorek G, Castel R, Breaux JK, Cheng TC, Berry CC, Los G, Howell SB. cDNA microarray-based identification of genes and pathways associated with oxaliplatin resistance. Cancer Chemother Pharmacol. 2005;55:1–11. doi: 10.1007/s00280-004-0819-9. [DOI] [PubMed] [Google Scholar]
- 14.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy KB, Nabha SM, Atanaskova N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003;22:395–403. doi: 10.1023/a:1023781114568. [DOI] [PubMed] [Google Scholar]
- 18.Mossman BT, Lounsbury KM, Reddy SP. Oxidants and signaling by mitogen-activated protein kinases in lung epithelium. Am J Respir Cell Mol Biol. 2006;34:666–669. doi: 10.1165/rcmb.2006-0047SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burdick AD, Davis JW, Liu KJ, Hudson LG, Shi H, Monske ML, Burchiel SW. Benzo(a)pyrene quinones increase cell proliferation, generate reactive oxygen species, and transactivate the epidermal growth factor receptor in breast epithelial cells. Cancer Res. 2003;63:7825–7833. [PubMed] [Google Scholar]
- 20.Demirjian L, Abboud RT, Li H, Duronio V. Acute effect of cigarette smoke on TNF-alpha release by macrophages mediated through the erk1/2 pathway. Biochim Biophys Acta. 2006;1762:592–597. doi: 10.1016/j.bbadis.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Rasiah KK, Kench JG, Gardiner-Garden M, Biankin AV, Golovsky D, Brenner PC, Kooner R, O'neill GF, Turner JJ, Delprado W, et al. Aberrant neuropeptide Y and macrophage inhibitory cytokine-1 expression are early events in prostate cancer development and are associated with poor prognosis. Cancer Epidemiol Biomarkers Prev. 2006;15:711–716. doi: 10.1158/1055-9965.EPI-05-0752. [DOI] [PubMed] [Google Scholar]
- 22.Amlal H, Faroqui S, Balasubramaniam A, Sheriff S. Estrogen up-regulates neuropeptide Y Y1 receptor expression in a human breast cancer cell line. Cancer Res. 2006;66:3706–3714. doi: 10.1158/0008-5472.CAN-05-2744. [DOI] [PubMed] [Google Scholar]
- 23.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 24.Joshi N, Johnson LL, Wei WQ, Abnet CC, Dong ZW, Taylor PR, Limburg PJ, Dawsey SM, Hawk ET, Qiao YL, et al. Gene expression differences in normal esophageal mucosa associated with regression and progression of mild and moderate squamous dysplasia in a high-risk Chinese population. Cancer Res. 2006;66:6851–6860. doi: 10.1158/0008-5472.CAN-06-0662. [DOI] [PubMed] [Google Scholar]
- 25.Chen BS, Wang MR, Xu X, Cai Y, Xu ZX, Han YL, Wu M. Transglutaminase-3, an esophageal cancer-related gene. Int J Cancer. 2000;88:862–865. doi: 10.1002/1097-0215(20001215)88:6<862::aid-ijc4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 26.Jiang WG, Ablin R, Douglas-Jones A, Mansel RE. Expression of transglutaminases in human breast cancer and their possible clinical significance. Oncol Rep. 2003;10:2039–2044. [PubMed] [Google Scholar]
- 27.Lewis TE, Milam TD, Klingler DW, Rao PS, Jaggi M, Smith DJ, Hemstreet GP, Balaji KC. Tissue transglutaminase interacts with protein kinase A anchor protein 13 in prostate cancer. Urol Oncol. 2005;23:407–412. doi: 10.1016/j.urolonc.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Polley AC, Mulholland F, Pin C, Williams EA, Bradburn DM, Mills SJ, Mathers JC, Johnson IT. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal neoplasia. Cancer Res. 2006;66:6553–6562. doi: 10.1158/0008-5472.CAN-06-0534. [DOI] [PubMed] [Google Scholar]
- 29.Shiraishi T, Mori M, Tanaka S, Sugimachi K, Akiyoshi T. Identification of cystatin B in human esophageal carcinoma, using differential displays in which the gene expression is related to lymph-node metastasis. Int J Cancer. 1998;79:175–178. doi: 10.1002/(sici)1097-0215(19980417)79:2<175::aid-ijc13>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Ding F, Zhang L, Liu Z, Wu Y, Luo A, Wu M, Wang M, Zhan Q, Liu Z. Overexpression of stefin A in human esophageal squamous cell carcinoma cells inhibits tumor cell growth, angiogenesis, invasion, and metastasis. Clin Cancer Res. 2005;11:8753–8762. doi: 10.1158/1078-0432.CCR-05-0597. [DOI] [PubMed] [Google Scholar]
- 31.Yoo YA, Kim MJ, Park JK, Chung YM, Lee JH, Chi SG, Kim JS, Yoo YD. Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol Cell Biol. 2005;25:6603–6616. doi: 10.1128/MCB.25.15.6603-6616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida A, Urasaki Y, Waltham M, Bergman AC, Pourquier P, Rothwell DG, Inuzuka M, Weinstein JN, Ueda T, Appella E, et al. Human apurinic/apyrimidinic endonuclease (Ape1) and its N-terminal truncated form (AN34) are involved in DNA fragmentation during apoptosis. J Biol Chem. 2003;278:37768–37776. doi: 10.1074/jbc.M304914200. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Lin H, Li Z, Wang M, Luo J. Modulation of expression of ribosomal protein L7a (rpL7a) by ethanol in human breast cancer cells. Breast Cancer Res Treat. 2001;69:29–38. doi: 10.1023/a:1012293507534. [DOI] [PubMed] [Google Scholar]
- 34.Kumar A, Chatopadhyay T, Raziuddin M, Ralhan R. Discovery of deregulation of zinc homeostasis and its associated genes in esophageal squamous cell carcinoma using cDNA microarray. Int J Cancer. 2007;120:230–242. doi: 10.1002/ijc.22246. [DOI] [PubMed] [Google Scholar]
- 35.Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, Ji J, Wang T, Zhi H, Wu M, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene. 2004;23:1291–1299. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Zhao LQ, Xiong MM, Wang XQ, Yang GR, Qiu ZL, Wu M, Liu ZH. Gene expression profiles at different stages of human esophageal squamous cell carcinoma. World J Gastroenterol. 2003;9:9–15. doi: 10.3748/wjg.v9.i1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]


