Abstract
AIM: To study milk consumption and subjective milk-related symptoms in adults genotyped for adult-type hypolactasia.
METHODS: A total of 1900 Finnish adults were genotyped for the C/T-13910 variant of adult-type hypolactasia and filled in a structured questionnaire concerning milk consumption and gastrointestinal problems.
RESULTS: The C/C-13910 genotype of adult-type hypolactasia was present in 18% of the study population. The prevalence of the C/C-13910 genotype was higher among subjects who were undergoing investigations because of abdominal symptoms (24%, P < 0.05). Those with the C/C-13910 genotype drank less milk than subjects with either the C/T-13910 or the T/T-13910 genotype of lactase persistence (18% vs 38%; 18% vs 36%, P < 0.01). Subjects with the C/C-13910 genotype had experienced more gastrointestinal symptoms (84%) during the preceding three-month period than those with the C/T-13910 (79%, P < 0.05) or the T/T-13910 genotype (78 %, P < 0.05). Only 9% (29/338) of the subjects with the C/C-13910 genotype consumed milk and reported no symptoms from it.
CONCLUSION: Gastrointestinal symptoms are more common among adults with the C/C-13910 genotype of adult-type hypolactasia than in those with genotypes of lactase persistence.
Keywords: Lactase persistence, Lactose malabsorption, C/T-13910 genotype, Abdominal symptoms, Milk consumption
INTRODUCTION
Gastrointestinal symptoms are a frequent cause for seeking medical advice in adult working age population[1]. Adult-type hypolactasia, characterised by the down regulation of lactase enzyme activity in the intestine during development is the most common enzyme deficiency in humans[2]. The symptoms caused by undigested lactose, bloating, diarrhoea and bowel gas are unspecific making the assessment of the diagnosis of adult-type hypolactasia a challenge to clinical practice. Symptoms vary greatly in severity and depend on the amount of lactose ingested and on individual sensitivity and may overlap with those of other gastrointestinal diseases such as irritable bowel syndrome (IBS) or diseases presenting with secondary lactose malabsorption, i.e. celiac disease[3]. Cases of individuals suspecting they have lactose intolerance are more common than the true prevalence of adult-type hypolactasia[4-9]. It has been shown that lactose-restricted diets improve symptoms markedly for example in IBS patients with lactose malabsorption and reduce the number of visits to the outpatient clinics[10]. Accurate diagnosis of lactose malabsorption, which is easily treatable by diet modification, would therefore be cost effective and time saving.
Diagnosis of adult-type hypolactasia has been based on indirect methods, the lactose tolerance test (LTT) or breath hydrogen test (BHT). Specificity of the LTT ranges from 77% to 96%, and that of BHT from 89% to 100%. Sensitivities vary from 76% to 94% for LTT and from 69% to 100% for BHT[11]. These methods are time consuming for the patient and need substantial assistance by medical personnel. The determination of disaccharidase activities and lactase/sucrase ratios from the intestinal biopsy specimen would be the most accurate diagnostic method[12] but this invasive method is not suitable for everyday clinical practice.
It has been demonstrated that lactase non-persistence is inherited as an autosomal recessive trait. A proportion of the human population can digest lactose (lactase persistence) due to a mutation retaining lactase activity[13]. A single nucleotide polymorphism C to T located 13 910 base pairs upstream the lactase gene is associated with the persistence/non-persistence trait in the Finnish families[14]. Analyses of several hundreds of intestinal biopsies have demonstrated that the C/C-13910 genotype is associated with low lactase activity (< 10 U/g/protein) and the C/T-13910 and T/T-13910 genotypes with high activity[14-16]. The prevalence of the C/T-13910 variant is compatible with the previously published figures for adult-type hypolactasia in European, Asian, African-American and Northern African populations[14-19]. It has been suggested, however, that in some African tribes the frequency of T-13910 allele does not parallel with the prevalence of lactase persistence[18]. Lactase mRNA transcribed from the C-13910 allele declines in the intestinal mucosa of children around five years of age[20], paralleling with the age of developmental down regulation of lactase enzyme activity[16]. In vitro -studies of the C/T-13910 variant have demonstrated greater increase in lactase promoter activity by the T-13910 variant[21,22]. This could possibly be explained by the recent finding of the T-13910 allele binding more strongly the transcription factor Oct-1 compared to the C-13910 allele[23]. Thus, the obtained functional data has given evidence for the use of the C/T-13910 variant as a robust marker for adult-type hypolactasia.
Here we have genotyped 1900 working-age people attending primary health care for the C/T-13910 variant associating with adult-type hypolactasia, and addressed the question about the prevalence and frequency of gastrointestinal symptoms as well as consumption of milk products.
MATERIALS AND METHODS
Participants
1902 adults from the capital area of Finland attending laboratory investigations in primary health care were asked to give a blood sample for genotyping of the C/T-13910 variant of adult-type hypolactasia and to fill in a questionnaire concerning the daily consumption of dairy products (normal and delactated milk, sour milk, yoghurt or ice cream, cheese) and possible milk related symptoms. The questions asked concerned whether or not a specific dairy product is included in the diet. Each participant was asked whether he/she had experienced any gastrointestinal complaints during the preceding three-month period. Frequency of gastrointestinal symptoms was also evaluated on daily and weekly basis. The questions on gastrointestinal symptoms covered: (1) the frequency of gastrointestinal symptoms such as flatulence, bloating, diarrhoea, heartburn, and constipation (2) the relation of their symptoms to meals, (3) correlation of the symptoms to different types of milk products or other foods, (4) previous diagnosis of lactose intolerance or a GI-disease such as colon cancer, celiac disease, or Helicobacter pylori infection. The age of the participants was ticked in questionnaire in three age groups (18-35 years, 36-51 years, and 52-64 years) due to the extensive number of study population. The formulated questionnaire was pre-tested on a small group of healthy adults. The collection of the questionnaires and blood samples occurred during a three-month period from February to May 2004. The study was approved by the Ethical Committee of the Helsinki University Central Hospital. All the subjects signed their written informed consent.
Genotyping
DNA of the study subjects (n = 1900) was isolated from peripheral blood samples by submerging a blank strip (Merck, Darmstadt, Germany) into EDTA blood. Strips were dried and heated in 1 × PCR reaction buffer (Dynazyme; Finnzymes, Espoo, Finland) containing 10 mmol/L Tris HCl (pH 8.8 at 25°C), 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 0.1% Triton X-100 to degrade proteins. After centrifuging 10 μL of the supernatant containing the DNA, the sample was amplified in a total volume of 50 μL, containing primers (one biotin-labeled 5’-(Biotin) CCTCGTTAATACCCACTGACCTA-3’) (5 μmol/L) and one unmodified 5’-GTCACTTTGATATGATGAGAGCA-3’ (50 μmol/L), dNTPs (1000 μmol/L), 0.5 U of Taq polymerase (Dynazyme, Finnzymes) in a standard buffer. The PCR cycle conditions used were as following: an initial round of denaturation at 94°C for 4 min, then 35 cycles at 94°C for 30 s, 53°C for 30 s, 72°C for 1.15 min, and a final extension of 72°C for 10 min. The resulting PCR products were analyzed by 1.5% agarose gel electrophoresis to verify the amplification and the size of the PCR product.
The C/T-13910 single nucleotide polymorphism was analysed using the solid-phase mini-sequencing method[24] that is based on the detection of tritium-labeled T-13910 and C-13910 alleles in the PCR reaction and measurement of their ratio using scintillation counter that directly reflects the ratio between the two sequences in the original sample. Briefly, in the mini-sequencing of the C/T-13910 single nucleotide polymorphism, two 10-μL aliquots of the biotin-labeled PCR product were captured to streptavidin coated microtitre wells (Thermo Electron, Helsinki, Finland). The reaction mixture contained 10 pmol of the detection primer (5’-GGCAATACAGATAAGATAATGTAG-3’), 0.1 μL of either 3H-dCTP or 3H-dTTP (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) and 0.05 U of DNA polymerase (Dynazyme II, Finnzymes, Espoo, Finland). The reactions were allowed to occur for 15 min at 56°C before washing off the unattached label. Finally, the attached detection primer was eluted by NaOH treatment and the radioactivity measured in a liquid scintillation counter (Rackbeta 1209; Wallac, Turku, Finland).
Two samples of the 1902 were disqualified after being found infected with hepatitis B.
Statistical analyses
Statistical analyses were conducted using Tixel (version 8.1), which is a VBA-program for Excel. Descriptive analyses were conducted with simple logistic regression. Proportions were compared by using Chi-squared tests with continuity correction or Fisher’s exact test when appropriate. Two-sided significance tests were used throughout.
RESULTS
The frequency of the C/C-13910 genotype associated with lactase non-persistence among the participants was 18% (Table 1). This figure corresponds to the earlier published prevalence of adult-type hypolactasia in the Finnish population[14-16], implying that the study population is representative for the general population. There was no difference in genotype distribution among male and female subjects, and no difference in separate age groups (data not shown).
Table 1.
Frequency of the C/T-13910 genotypes
| Genotype | % (n) | GI-complaints % (n) | P |
| C/C-13910 | 18 (341) | 24 (84) | < 0.05 |
| C/T-13910 | 47 (901) | 43 (148) | NS |
| T/T-13910 | 35 (658) | 33 (116) | NS |
| Total | 100 (1900) | 100 (348) |
The response rate for the questionnaire was high; a total of 99% (n = 1885) of the participants returned the questionnaire. A total of 42% of the patients reported a new, not gastrointestinal related disease as the reason for their laboratory visit. Earlier diagnosed diseases (25%) were the second most common cause for the visit, followed by gastrointestinal symptoms that were reported by 19% (348/1787) of the participants. Of these 348 subjects, the adult-type hypolactasia genotype C/C-13910 was observed in 24% (84/348), which was significantly more than in the study population (P < 0.05, Table 1).
Subjects with the C/C-13910 genotype had experienced more gastrointestinal symptoms (84%; 280/330) during the preceding three-month period than those with the C/T-13910 (79%; 698/885; P < 0.05) or the T/T-13910 genotype (78%; 501/641; P < 0.05, Table 2). Those with the C/C-13910 genotype reported more frequently gastrointestinal symptoms compared to C/T-13910 or T/T-13910 genotypes, but the difference was not significant (Table 2).
Table 2.
The frequency of gastrointestinal symptoms according to the three genotype groups of lactase persistence/nonpersistence during previous three months
| Abdominal complaints | C/C % (n) | C/T % (n) | T/T % (n) | OR C/C vs C/T | P | OR C/C vs T/T | P |
| (95% CI) | (95% CI) | ||||||
| During previous three months | 84 (280/332) | 79 (698/885) | 78 (501/641) | 1.44 (1.03-2.02) | < 0.05 | 1.50 (1.06-2.14) | < 0.05 |
| Daily | 23 (78/338) | 22 (199/891) | 19 (127/652) | 1.04 (0.77-1.41) | NS | 1.24 (0.90-1.71) | NS |
| Every other day | 21 (72/338) | 16 (145/891) | 20 (129/652) | 1.39 (1.02-1.91) | < 0.05 | 1.10 (0.79-1.52) | NS |
| Once a week | 22 (74/338) | 20 (176/891) | 18 (115/652) | 1.14 (0.84-1.55) | NS | 1.31 (0.94-1.82) | NS |
| More seldom than once a week | 17 (58/338) | 20 (177/891) | 21 (138/652) | 0.84 (0.60-1.16) | NS | 0.77 (0.55-1.08) | NS |
| No complaints | 16 (52/332) | 21 (187/885) | 22 (140/641) | 1.44 (1.03-2.02) | < 0.05 | 1.50 (1.06-2.13) | < 0.05 |
Gastrointestinal symptoms associated with lactose intolerance, namely flatulence, bloating and diarrhoea, were frequent among all genotype groups (Table 3). Flatulence was the only symptom significantly more frequent among the subjects with lactase non-persistent genotype compared to those with genotypes of lactase persistent (P < 0.05). Those with the C/C-13910 genotype experienced more bloating (61%) than those with the T/T-13910 genotype (55%) and C/T-13910 genotype (58%) but the difference did not reach statistical significance (Table 3).
Table 3.
The type of gastrointestinal symptoms among three genotype groups of lactase persistence/nonpersistence
| Gastrointestinal symptoms | C/C % (n = 294) | C/T % (n = 725) | T/T % (n = 524) | OR C/C vs C/T (95% CI) | P | OR C/C vs T/T (95% CI) | P |
| Flatulence | 79 (232) | 73 (531) | 73 (380) | 1.37 (0.99-1.89) | < 0.05 | 1.42 (1.01-1.99) | < 0.05 |
| Diarrhoea | 40 (119) | 38 (275) | 40 (207) | 1.11 (0.84-1.47) | NS | 1.04 (0.78-1.39) | NS |
| Constipation | 24 (72) | 21 (152) | 20 (107) | 1.22 (0.89-1.68) | NS | 1.26 (0.90-1.78) | NS |
| Bloating | 61 (178) | 58 (423) | 55 (287) | 1.10 (0.83-1.44) | NS | 1.27 (0.94-1.69) | NS |
| Heartburn | 32 (94) | 37 (265) | 38 (198) | 0.82 (0.61-1.09) | NS | 0.77 (0.57-1.04) | NS |
As many as 45% of the participants informed having experienced gastrointestinal symptoms after drinking milk (Figure 1) and 25% of the participants reported symptoms from food containing milk. It is notable, that only 18% (60/338; P < 0.01)) of the subjects with the C/C-13910 genotype of adult-type hypolactasia informed drinking milk with meals, which is significantly less (P < 0.01) than those with the lactase persistent genotypes C/T-13910 (38%; 333/894) and T/T-13910 (36%; 236/653; Figure 1). The fat content of the milk was not questioned, but in Finland the vast majority of people use low-fat or fat-free milk as a drink. The number of people using low lactose containing milk as a drink was marginal according the questionnaire.
Figure 1.
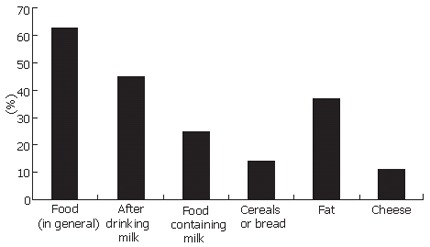
Frequency of gastrointestinal symptoms experienced by various foods in the study population.
Only 9% (29/338) with the C/C-13910 genotype of adult-type hypolactasia consumed milk daily and reported no symptoms from milk. However, one third (18/60) of the milk drinkers with the C/C-13910 genotype did not answer the question about milk related symptoms. Most people with C/C-13910 genotype reporting GI-problems from milk (69%; 190/274) did not drink milk. The respective number of people with lactase persistence genotypes who reported milk-related problems and did not drink milk was 54% (299/554) for the C/T-13910 genotype and 50% (207/411) for the T/T-13910 genotype.
Cheese caused gastrointestinal symptoms for 11% of the participants according to their own judgment: for 17% of those with the C/C-13910 genotype, 10% of those with the C/T-13910 genotype and 9% of the ones with the T/T-13910 genotype (P < 0.05, Figure 2). Among all participants 14% experienced symptoms from cereal or bread, and 37% from ingested fat (Figure 1) and these were not related to the genotype of adult-type hypolactasia.
Figure 2.
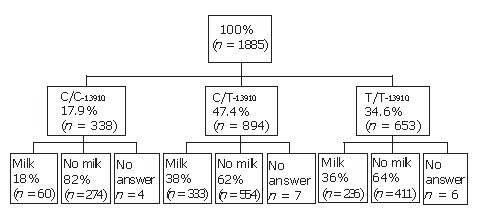
Flow-chart of the use of cow’s milk as a drink in different genotype groups.
Among the study population, 15% (245/1649) reported having had a pathological LTT earlier: 36% (109/299) of those with the C/C-13910 genotype, 11% (82/777) with the C/T-13910 and 9% (54/573) with the T/T-13910 genotype.
A previous, pathological LTT was reported by 19% (64/341) of participants with the C/C-13910 genotype, by 10% (89/901) with the C/T-13910, and by 14% (91/658) with the T/T-13910 genotype. Five out of these 180 subjects with the C/T-13910 or T/T-13910 genotype with a pathological LTT reported a previously diagnosed possible secondary cause for hypolactasia i.e. celiac disease. An undiagnosed celiac disease in two other participants of these 180 subjects was suggested due to elevated level of transglutaminase antibodies in their sera[25].
DISCUSSION
The inability to absorb lactose is frequently suspected to underlie gastrointestinal symptoms in populations with high prevalence of adult-type hypolactasia and frequent dairy consumption. In this study we show that subjects with the C/C-13910 genotype, having low lactase activity in the intestinal wall[14-17,20] indeed do seek medical advice for abdominal symptoms more often than subjects with high lactase activity. This occurred although they already had self restricted their milk consumption.
The response rate in this study was extremely high, 99%, thus the data about GI-symptoms was comprehensive. This exceptionally high response rate was achieved by the sound motivation of both the study subjects and the laboratory personnel. As expected, gastrointestinal symptoms were very common (80%) during the previous three months comparable to the earlier findings[1]. Flatulence was the only symptom that in this study was significantly more common among subjects with low lactase (C/C-13910 genotype). Diarrhoea and bloating, which according to earlier published studies are more common among lactose malabsorbers[4] were not more frequent among those with adult-type hypolactasia. These symptoms, however, were very common in the study population (40%-60%) pointing out the high prevalence of functional GI-symptoms[26].
The great majority of the study subjects did not drink milk at all. Milk consumption was rarest in the group of lactase non-persistent subjects: only 18% reported drinking milk daily, which might indicate that a natural aversion of milk has been developed by the subjects with low lactase activity. This is in agreement with our findings in a paediatric population in which children with the C/C-13910 genotype consumed less milk than children with a non-C/C-13910 genotype[16,27]. The majority of the children with the lactase non-persistent genotype C/C-13910 reported that they never drank milk[16]. At the age of 8-9 years, more than 40% of the subjects with the C/C-13910 genotype reported drinking less than one dl of milk daily, compared to 20% of the subjects with the C/T-13910 and T/T-13910 genotype, respectively[26].
Only 9% of those with the C/C-13910 genotype used daily milk and did not experience symptoms from ingested milk. This is in line with other studies[6,8,9] showing that not all individuals with lactose malabsorption report symptoms from food containing lactose. The colonic micro biota is variable[28] and the individual sensitivity to feel distension of the colon and to sense discomfort varies[29]. Accordingly, the recognition of the possible link between milk consumption and abdominal symptoms is not always easy[30]. The meal content has an effect on emptying of the stomach and thereby affects the lactose load in the intestine[31,32]. Most lactose malabsorbers seem to tolerate small amounts of milk especially during meals[5,33]. Further studies are warranted to evaluate the amount of lactose tolerated by different genotype groups.
In this study population, 10% of those with the genotypes associated with lactase persistence (the C/T-13910 genotype or T/T-13910 genotype) reported that they had been earlier diagnosed lactose intolerant based on results of LTT. A possible cause for secondary hypolactasia, i.e. celiac disease, was found in 3% of these individuals. These findings imply that LTT, a commonly used method for diagnosis of lactose intolerance, produces high numbers of false positive test results as has been earlier observed[11]. One third of those with the C/C-13910 genotype associated with low lactase reported that they had previously had a positive LTT. Due to the way our question on earlier diagnosis of lactose intolerance was formulated, we do not know the extent of subjects with the C/C-13910 genotype who might have had a negative result for their LTT.
In conclusion, gastrointestinal symptoms are more common among adults with the C/C-13910 genotype of adult-type hypolactasia than in those with genotypes of lactase persistence. This was seen although individuals with the C/C-13910 genotype had restricted their milk consumption. Genotyping of the C/T-13910 polymorphism is a practical means for defining adult-type hypolactasia.
ACKNOWLEDGMENTS
We are grateful to the participants and the laboratory personnel of the outpatient clinics of the city of Espoo and the Central Military Hospital for their contribution.
Footnotes
Supported by the Sigrid Jusélius Foundation, Helsinki, Finland; the Foundation for Nutrition Research, Helsinki, Finland; the Research Foundation of Alfred Kordelin, Helsinki, Finland; Helsinki University Hospital Research Funding, Helsinki, Finland; the Foundation for Promoting Occupational Medicine in Finland, Helsinki, Finland; the Academy of Finland
S- Editor Liu Y L- Editor Zhu LH E- Editor Lu W
References
- 1.Drossman DA, Li Z, Andruzzi E, Temple RD, Talley NJ, Thompson WG, Whitehead WE, Janssens J, Funch-Jensen P, Corazziari E. U.S. householder survey of functional gastrointestinal disorders. Prevalence, sociodemography, and health impact. Dig Dis Sci. 1993;38:1569–1580. doi: 10.1007/BF01303162. [DOI] [PubMed] [Google Scholar]
- 2.Järvelä IE. Molecular genetics of adult-type hypolactasia. Ann Med. 2005;37:179–185. doi: 10.1080/07853890510007359. [DOI] [PubMed] [Google Scholar]
- 3.Tamm A. Management of lactose intolerance. Scand J Gastroenterol Suppl. 1994;202:55–63. doi: 10.3109/00365529409091744. [DOI] [PubMed] [Google Scholar]
- 4.Jussila J, Launiala K, Gorbatow O. Lactase deficiency and a lactose-free diet in patients with "unspecific abdominal complaints". Acta Med Scand. 1969;186:217–222. doi: 10.1111/j.0954-6820.1969.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 5.Vesa TH, Korpela RA, Sahi T. Tolerance to small amounts of lactose in lactose maldigesters. Am J Clin Nutr. 1996;64:197–201. doi: 10.1093/ajcn/64.2.197. [DOI] [PubMed] [Google Scholar]
- 6.Carroccio A, Montalto G, Cavera G, Notarbatolo A. Lactose intolerance and self-reported milk intolerance: relationship with lactose maldigestion and nutrient intake. Lactase Deficiency Study Group. J Am Coll Nutr. 1998;17:631–636. doi: 10.1080/07315724.1998.10718813. [DOI] [PubMed] [Google Scholar]
- 7.Saltzman JR, Russell RM, Golner B, Barakat S, Dallal GE, Goldin BR. A randomized trial of Lactobacillus acidophilus BG2FO4 to treat lactose intolerance. Am J Clin Nutr. 1999;69:140–146. doi: 10.1093/ajcn/69.1.140. [DOI] [PubMed] [Google Scholar]
- 8.de Vrese M, Stegelmann A, Richter B, Fenselau S, Laue C, Schrezenmeir J. Probiotics--compensation for lactase insufficiency. Am J Clin Nutr. 2001;73:421S–429S. doi: 10.1093/ajcn/73.2.421s. [DOI] [PubMed] [Google Scholar]
- 9.Johnson AO, Semenya JG, Buchowski MS, Enwonwu CO, Scrimshaw NS. Correlation of lactose maldigestion, lactose intolerance, and milk intolerance. Am J Clin Nutr. 1993;57:399–401. doi: 10.1093/ajcn/57.3.399. [DOI] [PubMed] [Google Scholar]
- 10.Böhmer CJ, Tuynman HA. The effect of a lactose-restricted diet in patients with a positive lactose tolerance test, earlier diagnosed as irritable bowel syndrome: a 5-year follow-up study. Eur J Gastroenterol Hepatol. 2001;13:941–944. doi: 10.1097/00042737-200108000-00011. [DOI] [PubMed] [Google Scholar]
- 11.Arola H. Diagnosis of hypolactasia and lactose malabsorption. Scand J Gastroenterol Suppl. 1994;202:26–35. doi: 10.3109/00365529409091742. [DOI] [PubMed] [Google Scholar]
- 12.Dahlqvist A. Assay of intestinal disaccharidases. Scand J Clin Lab Invest. 1984;44:169–172. doi: 10.3109/00365518409161400. [DOI] [PubMed] [Google Scholar]
- 13.Sahi T, Isokoski M, Jussila J, Launiala K, Pyörälä K. Recessive inheritance of adult-type lactose malabsorption. Lancet. 1973;2:823–826. doi: 10.1016/s0140-6736(73)90862-3. [DOI] [PubMed] [Google Scholar]
- 14.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 15.Kuokkanen M, Enattah NS, Oksanen A, Savilahti E, Orpana A, Järvelä I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003;52:647–652. doi: 10.1136/gut.52.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasinperä H, Savilahti E, Enattah NS, Kuokkanen M, Tötterman N, Lindahl H, Järvelä I, Kolho KL. A genetic test which can be used to diagnose adult-type hypolactasia in children. Gut. 2004;53:1571–1576. doi: 10.1136/gut.2004.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swallow DM. Genetics of lactase persistence and lactose intolerance. Annu Rev Genet. 2003;37:197–219. doi: 10.1146/annurev.genet.37.110801.143820. [DOI] [PubMed] [Google Scholar]
- 18.Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, Swallow DM, Bradman N, Thomas MG. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet. 2004;74:1102–1110. doi: 10.1086/421050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myles S, Bouzekri N, Haverfield E, Cherkaoui M, Dugoujon JM, Ward R. Genetic evidence in support of a shared Eurasian-North African dairying origin. Hum Genet. 2005;117:34–42. doi: 10.1007/s00439-005-1266-3. [DOI] [PubMed] [Google Scholar]
- 20.Rasinperä H, Kuokkanen M, Kolho KL, Lindahl H, Enattah NS, Savilahti E, Orpana A, Järvelä I. Transcriptional downregulation of the lactase (LCT) gene during childhood. Gut. 2005;54:1660–1661. doi: 10.1136/gut.2005.077404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 22.Troelsen JT, Olsen J, Møller J, Sjöström H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003;125:1686–1694. doi: 10.1053/j.gastro.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 23.Lewinsky RH, Jensen TG, Møller J, Stensballe A, Olsen J, Troelsen JT. T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet. 2005;14:3945–3953. doi: 10.1093/hmg/ddi418. [DOI] [PubMed] [Google Scholar]
- 24.Syvänen AC, Aalto-Setälä K, Harju L, Kontula K, Söderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]
- 25.Tikkakoski S, Savilahti E, Kolho KL. Undiagnosed coeliac disease and nutritional deficiencies in adults screened in primary health care. Scand J Gastroenterol. 2007;42:60–65. doi: 10.1080/00365520600789974. [DOI] [PubMed] [Google Scholar]
- 26.Hillilä MT, Färkkilä MA. Prevalence of irritable bowel syndrome according to different diagnostic criteria in a non-selected adult population. Aliment Pharmacol Ther. 2004;20:339–345. doi: 10.1111/j.1365-2036.2004.02034.x. [DOI] [PubMed] [Google Scholar]
- 27.Rasinpera H, Saarinen K, Pelkonen A, Jarvela I, Savilahti E, Kolho KL. Molecularly defined adult-type hypolactasia in school-aged children with a previous history of cow's milk allergy. World J Gastroenterol. 2006;12:2264–2268. doi: 10.3748/wjg.v12.i14.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egert M, de Graaf AA, Smidt H, de Vos WM, Venema K. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 2006;14:86–91. doi: 10.1016/j.tim.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Vesa TH, Seppo LM, Marteau PR, Sahi T, Korpela R. Role of irritable bowel syndrome in subjective lactose intolerance. Am J Clin Nutr. 1998;67:710–715. doi: 10.1093/ajcn/67.4.710. [DOI] [PubMed] [Google Scholar]
- 30.Suarez FL, Savaiano DA, Levitt MD. A comparison of symptoms after the consumption of milk or lactose-hydrolyzed milk by people with self-reported severe lactose intolerance. N Engl J Med. 1995;333:1–4. doi: 10.1056/NEJM199507063330101. [DOI] [PubMed] [Google Scholar]
- 31.Martini MC, Savaiano DA. Reduced intolerance symptoms from lactose consumed during a meal. Am J Clin Nutr. 1988;47:57–60. doi: 10.1093/ajcn/47.1.57. [DOI] [PubMed] [Google Scholar]
- 32.Vesa TH, Marteau PR, Briet FB, Boutron-Ruault MC, Rambaud JC. Raising milk energy content retards gastric emptying of lactose in lactose-intolerant humans with little effect on lactose digestion. J Nutr. 1997;127:2316–2320. doi: 10.1093/jn/127.12.2316. [DOI] [PubMed] [Google Scholar]
- 33.Vesa TH, Marteau P, Korpela R. Lactose intolerance. J Am Coll Nutr. 2000;19:165S–175S. doi: 10.1080/07315724.2000.10718086. [DOI] [PubMed] [Google Scholar]


