Abstract
Effective cell-to-cell communication is critical for the survival of both unicellular and multicellular organisms. In multicellular plants, direct cell coupling across the cell wall boundaries is mediated by long membrane-lined cytoplasmic bridges, the plasmodesmata. Exciting recent discoveries suggest that the occurrence of such membrane-lined intercellular channels is not unique to plant lineages but more prevalent across biological kingdoms than previously assumed. Striking functional analogies exist among those channels since they all facilitate the exchange of various forms of macromolecules, but at the same time allow some opportunistic pathogens to exploit those bridges to move from one host cell to another. However, host immune surveillance system may have also evolved strategies to offset such exploitation of the critical cellular infrastructure by the pathogen. Indeed, an emerging new paradigm illustrates that cellular connectivity via plasmodesmata plays an important role in innate immune responses. Preliminary hypothesis is proposed as to how the regulatory mechanism evolved by which plants integrated plasmodesmata into immune signaling pathways based on the key players identified in this process.
Keywords: plasmodesmata, tunneling nanotubes, plant immunity, cell-to-cell communication
1. Introduction
Plasmodesmata, which were first described by Edward Tangl some hundred years ago, are fundamental intercellular channels that allow plants to function as multicellular organisms. These channels form long, membrane-lined cytoplasmic bridges between neighboring cells, providing both membrane and cytoplasmic continuity among connected cells virtually throughout the whole plant body (Fig. 1A). Primitive forms of plasmodesmata found in multicellular algae mainly have plasma membrane-lined cytoplasmic openings embedded in the cell walls (Fig. 1B). The inner space of plasmodesmata in higher plants is occupied by the central core composed of a modified single endoplasmic reticulum (ER) tube [1]. Small molecules such as ions, hormones, photosynthates, and other nutrients pass through the cytoplasmic sleeve formed between the plasma membrane and the ER membranes. Plasmodesmata in higher plants have the capacity to gate and dilate, which facilitates trafficking of macromolecules such as transcription factors and RNAs [2, 3]. Intriguingly, plasmodesmata-like channels are not unique to plant systems as had long been assumed (Fig. 1). Until recently, gap junctions, which form cytoplasmic channels at plasma membrane contact sites and allow the diffusion of molecules smaller than 1 kDa, were considered the primary form of intercellular communication channels occurring in animals. However, exciting new progress in both animal and bacterial fields has shown that cell-to-cell communication through plasmodesmata-like membrane-lined connections is more common than previously thought [4, 5].
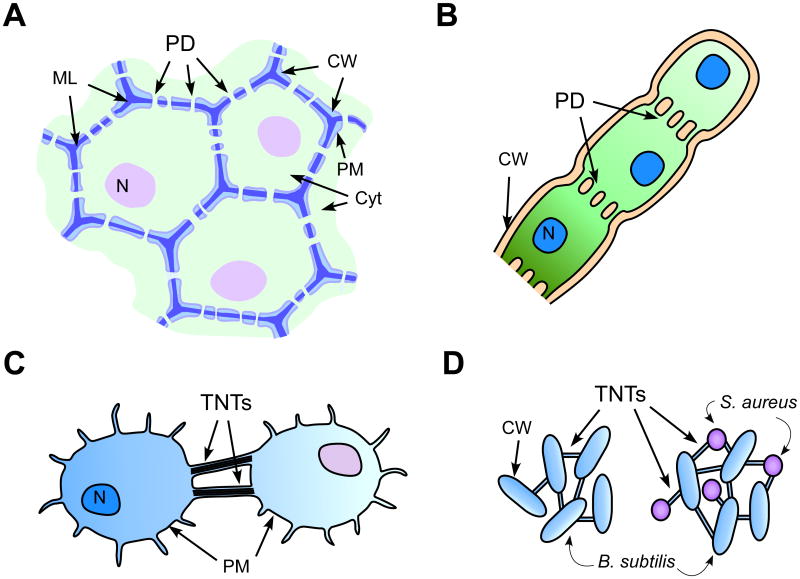
Figure 1. Membrane-lined cell-to-cell communication channels found across biological kingdoms.
A. Plasmodesmata in higher plants provide symplasmic and membrane continuity among neighboring cells. Detailed structural features of plasmodesmata are not illustrated in this diagram. CW, cell wall (light blue); ML, middle lamella (dark blue); PM, plasma membrane (grey line); N, nucleus (purple); Cyt (cytoplasm, light green).
B. Plasmodesmata in filamentous algae form plasma membrane-lined simple pores, which support diffusion of small molecules and nutrients.
C. Mammalian tunneling nanotubes can form de novo in cultured cells.
D. Bacterial tunneling nanotubes can form among cells of the same or different species. Illustration shows tunneling nanotubes formed within B. subtilis (left) and those between B. subtilis and other bacterial species such as Staphylococcus aureus or Escherichia coli (right). PD, plasmodesmata; N, nucleus; CW, cell wall; ML, middle lamella.
Plasmodesmata form from ER membranes at the point where they are encased by plasma membrane within the newly forming cell wall between two daughter cells during cell division. In this regard, all plant cells have plasmodesmal connections at their birth, which are referred as primary plasmodesmata. However, the plasmodesmal density and functional connectivity undergo substantial changes in both permanent and temporary ways while cells grow, develop, and/or differentiate [1, 6]. Some cells retain plasmodesmal connections during development, while others lose them. However, the loss of plasmodesmal structures is not necessarily an irreversible modification: plasmodesmata can be also inserted across existing cell walls via de novo biogenesis. For example, in the shoot apical meristem of Sinapis alba, plasmodesmal frequency was transiently increased at 28 to 48 h after the exposure to a single floral inductive long-day treatment [7]. This increase in plasmodesmal frequency was found not only in relatively young anticlinal walls but also in periclinal walls, which are cytokinetically inactive. Therefore, the newly formed plasmodesmata in the shoot meristem during floral transition were concluded to be secondary in their origin.
The presence of plasmodesmata in a given cellular boundary does not necessarily mean that those plasmodesmata are functionally open all the time. Higher plants have evolved a mechanism that restricts plasmodesmal opening by building a cell wall scaffold composed of β-1,3 glucan called callose within the cell wall space immediately surrounding plasmodesmata [8]. This process achieved by the activities of callose synthases can be reversed enzymatically via β-1,3 glucan hydrolases. Collectively, plasmodesmal function is under a multitude of regulatory modes, e.g., permanent and transient changes in plasmodesmal frequency and permeability, which allows for dynamic intercellular connectivity. More importantly, it is conceivable that a higher order of supracellular regulation through plasmodesmata could be expected in plants if molecular mechanisms exist that link various cellular signaling pathways to the control of plasmodesmal permeability. Indeed, recent advances in the field demonstrated that cellular connectivity through plasmodesmata is tightly controlled by plant defense pathways [9-12]. It is tempting to speculate that this integration may have originated via a progressive acquisition of genetic modules during the evolution of higher plants.
2. Membrane-lined cytoplasmic connections: a universal cellular bridging theme?
2.1. Plasmodesmata
Plasmodesmata are specialized, intercellular channels that establish cytoplasmic and endomembrane continuities between neighboring cells in plants [13-15]. The plasmodesmata in higher plants have evolved a complex structure, containing appressed ER within the plasma membrane-lined cytoplasmic cylinder (see Supplemental Table 1 and references therein). The substructure of plasmodesmata in cryofixed, freeze-substituted tobacco leaves was revealed by computer-enhanced high resolution electron microscopy [16]. According to the schematic model based on this study, the cytoplasmic space between the plasma membrane and ER is divided into discrete microchannels by the crosslinking of proteinacious materials embedded within the plasma membrane and ER membranes. The plasmodesmata in higher plants overall may be more complex than those found in the green algae lineage, Charophytes [6, 17]. For example, in charophycean green alga Chara zeylanica, primary plasmodesmata contain an internal core structure that appears less specialized and may be associated with ER in rare cases [18]. Some species of fungi such as Endomyces geotrichum produce plasmodesmata that strikingly resemble the structure of plasmodesmata of higher plants, e.g., they have an inner core structure connected to cortical ER [19, 20]. Endomyces produce plasmodesmata in addition to septa, which are the more typical form of cytoplasmic conduits in fungi. Unfortunately, no further studies regarding the plasmodesmata in this fungus have been documented other than the two describing their ultrastructural features. Many filamentous cyanobacteria also form cytoplasmic bridges that seem to be a more primitive form of plasmodesmata in that they are plasma membrane-lined cytoplasmic strands without additional structural features (Fig. 1B).
There are several fundamental cellular capacities required to establish a multicellular body in plants, and certainly the creation of plasmodesmata is considered one of the most critical events in the evolution of higher plants [18, 21, 22]. Moreover, it is now well accepted that plasmodesmata have essential roles in symplasmic intercellular signaling in higher plants by facilitating various types of regulatory molecules including transcription factors and large and small RNA species [23, 24].
2.2. Tunneling nanotubes
The existence in animals of membrane-lined cytoplasmic connections that can function in intercellular molecular transfer was only recently discovered. These intercellular connections were initially observed as about 50-200 nm wide membrane tubes in the rat pheochromocytoma cultured cells (PC12) and named tunneling nanotubes (also called membrane nanotubes) (Fig. 1C) [4]. Following this report, various cell types have been shown to form thin membrane tubes extending from the plasma membrane that directly connect neighboring cells. Tunneling nanotubes contain filamentous actin (f-actin) as the core structure. This f-actin core functions as a cytoplasmic cargo transfer system through tunneling nanotubes but may also have an important role for the protrusion of plasma membrane during the initial stage of tunneling nanotube formation. Tunneling nanotubes were found to form de novo in neuronal, immune and epithelial cells cultured in vitro [25, 26]. Several imaging studies using embryonic tissues of both vertebrate and invertebrate species have now confirmed that tunneling nanotube-like structures do form in vivo as well [27]. Tunneling nanotubes observed in different cell types are quite diverse in terms of the timing, condition, and mode of their formation as well as their morphology, structure, and dimension (see Supplemental Table 1 and references therein).
The original discovery of tunneling nanotubes was striking for two major reasons. First, the finding showed that animal cells can form membrane-lined intercellular connections. Until then, protein-based gap junctions were thought to be the canonical form of intercellular channels allowing for direct cytoplasmic coupling across the plasma membrane contacts of neighboring animal cells. Second, tunneling nanotubes facilitated the transfer of large cellular cargoes, which is impossible through gap junctions, and hence acted more like plasmodesmata in plant cells. Notably, tunneling nanotubes facilitated the intercellular transfer of membrane-bound macromolecules as well as vesicles and even organelles. However, they were selective against passive diffusion of soluble cytoplasmic molecules. For example, although the intercellular transfer of fluorescently-tagged, cytoplasmically expressed actin was facilitated by tunneling nanotubes, diffusion through tunneling nanotubes of cytoplasmically-expressed, free green fluorescent protein or the small fluorescent dye calcein was not observed [4].
The most recent and unexpected finding regarding tunneling nanotube-mediated cellular connections is that similar intercellular nanotubes are also formed among bacterial cells [5]. The bacterial membrane nanotubes found in Bacillus subtilis were 30 to 130 nm wide and extended up to 1 μm. These tunneling nanotubes can form among the progenies of a bacterial strain but also between cells of different strains (Fig. 1D). Moreover, the tunneling nanotubes formed among bacterial cells can transfer not only cytoplasmic material such as ectopically expressed green fluorescent protein, but also genetic materials encoded within non-conjugative plasmids. Bacterial tunneling nanotubes resemble plant plasmodesmata more than mammalian tunneling nanotubes as they are encased by the cell wall, suggesting also that the cell wall matrix did not impose any hindrance to the extension of membrane tubes between cells. The uncovering of the genetic requirements and mechanisms of nanotube formation will be interesting, as this information would allow insights not only into the evolutionary aspects of tunneling nanotubes (and perhaps also plasmodesmata), but also into potential genetic manipulation for future organismal bioengineering.
The occurrence of both tunneling nanotubes and plasmodesmata across phylogenetic kingdoms suggests that direct cell-to-cell communication mediated by membrane-lined channels may represent the most prevalent and fundamental means of intercellular coordination and signaling. Below, I will discuss recent findings that place plasmodesmata and tunneling nanotubes in the spotlight in terms of the old and new roles they play in intercellular communication, especially during pathogen infection.
3. Intercellular bridges: infection highways and escape routes of opportunistic pathogens?
3.1. Plasmodesmata as conduits of pathogen spread
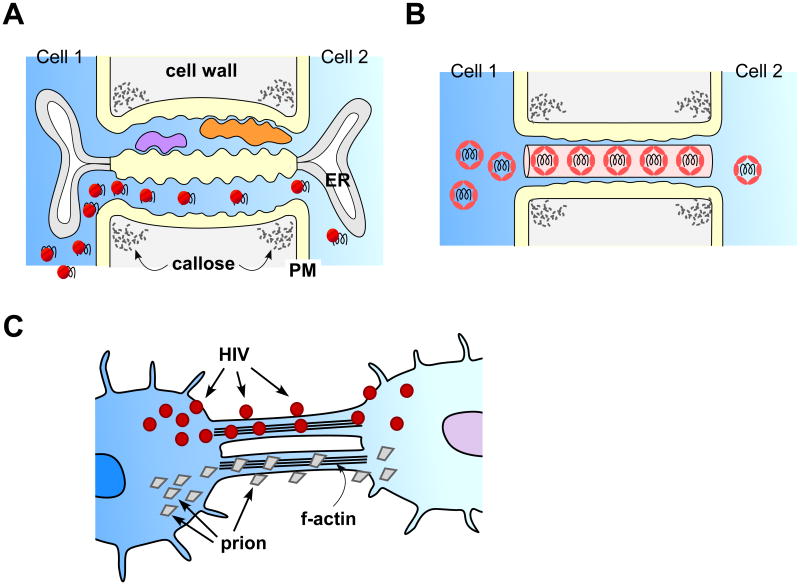
Studies over the past four decades on viral movement through plasmodesmata have established the concept that viruses “hijack” plasmodesmata when spreading their infectious materials from one cell to another in plants [28, 29] (Fig. 2A). Plant viruses encode movement proteins, which are non-structural viral proteins but essential for viral spread. The non-tubule forming movement proteins encoded by viruses, such as Tobacco mosaic virus, Cucumber mosaic virus, Potato virus X, etc., associate with plasmodesmata and facilitate trafficking of infectious materials through the channels [9]. Movement proteins encoded by another group of plant viruses, such as Cowpea mosaic virus and Grapevine fanleaf virus, assemble into tubular structures through plasmodesmata by removing the ER core (Fig. 2B). Supporting the view that plasmodesmata are likely targets for microbial pathogens which require cell-to-cell spread, a hemibiotropic fungus of rice, Magnaporthe oryzae, forms infection hyphae that appear to grow towards the location of plasmodesmata before passing through the host cell wall [30]. This fungal pathogen is capable of digesting the host cell wall without inducing host immunity or membrane disintegration. By contrast, the cellulosic walls encasing each plant cell impose structural barriers that plant viruses must pass.
Figure 2. Plasmodesmata and tunneling nanotubes are exploited by opportunistic microbial pathogens.
A & B. Plant viruses hijack plasmodesmata and spread infectious materials cell to cell. Some viruses encoding non-tubule forming movement proteins enhance plasmodesmal opening and pass through plasmodesmata as macromolecular complexes (A). Viruses encoding movement proteins that form tubules move cell to cell via modifying plasmodesmal structure (B). This process involves a replacement of the appressed ER with the tubules. In these diagrams, callose accumulated within the cell walls surrounding the neck regions of each plasmodesmal channel is illustrated as a structural constituent of plasmodesma.
C. HIV or prion proteins spread cell to cell via moving on the membrane surface or through tunneling nanotubes along the f-actin core complex.
If viral pathogens had to use an extracellular pathway for infection, it would involve some type of secretion of their infectious materials or virions into the cell wall matrices and a mechanism of reentry into the cytoplasm of adjacent cells. This path may be possible given that viral particles have dimensions in a nano scale and plant cell walls are porous enough (3-4 nm) [31] to accommodate the passage of viral particles. However, this pathway would significantly impede the speed of viral spread in at least three ways. First, their spread would rely heavily on diffusion into the cell wall matrices, diluting the viral concentration. Second, viruses may not pass through certain cell wall boundaries that are highly water-impermeable due to lignin and suberin deposits. Third, the systemic spread of viruses through phloem would be impossible in the absence of targeted or guided penetration through other tissues into the phloem, unless their infection starts in the phloem. Considering these points, one can safely assume that the presence of plasmodesmata in higher plants does provide a fast track cytoplasmic transfer for plant viruses, as well as a means of avoiding exposure to an extracellular environment that might be adverse to their structural stability.
While it is now well established that viruses exploit plasmodesmal gating properties that allow macromolecular trafficking for their cell-to-cell infectivity, pathogen and host co-evolution theory [32] predicts that plants would evolve counteracting mechanisms against the exploitation of plasmodesmata by viral pathogens. Recent studies illustrate that higher plants may, in fact, have evolved defense mechanisms using innate immune responses that could help guarding plasmodesmal passageways from microbial intruders [9-12, 33].
3.2. Mammalian tunneling nanotubes as cellular bridges of pathogen spread
Since the very first report, tunneling nanotubes have been found to be pervasive and highly versatile in terms of providing both a structural foundation that bridges various mammalian cell types and a mechanism that couples cells in physiological signaling (see Supplemental Table 1 and references therein). They facilitate the transfer of cellular components, ranging from proteins associated with the plasma membrane to vesicles/organelles, and to Ca2+ and electrical signals [25, 26]. Their function has also been linked to the cell-to-cell spread of diseases as well as intercellular exchange of apoptotic signals by trafficking of both cytoplasmic and membranous cellular contents. In addition, the bridging system that tunneling nanotubes provide can be used by cargos both internally through the channels and externally along the surface of the channels (Fig. 2 B). In contrast, neither trafficking of organelles nor cargo transfer along the plasma membrane leaflet of plasmodesmata has been reported. However, it is fascinating to find many analogies between plasmodesmata and tunneling nanotubes including the interactions with opportunistic pathogens (see Supplemental Table 1 and references therein). Justas plant plasmodesmata facilitate movement of viral and fungal pathogens, so tunneling nanotubes now appear to carry viral pathogens as well as obligate intracellular bacterial pathogens.
Human immunodeficiency virus (HIV), prions, and bacterial pathogens utilize tunneling nanotubes for cell-to-cell movement. HIV particles in primary macrophages move both inside and along the surface of tunneling nanotubes [34] (Fig. 2C). This movement is thought to provide not only a rapid infection of neighboring cells but also an escape route for the virus from the immune surveillance system of the host cells. Interestingly, HIV stimulates formation of tunneling nanotubes in the infected macrophages prior to viral transfer into the recipient cells, potentially manipulating the host cell system, allowing effective viral spread [35]. HIV components then pass intracellularly through the ER and Golgi network to endosomes, and intercellularly via tunneling nanotubes from infected to non-infected cells. Prions and prion-like protein aggregates that cause neurodegenerative diseases are other examples of pathogen transfer from infected cells, resulting in pathogen propagation in recipient cells [36]. They are thought to traffic also either along the surface or through tunneling nanotubes by using endocytic vesicles as shuttling vehicles (Fig. 2C). Prions can associate with the cellular membrane via post-translational attachment of glycosylphosphatidylinositol, which may allow the protein to gain access to tunneling nanotubes [37]. The bacterium Ehrlichia chaffeensis passes from an infected host cell to a non-infected cell via tunneling nanotube-like membrane intrusions, called filopodia [38]. Ehrlichia is an obligatory intracellular bacterium that causes a fatal infectious disease affecting immune cells in humans. Interestingly, Ehrlichia infection promotes the formation of filopodia through which the bacteria are transported into the uninfected cells during the early stages of infection [38].
4. Plasmodesmata in innate immunity
4.1. Plant defense systems
Plants do not have dedicated immune cells such as macrophages, natural killer cells, or lymphocytes that mammalian systems produce. However, plants can cope with invading microbial pathogens through unique immune signaling pathways that sense the arrival of the pathogens and direct nuclear expression to switch from a growth to a defense mode [39]. Perception of microbial pathogens is mediated by various types of pattern recognition receptors expressed at the cell surface, which detect the presence of specific pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs, respectively) [40, 41]. Activation of these receptors by a cognate PAMP or MAMP brings about membrane de- or hyper-polarization, ion and H+ influx, rise in cytosolic [Ca2+], changes in redox states, and protein phosphorylation cascades stimulated by mitogen-activated protein kinases. These multilayered signaling events are part of pathogen-triggered immunity and mount fast and effective defense responses against intruders. Plants can also recognize the microbial pathogens that evade pathogen-triggered immunity through a defense system named effector-triggered immunity [42].
Basal and induced immune responses require an accumulation of the phytohormone salicylic acid, which boosts hypersensitive cell death responses of infected cells in local tissues and induces systemic acquired resistance in distal tissues [43]. The major genetic components of salicylic acid biosynthesis and signaling include the upstream regulator ENHANCED DESEASE RESISTANCE 1 (EDS1), the SA biosynthetic enzyme ISOCHORISMATE SYNTHASE 1 (ICS1), and the downstream regulator NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1). As a transcriptional coactivator, NPR1 has a major role in shifting nuclear gene expression patterns including the induction of pathogenesis-related genes and secretion pathways [43]. All of these salicylic acid signaling components play a critical role in the regulation of cellular connectivity via plasmodesmata as part of the immune response [11] (discussed below).
4.2. Integration of plasmodesmata into the defense signaling pathways
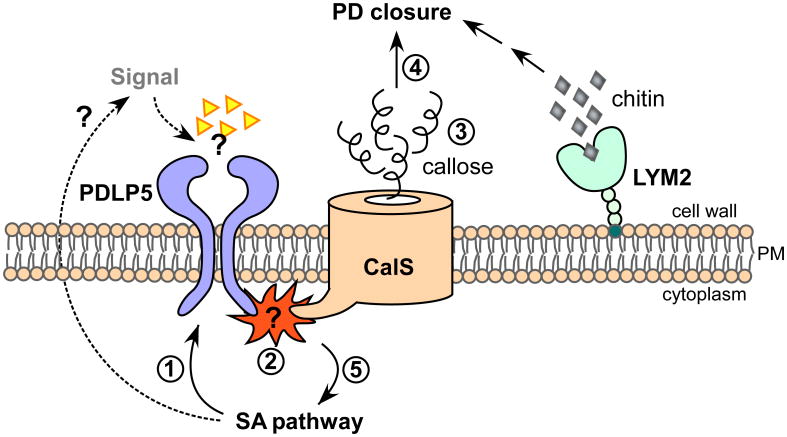
As a majority of bacterial pathogens are epiphytic in plants, plant responses to these microbes were not thought to involve the modulation of plasmodesmal gating [9]. However, the findings that plasmodesmata close in response to bacterial infection, and that lack of this closure renders Arabidopsis thaliana susceptible to pathogen spread, demonstrated that the regulation of plasmodesmata is critical to plant innate immunity [10]. The first hint for such integrative signaling was provided by the study of PLASMODESMATA-LOCATED PROTEIN 5 (PDLP5) [10]. PDLP5 functions as the molecular link between the regulation of plasmodesmal permeability and the salicylic acid signaling pathway, and is required for both plasmodesmal closure and basal immunity (Fig. 3). PDLP5 as a plasmodesmal resident protein is expressed in plants at a very low level under normal growth conditions in the absence of pathogen challenges. However, its expression is elevated during natural senescence and upon microbial infection, which induces endogenous salicylic acid accumulation, as well as by direct applications of exogenous salicylic acid. Using both overexpression and loss-of-function mutants, PDLP5 was shown to block plasmodesmal permeability by stimulating callose deposition at plasmodesmata. Although the mechanism underlying this process is under active investigation, two novel callose synthase isoforms have just been found that function directly downstream of PDLP5 (W. Cui and J.-Y. Lee, unpublished data), indicating that PDLP5 either recruits or activates those callose synthases. Callose deposition at plasmodesmata is assumed to impose physical constraint to the outer membrane leaflet of plasmodesmata, narrowing the cytoplasmic sleeves, and hence restricting the molecular traffic through the channels [44, 45].
Figure 3. Regulation of plasmodesmata during plant defense.
A model is presented illustrating how the closure of plasmodesmata is mediated by PDLP5 during immune responses: PDLP5 induced by the salicylic acid (SA) signaling pathway (1) stimulates the enzymatic activity of yet-to-be-identified callose synthase (2), which enhances callose accumulation at plasmodesmata (PD) (3). How this activation occurs remains to be discovered. Accumulation of plasmodesmal callose leads to the closure of plasmodesmata (4). PDLP5 contains an extracellular domain that may function in perceiving a putative signal or ligands (triangles), generation of which is dependent on salicylic acid pathway (SA) pathway (dashed arrows). The signal perception by PDLP5 may be required for the PDLP5 function in activating callose synthase (2) and/or feedback regulation of the SA pathway (5). LYM2, anchored to the membrane via glycosylphosphatidylinositol modification, binds to chitin derived from a fungal pathogen, which by an unknown mechanism closes plasmodesmata.
Hyperaccumulation of callose at plasmodesmata was induced upon either bacterial infection or exogenous salicylic acid treatment. This response was consistent with the role of the callose in reducing plasmodesmal permeability [11]. However, the induction of both plasmodesmal closure and hyperaccumulation of callose in response to either pathogen challenge or exogenous application of salicylic acid was impaired in pdlp5-1 mutant [11]. Moreover, the basal level of plasmodesmal callose in pdlp5-1 was substantially lowered. Thus, the plasmodesmata remained constitutively more permeable in pdlp5-1 than in normal wild type Arabidopsis. Surprisingly, the effect of salicylic acid on the plasmodesmal closure response was not solely due to its role in the transcriptional activation of PDLP5. Rather, the salicylic acid pathway was also required for the molecular function of PDLP5 itself. For example, PDLP5 was not able to stimulate plasmodesmal callose deposition or plasmodesmal closure in the absence of salicylic acid accumulation or functional NPR1, inferring that there is multilevel crosstalk between the salicylic acid signaling pathway and plasmodesmal closure. Together, the state of open plasmodesmata and the inability to activate plasmodesmal closure in pdlp5-1 compromised the innate immunity of the mutant plants [10, 11]. Collectively, it appears that the capacity of the cells to switch from a non-cell autonomous state to a cell-autonomous state via dynamic regulation of plasmodesmata via PDLP5 is a prerequisite to achieve full defense responses. In addition, given that PDLP5 functions in conjunction with the salicylic acid signaling pathway, PDLP5-mediated plasmodesmal closure is unlikely limited to defense against bacterial infection but rather is probably also involved in broad-spectrum disease resistance that accompanies salicylic acid accumulation.
An additional example supporting the concept that plasmodesmal closure is integrated into defense signaling is recently provided by the study of the LYSIN MOTIF DOMAIN-CONTAINING GLYCOSYLPHOSPHATIDYLINOSITOL-ANCHORED PROTEIN 2 (LYM2) [12]. LYM2 localizes in plasma membrane and binds to chitin, a PAMP derived from fungal pathogens, and is required for the chitin-mediated plasmodesmal closure upon fungal infection (Fig. 3). How this PAMP perception by LYM2 signals and alters plasmodesmata permeability is not yet known. However, the studies of LYM2 and PDLP5 suggest that multiple players and mechanisms of defense are likely linked to plasmodesmal closure during defense against various types of plant pathogens. It is tempting to speculate about the existence of a hypothetical signalosome, made up of various signal receptors and transducers that integrate immune signals induced by pathogens into plasmodesmal closure and vice versa. It is conceivable that this hypothetical sigmalosome may exist at or near plasmodesmal channels. On one hand, plasmodesmal office, where callose mostly accumulates, or the central region, where PDLP5 locates could be a possibility. On the other hand, along the plasma membrane lining of plasmodesmal channel, or plasma membrane and/or ER at the vicinity of plasmodesmal opening could be another good possibility.
4.3. An evolutionary peek into the regulation of plasmodesmata
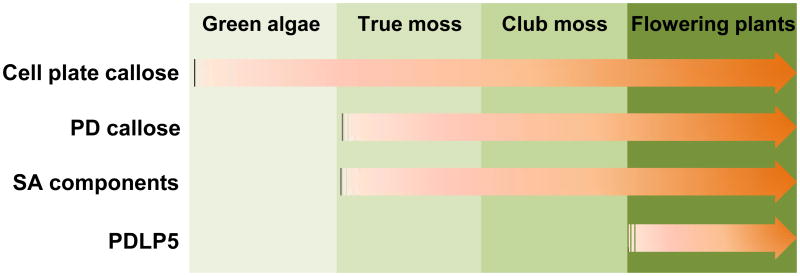
In an attempt to get a glimpse into the evolutionary path, through which plants have potentially built the molecular machinery that regulates plasmodesmata, I have performed a phylogenetic analysis based on genomic information available for different plant species. For this task, orthologs of the key Arabidopsis genes known to regulate plasmodesmata during defense were searched using Phytosome (http://www.phytozome.net/) and KEGG ORTHOLOHY (http://www.genome.jp) databases. The protein coding sequences of PDLP5 (At1g70690), ICS1 (At1g74710), and NPR1 (At1g64280) were used for a BLAST search against all green plant sequences. The result of this analysis, which is summarized as an illustration in Fig. 4, revealed that the orthologs of ICS1 and NPR1 were found in an evolutionary primitive moss, Physcomitrella, and also in the seedless vascular plant Selaginella, besides being found in flowering plants. Moreover, the BLAST search showed that Physcomitrella contains putative orthologs for all twelve members of Arabidopsis callose synthases [46]. Interestingly, PDLP5 orthologues were only found in the flowering plants, suggesting that the mechanism to close plasmodesmata mediated by PDLP5 may have developed after the evolution of the seed habit.
Figure 4. A hypothetical progression underlying the regulation of plasmodesmata.
The capacity to produce cell plate callose may have evolved in multicellular green algae, followed by the capacity to accumulate plasmodesmal callose and to adopt the salicylic acid (SA) pathway in lower land plants including true mosses. PDLP5, a molecular switch for callose-mediated plasmodesmal closure may have evolved more recently in flowering plants to integrate the regulatory mechanism of plasmodesmal permeability into a defense system.
A recent study on plasmodesmal structure and distribution in true mosses indicates that the capacity to deposit callose at plasmodesmata evolved much earlier than the evolution of PDLP5. This study, in which immuno-electron microscopy was performed using monoclonal β-1,3 glucan antibody, showed that callose accumulates around the plasmodesmata in apical cells of Physcomitrella gametophytes [47]. In fact, the genome of Physcomitrella encodes twelve putative callose synthase genes like Arabidopsis, although eight Physcomitrella isoforms are clustered in a single clade which contains one Arabidopsis gene encoding CALS9 [46]. Identifying callose synthase isoform(s) that is responsible for plasmodesmal callose accumulation in Physcomitrella would be very helpful in terms of gaining insight into the evolution of this important feature of plasmodesmal structure and function. Although it is not yet known whether callose is also associated with the plasmodesmata in filamentous algae, multicellular green algae appear to have the capacity to produce cell plate-associated callose (see Fig. 4). According to a study in which fluorescent microscopic examinations were used to detect callose accumulation by aniline blue staining in various organisms representing green algae, bryophytes, ferns and seed plants, cytokinesis-associated callose was found in multicellular green algae and land plants [48]. In unicellular algae, callose deposition was observed in response to wounding but not in dividing cells. Examination of the multicellular alga Chara corallina by immunofluorescence using monoclonal β-1,3 glucan antibody suggested that callose deposits at plasmodesmata [49]. As this evolutionary time frame coincides with the appearance of plasmodesmata in plants [22], important questions to address in future studies include if callose deposition occurs at the algal plasmodesmata; what callose synthases are involved in this process; and whether it is necessary for plasmodesmal formation or other function.
Hypothetically, it appears that the occurrence of the genes required for plasmodesmal callose deposition may coincide with the early development of vascular plants or multicellular algae (see Fig. 4). The genes required for the salicylic acid pathway may have been selected during the early development of vascular plants. PDLP5-like genes, which are assumed to confer the regulation of plasmodesmal callose deposition as shown in Arabidopsis, may have been only acquired very recently during the evolution of seed plants. It is tempting to speculate whether higher plants recruited specific callose synthases that perform spatiotemporal regulation of plasmodesmal permeability under the control of PDLP5 and PDLP5-like proteins. This regulatory module may have been selected to link salicylic acid-dependent defense responses to plasmodesmal callose deposition as part of a surveillance system, under which intercellular connectivity is tightly linked to plant immunity.
5. Concluding remarks
Recent advances in the animal and bacterial fields have revealed a surprising mechanism that provides an evolutionary insight into the survival strategy as a cellular community, whether that conforms to one body or not, and whether that is originated from animal or plant lineages. Bacteria, mammals, fungi, green algae, and plants all have evolved a means for cells-to-cell communication through membrane-lined intercellular channels. The presence of these types of channels, tunneling nanotubes in mammalian and plasmodesmata in plant cells, furnished opportunities to certain pathogens to spread infection between host cells. However, plant biologists are just beginning to realize that plasmodesmata are closely controlled by the immune surveillance system. It is conceivable that a parallel control mechanism may also operate for tunneling nanotubes in animals. However, currently not much is known as to how the molecular transfer across tunneling nanotubes is controlled and whether animal cells have evolved mechanisms by which tunneling nanotubes are guarded from exploitation by pathogens and/or are utilized by bridged healthy cells to rescue infected cells from pathogenicity. It will be truly exciting to have the molecular compositions of tunneling nanotubes and plasmodesmata revealed soon via technical advances in genomics and proteomics. Once achieved, molecular understanding as to how these bridges between basic units of life could form, develop, and integrate fundamental cellular processes will help solve many challenging issues related both human and plant health.
Supplementary Material
Acknowledgments
The author thanks R. Sager for assisting with supplemental information and T. D. Pizzolato for helpful comments and edits of the manuscript. The support for this work is provided by the National Science Foundation (IOS-0954931 and ABI-1062520).
References
- 1.Robards AW, Lucas WJ. Plasmodesmata. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:369–419. [Google Scholar]
- 2.Lucas WJ, Lee JY. Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol. 2004;5:712–726. doi: 10.1038/nrm1470. [DOI] [PubMed] [Google Scholar]
- 3.Lee JY, Cui WE. Non-cell autonomous RNA trafficking and long-distance signaling. J Plant Biol. 2009;52:10–18. [Google Scholar]
- 4.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 5.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Lee JY, Cho S, Sager R. Plasmodesmata dn non-cell autonomous signling in plants. In: Murphy AS, editor. The plant plasma membrane. Springer-Verlag; 2011. pp. 87–106. [Google Scholar]
- 7.Ormenese S, Havelange A, Deltour R, Bernier G. The frequency of plasmodesmata increases early in the whole shoot apical meristem of sinapis alba l. During floral transition. Planta. 2000;211:370–375. doi: 10.1007/s004250000294. [DOI] [PubMed] [Google Scholar]
- 8.Sevilem I, Miyashima S, Helariutta Y. Cell-to-cell communication via plasmodesmata in vascular plants. Cell Adh Migr. 2013;7:27–32. doi: 10.4161/cam.22126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JY, Lu H. Plasmodesmata: The battleground against intruders. Trends Plant Sci. 2011;16:201–210. doi: 10.1016/j.tplants.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Lee JY, et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in arabidopsis. Plant Cell. 2011;23:3353–3373. doi: 10.1105/tpc.111.087742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. Salicylic acid regulates plasmodesmata closure during innate immune responses in arabidopsis. Plant Cell. 2013;25:2315–2329. doi: 10.1105/tpc.113.110676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner C, et al. Lym2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci U S A. 2013;110:9166–9170. doi: 10.1073/pnas.1203458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maule AJ. Plasmodesmata: Structure, function and biogenesis. Curr Opin Plant Biol. 2008;6:680–686. doi: 10.1016/j.pbi.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Lucas WJ, Ham BK, Kim JY. Plasmodesmata - bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Burch-Smith TM, Stonebloom S, Xu M, Zambryski PC. Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma. 2011;248:61–74. doi: 10.1007/s00709-010-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding B, Turgeon R, Parthasarathy MV. Substructure of freeze-substituted plasmodesmata. Protoplasma. 1992;169:28–41. [Google Scholar]
- 17.Lucas WJ, Ding B, Vanderschoot C. Plasmodesmata and the supracellular nature of plants. New Phytologist. 1993;125:435–476. doi: 10.1111/j.1469-8137.1993.tb03897.x. [DOI] [PubMed] [Google Scholar]
- 18.Cook ME, Graham LE, Botha CEJ, Lavin CA. Comparative ultrastructure of plasmodesmata of chara and selected bryophytes: Toward an elucidation of the evolutionary origin of plant plasmodesmata. Am J Bot. 1997;84:1169–1178. [PubMed] [Google Scholar]
- 19.Kirk BT, Sinclair JB. Plasmodesmata between hyphal cells of geotrichum candidum. Science. 1966;153:1646. doi: 10.1126/science.153.3744.1646. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto T, Morgan J, Conti SF. Morphogenesis and ultrastructure of geotrichum candidum septa. J Bacteriol. 1973;116:447–455. doi: 10.1128/jb.116.1.447-455.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham LE, Cook ME, Busse JS. The origin of plants: Body plan changes contributing to a major evolutionary radiation. Proc Natl Acad Sci U S A. 2000;97:4535–4540. doi: 10.1073/pnas.97.9.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- 23.Oparka KJ. Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 2004;9:33–41. doi: 10.1016/j.tplants.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Gallagher KL, Benfey PN. Not just another hole in the wall: Understanding intercellular protein trafficking. Genes Dev. 2005;19:189–195. doi: 10.1101/gad.1271005. [DOI] [PubMed] [Google Scholar]
- 25.Marzo L, Gousset K, Zurzolo C. Multifaceted roles of tunneling nanotubes in intercellular communication. FPHYS. 2012;3:72. doi: 10.3389/fphys.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abounit S, Zurzolo C. Wiring through tunneling nanotubes--from electrical signals to organelle transfer. J Cell Sci. 2012;125:1089–1098. doi: 10.1242/jcs.083279. [DOI] [PubMed] [Google Scholar]
- 27.Gerdes HH, Rustom A, Wang X. Tunneling nanotubes, an emerging intercellular communication route in development. Mech Dev. 2013;130:381–387. doi: 10.1016/j.mod.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann C, Sambade A, Heinlein M. Plasmodesmata and intercellular transport of viral rna. Biochem Soc Trans. 2007;35:142–145. doi: 10.1042/BST0350142. [DOI] [PubMed] [Google Scholar]
- 29.Schoelz JE, Harries PA, Nelson RS. Intracellular transport of plant viruses: Finding the door out of the cell. Mol Plant. 2011;4:813–831. doi: 10.1093/mp/ssr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpita N, Sabularse D, Montezinos D, Delmer DP. Determination of the pore size of cell walls of living plant cells. Science. 1979;205:1144–1147. doi: 10.1126/science.205.4411.1144. [DOI] [PubMed] [Google Scholar]
- 32.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 33.Lockhart J. Surviving the onslaught: Salicylic acid regulates plasmodesmata closure during pathogen attack in arabidopsis. Plant Cell. 2013;25:1911. doi: 10.1105/tpc.113.250610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowinski S, et al. Membrane nanotubes physically connect t cells over long distances presenting a novel route for hiv-1 transmission. Nat Cell Biol. 2008;10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- 35.Kadiu I, Gendelman HE. Macrophage bridging conduit trafficking of hiv-1 through the endoplasmic reticulum and golgi network. J Proteome Res. 2011;10:3225–3238. doi: 10.1021/pr200262q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gousset K, et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol. 2009;11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- 37.Miyazawa K, Emmerling K, Manuelidis L. Proliferative arrest of neural cells induces prion protein synthesis, nanotube formation, and cell-to-cell contacts. J Cell Biochem. 2010;111:239–247. doi: 10.1002/jcb.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas S, Popov VL, Walker DH. Exit mechanisms of the intracellular bacterium ehrlichia. PLoS ONE. 2010;5:e15775. doi: 10.1371/journal.pone.0015775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 40.Schwessinger B, Ronald PC. Plant innate immunity: Perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- 41.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 42.Gohre V, Robatzek S. Breaking the barriers: Microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- 43.Fu ZQ, Dong X. Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol. 2013 doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 44.Radford JE, Vesk M, Overall RL. Callose deposition at plasmodesmata. Protoplasma. 1998;201:30–37. [Google Scholar]
- 45.Zavaliev R, Ueki S, Epel BL, Citovsky V. Biology of callose (beta-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–130. doi: 10.1007/s00709-010-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuette S, et al. Novel localization of callose in the spores of physcomitrella patens and phylogenomics of the callose synthase gene family. Ann Bot. 2009;103:749–756. doi: 10.1093/aob/mcn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansouri K. Plant Biology. Southern Illinois University; Carbondale: 2012. Comparative ultrastructure of apical cells and derivatives in bryophytes, with special reference to plasmodesmata. [Google Scholar]
- 48.Scherp P, Grotha R, Kutschera U. Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns and seed plants. Plant Cell Rep. 2001;20:143–149. doi: 10.1007/s002990000301. [DOI] [PubMed] [Google Scholar]
- 49.Faulkner CR, Blackman LM, Collings DA, Cordwell SJ, Overall RL. Anti-tropomyosin antibodies co-localise with actin microfilaments and label plasmodesmata. Eur J Cell Biol. 2009;88:357–369. doi: 10.1016/j.ejcb.2009.02.184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.